Extensive Reliability Evaluation of Docking-Based Target-Fishing Strategies
Abstract
1. Introduction
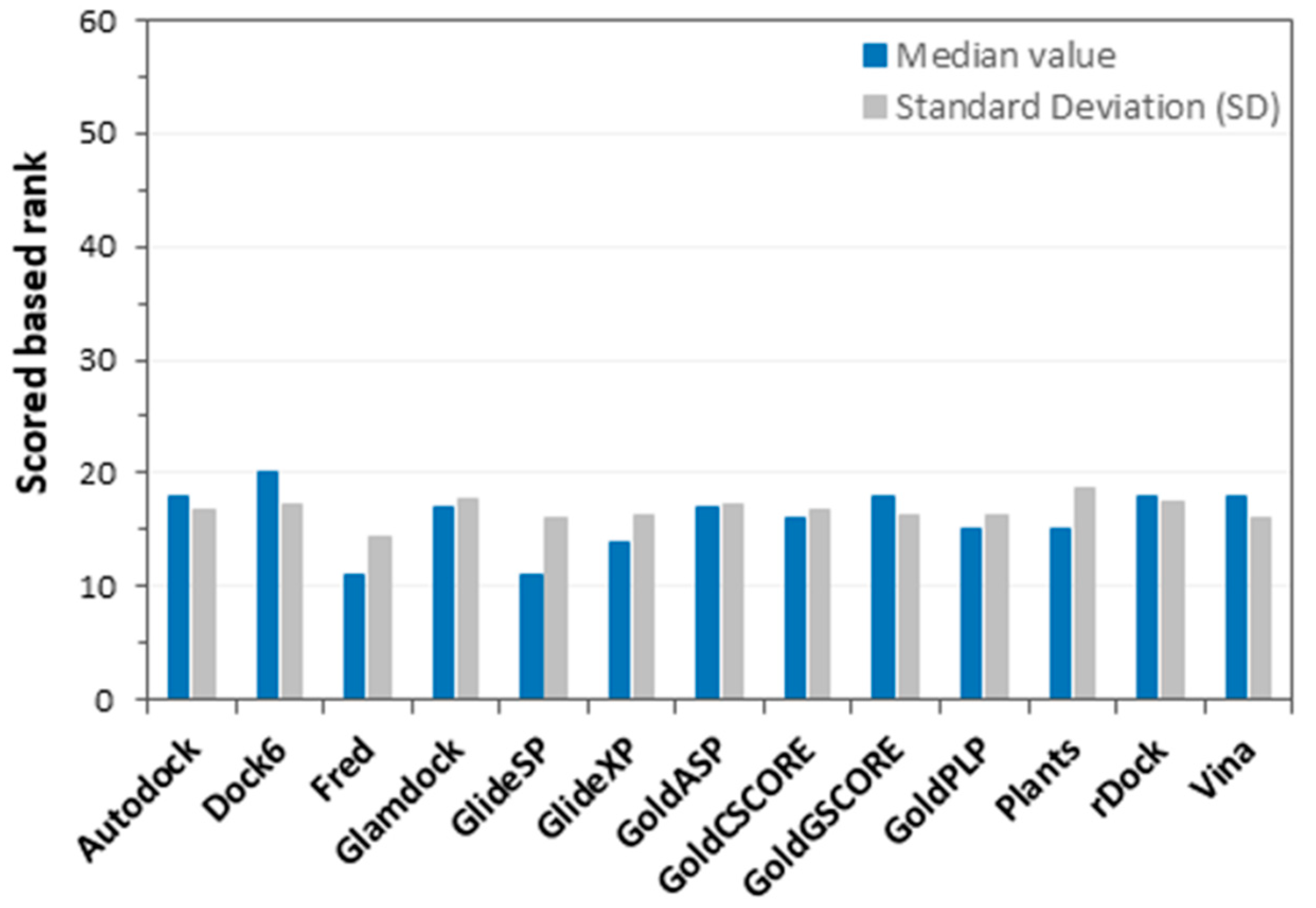
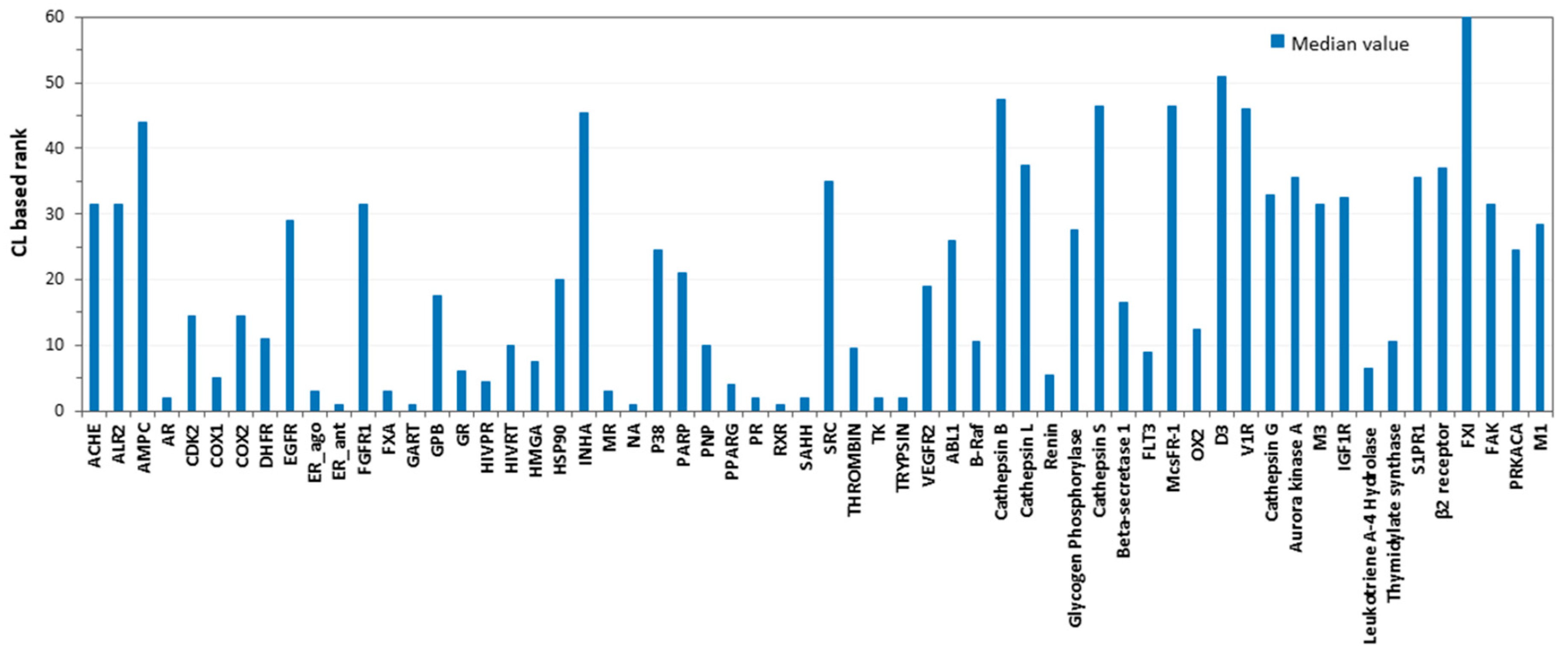
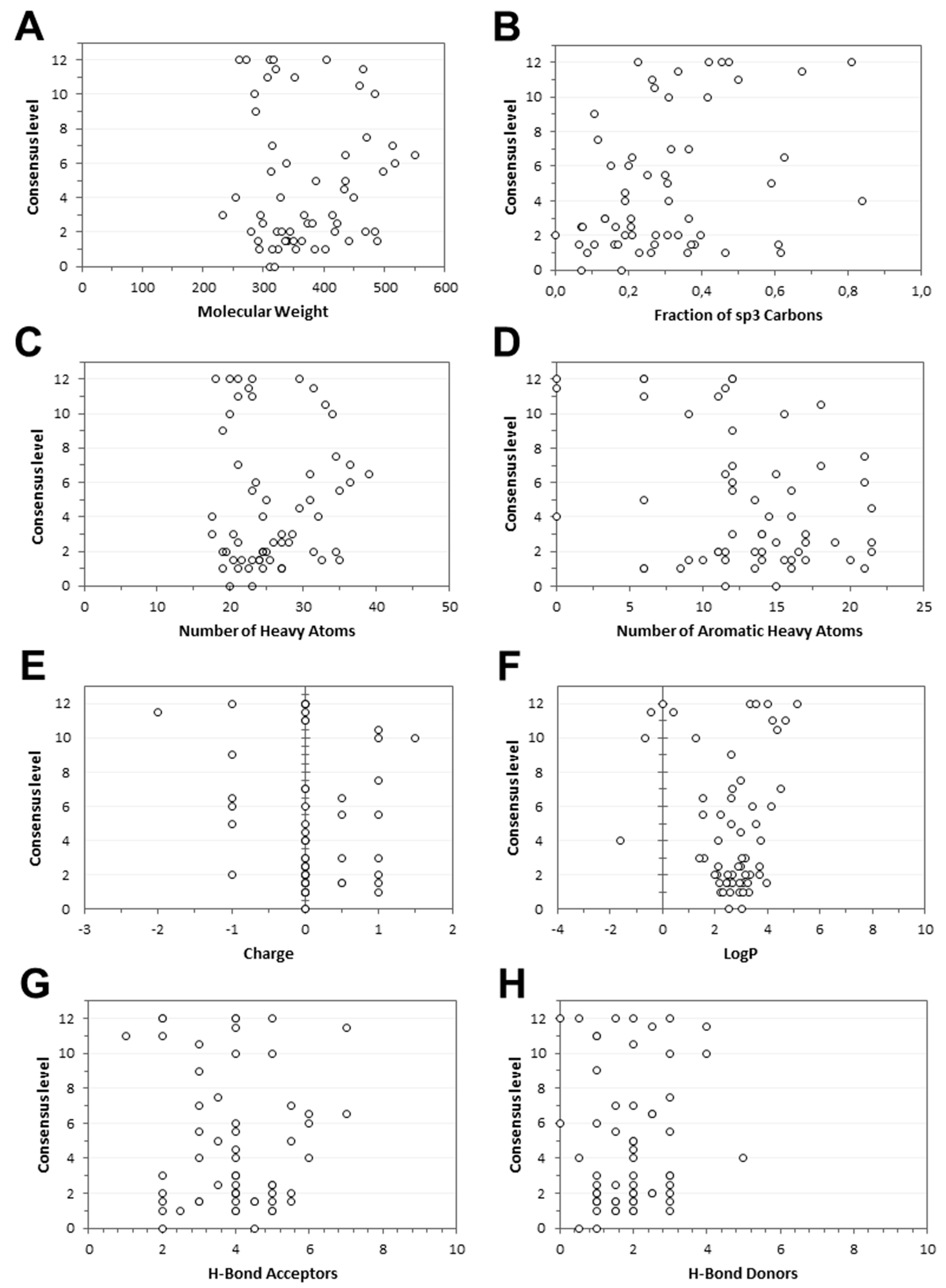
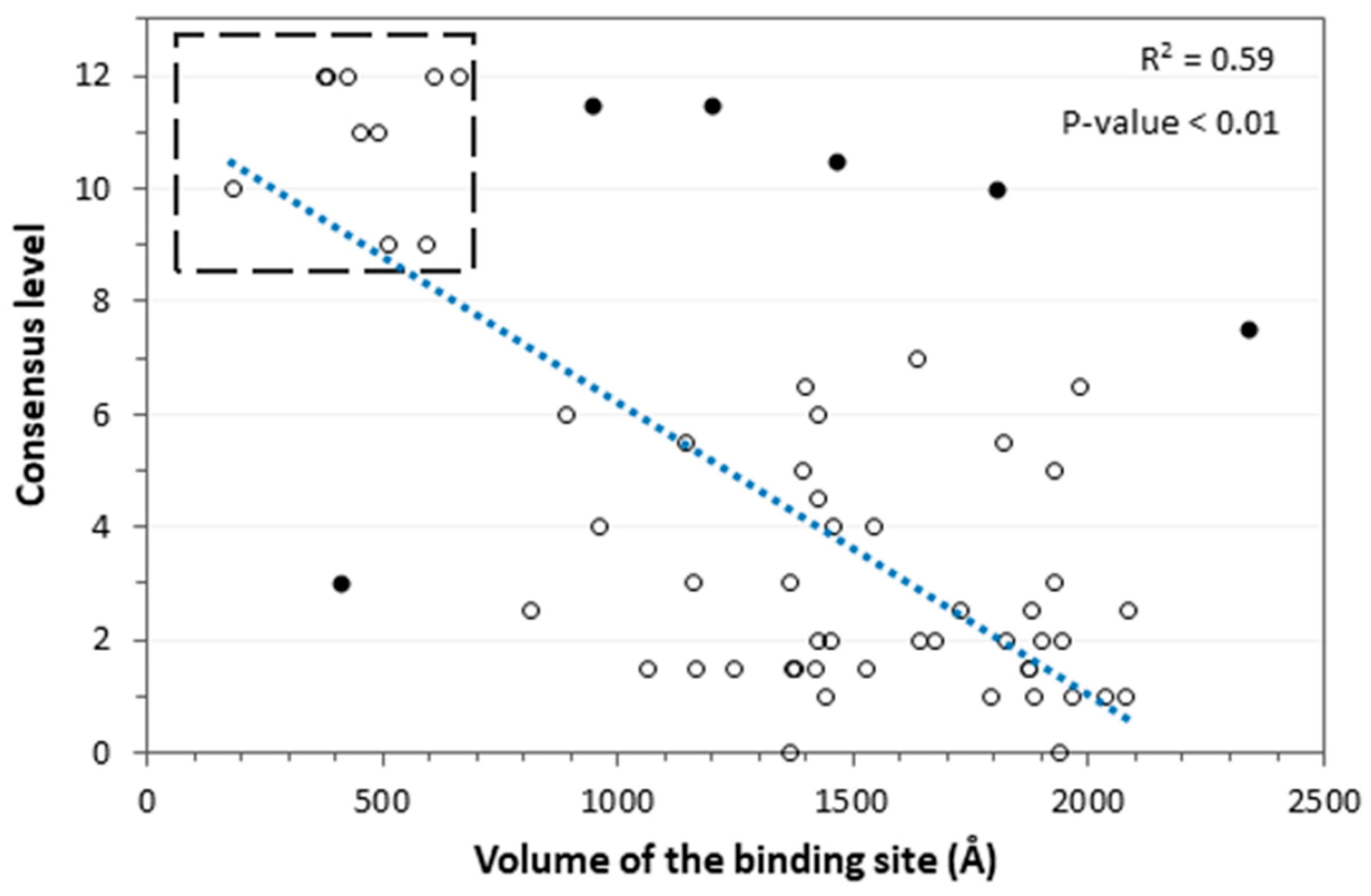
2. Results and Discussion
3. Materials and Methods
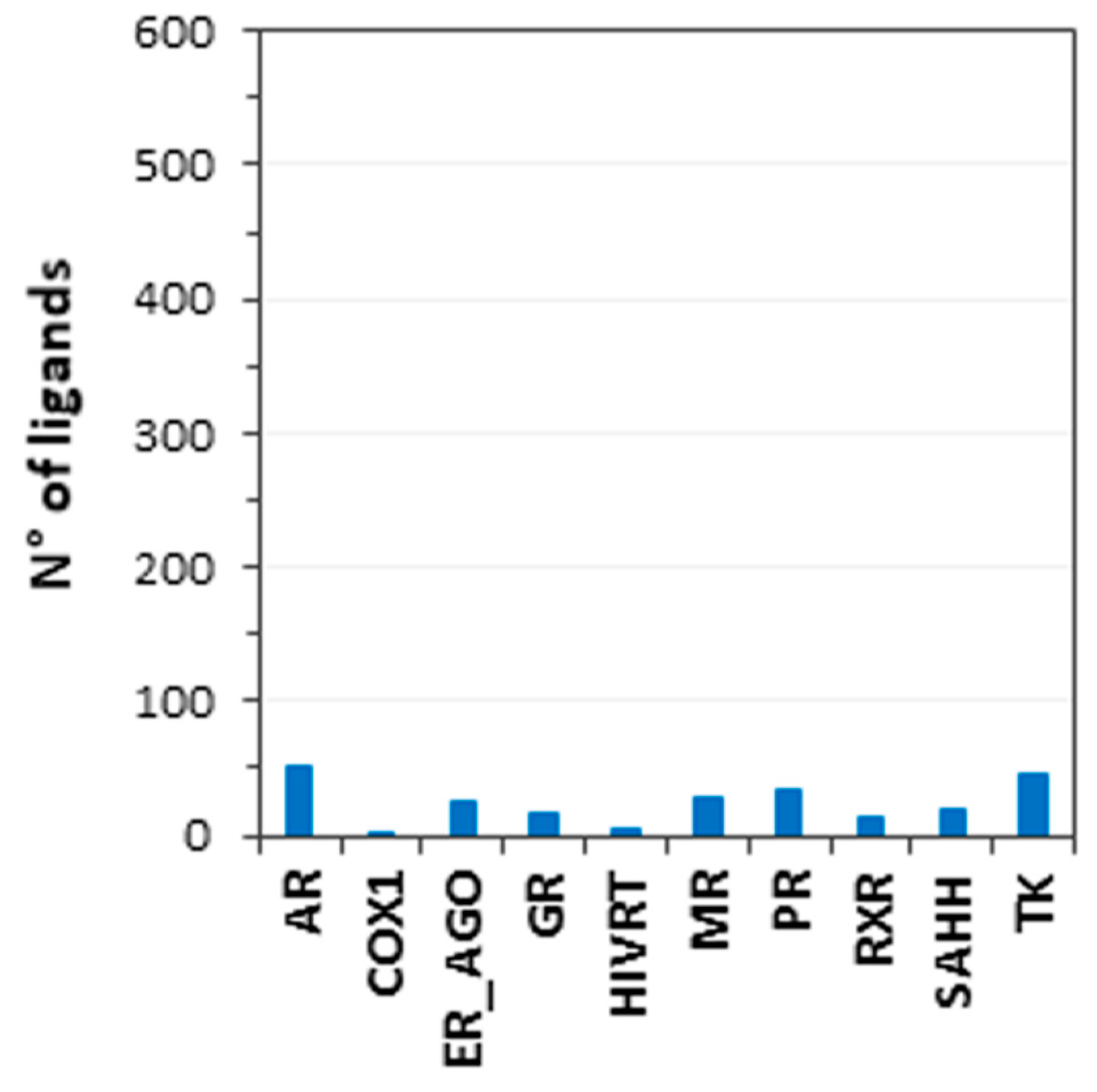
3.1. Database Generation
3.2. Protein Structure Alignment
3.3. Docking Procedures
3.4. Docking Score Evaluation
3.5. Consensus Docking Analysis
3.6. Evaluation of True Positive Rate and False Discovery Rate
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shoichet, B.K. Virtual screening of chemical libraries. Nature 2004, 432, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Tuccinardi, T.; Poli, G.; Corchia, I.; Granchi, C.; Lapillo, M.; Macchia, M.; Minutolo, F.; Ortore, G.; Martinelli, A. A Virtual Screening Study for Lactate Dehydrogenase 5 Inhibitors by Using a Pharmacophore-based Approach. Mol. Inform. 2016, 35, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Rognan, D. Structure-Based Approaches to Target Fishing and Ligand Profiling. Mol. Inform. 2010, 29, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, L.; Lee, Y.M.; Kalesh, K.A.; Ong, Y.S.; Lim, J.; Jee, J.-E.; Sun, H.; Lee, S.S.; Hua, Z.-C.; et al. Target identification of natural and traditional medicines with quantitative chemical proteomics approaches. Pharmacol. Ther. 2016, 162, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Lucas, X.; Bendik, I.; Günther, S.; Merfort, I. Target Fishing by Cross-Docking to Explain Polypharmacological Effects. ChemMedChem 2015, 10, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.R.; Sullivan, D.J. New uses for old drugs. Nature 2007, 448, 645–646. [Google Scholar] [CrossRef] [PubMed]
- Dudley, J.T.; Deshpande, T.; Butte, A.J. Exploiting drug-disease relationships for computational drug repositioning. Brief. Bioinform. 2011, 12, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Bender, A.; Scheiber, J.; Glick, M.; Davies, J.W.; Azzaoui, K.; Hamon, J.; Urban, L.; Whitebread, S.; Jenkins, J.L. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. ChemMedChem 2007, 2, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Lounkine, E.; Keiser, M.J.; Whitebread, S.; Mikhailov, D.; Hamon, J.; Jenkins, J.L.; Lavan, P.; Weber, E.; Doak, A.K.; Côté, S.; et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 2012, 486, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Mestres, J.; Gregori-Puigjané, E.; Valverde, S.; Solé, R.V. The topology of drug–target interaction networks: Implicit dependence on drug properties and target families. Mol. Biosyst. 2009, 5, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Pujadas, G.; Garcia-Vallve, S. Tools for in silico target fishing. Methods 2015, 71, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Drews, J. Drug discovery: A historical perspective. Science 2000, 287, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Henrich, S.; Salo-Ahen, O.M.H.; Huang, B.; Rippmann, F.; Cruciani, G.; Wade, R.C. Computational approaches to identifying and characterizing protein binding sites for ligand design. J. Mol. Recognit. 2010, 23, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Schuffenhauer, A.; Floersheim, P.; Acklin, P.; Jacoby, E. Similarity metrics for ligands reflecting the similarity of the target proteins. J. Chem. Inf. Comput. Sci. 2003, 43, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-B.; Yang, L.-L.; Xu, Y.; Wang, W.-J.; Li, L.-L.; Yang, S.-Y. A combined molecular docking-based and pharmacophore-based target prediction strategy with a probabilistic fusion method for target ranking. J. Mol. Gr. Model. 2013, 44, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Zhi, D.G. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins 2001, 43, 217–226. [Google Scholar] [CrossRef]
- Wang, J.-C.; Chu, P.-Y.; Chen, C.-M.; Lin, J.-H. idTarget: A web server for identifying protein targets of small chemical molecules with robust scoring functions and a divide-and-conquer docking approach. Nucleic Acids Res. 2012, 40, W393–W399. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, S.G.; Baumann, K. Maximum Unbiased Validation (MUV) Data Sets for Virtual Screening Based on PubChem Bioactivity Data. J. Chem. Inf. Model. 2009, 49, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Willighagen, E.L.; Waagmeester, A.; Spjuth, O.; Ansell, P.; Williams, A.J.; Tkachenko, V.; Hastings, J.; Chen, B.; Wild, D.J. The ChEMBL database as linked open data. J. Cheminform. 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Robertson, T.A.; Varani, G. An all-atom, distance-dependent scoring function for the prediction of protein-DNA interactions from structure. Proteins Struct. Funct. Bioinform. 2006, 66, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.H.J.; Kraus, J.; Kramer, B. Virtual high-throughput screening of molecular databases. Curr. Opin. Drug Discov. Dev. 2007, 10, 298–307. [Google Scholar]
- Tuccinardi, T.; Poli, G.; Romboli, V.; Giordano, A.; Martinelli, A. Extensive Consensus Docking Evaluation for Ligand Pose Prediction and Virtual Screening Studies. J. Chem. Inf. Model. 2014, 54, 2980–2986. [Google Scholar] [CrossRef] [PubMed]
- Tuccinardi, T.; Poli, G.; Dell’Agnello, M.; Granchi, C.; Minutolo, F.; Martinelli, A. Receptor-based virtual screening evaluation for the identification of estrogen receptor β ligands. J. Enzyme Inhib. Med. Chem. 2015, 30, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Russo Spena, C.; De Stefano, L.; Poli, G.; Granchi, C.; El Boustani, M.; Ecca, F.; Grassi, G.; Grassi, M.; Canzonieri, V.; Giordano, A.; et al. Virtual screening identifies a PIN1 inhibitor with possible antiovarian cancer effects. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Martinelli, A.; Tuccinardi, T. Reliability analysis and optimization of the consensus docking approach for the development of virtual screening studies. J. Enzyme Inhib. Med. Chem. 2016, 31, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Lapillo, M.; Granchi, C.; Caciolla, J.; Mouawad, N.; Caligiuri, I.; Rizzolio, F.; Langer, T.; Minutolo, F.; Tuccinardi, T. Binding investigation and preliminary optimisation of the 3-amino-1,2,4-triazin-5(2H)-one core for the development of new Fyn inhibitors. J. Enzyme Inhib. Med. Chem. 2018, 33, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Chiarelli, L.R.; Mori, M.; Barlocco, D.; Beretta, G.; Gelain, A.; Pini, E.; Porcino, M.; Mori, G.; Stelitano, G.; Costantino, L.; et al. Discovery and development of novel salicylate synthase (MbtI) furanic inhibitors as antitubercular agents. Eur. J. Med. Chem. 2018, 155, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Bononi, G.; Granchi, C.; Lapillo, M.; Giannotti, M.; Nieri, D.; Fortunato, S.; Boustani, M.E.; Caligiuri, I.; Poli, G.; Carlson, K.E.; et al. Discovery of long-chain salicylketoxime derivatives as monoacylglycerol lipase (MAGL) inhibitors. Eur. J. Med. Chem. 2018, 157, 817–836. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Allen, W.J.; Balius, T.E.; Mukherjee, S.; Brozell, S.R.; Moustakas, D.T.; Lang, P.T.; Case, D.A.; Kuntz, I.D.; Rizzo, R.C. DOCK 6: Impact of new features and current docking performance. J. Comput. Chem. 2015, 36, 1132–1156. [Google Scholar] [CrossRef] [PubMed]
- McGann, M. FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Tietze, S.; Apostolakis, J. GlamDock: Development and Validation of a New Docking Tool on Several Thousand Protein–Ligand Complexes. J. Chem. Inf. Model. 2007. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Monecke, P.; Hessler, G.; Stützle, T.; Exner, T.E. pharmACOphore: Multiple Flexible Ligand Alignment Based on Ant Colony Optimization. J. Chem. Inf. Model. 2010, 50, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical Scoring Functions for Advanced Protein−Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carmona, S.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A.B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R.E.; Morley, S.D. rDock: A fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput. Biol. 2014, 10, e1003571. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
| Type | PDB Code | Source |
|---|---|---|
| Acetylcholinesterase (ACHE) | 1EVE | DUD |
| Aldose Reductase (ALR2) | 1AH3 | |
| AmpC beta-lactamase (AmpC) | 1XGJ | |
| Androgen Receptor (AR) | 1XQ2 | |
| Cyclic dependent Kinase 2 (CDK2) | 1CKP | |
| Cyclooxigenase 1 (COX1) | 1P4G | |
| Cyclooxigenase 2 (COX2) | 1CX2 | |
| Dihidrofolate Reductase (DHFR) | 2DFR | |
| Epidermal Growth Factor Receptor (EGFr) | 1M17 | |
| Estrogen Receptor Alpha (ERagonist) | 1L2I | |
| Estrogen Receptor Alpha (ERantagonist) | 3ERT | |
| Fibroblast Growth Factor Receptor 1 (FGFr1) | 1AGW | |
| Coagulation Factor XA (FXa) | 1F0R | |
| GAR Transformylase (GART) | 1C2T | |
| Glycogen Phosphorylase (GPB) | 1AI8 | |
| Glucocorticoid Receptor (GR) | 1M2Z | |
| HIV-1 protease (HIVPR) | 1HPX | |
| HIV-1 reverse transcriptase (HIVRT) | 1RT1 | |
| HMG-CoA reductase (HMGR) | 1HW8 | |
| Heat shock protein 90 (HSP90) | 1UY6 | |
| Enoyl reductase (InhA) | 1P44 | |
| Mineralocorticoid Receptor (MR) | 2AA2 | |
| Neuraminidase (NA) | 1A4G | |
| p38 MAP Kinase (P38 MAP) | 1KV2 | |
| poly(ADP-ribose) Polymerase (PARP) | 1EFY | |
| Purine Nucleoside Phosphorylase (PNP) | 1B8O | |
| Peroxisome proliferator-activated receptor gamma (PPARg) | 1FM9 | |
| Progesterone receptor (PR) | 1SR7 | |
| Retinoic acid receptor RXR-alpha (RXRa) | 1MVC | |
| S-adenosylhomocysteine Hydrolase (SAHH) | 1A7A | |
| Tyrosine-protein kinase Src (SRC) | 2SRC | |
| Thrombin | 1BA8 | |
| Thymidine Kinase (TK) | 1KIM | |
| Beta-Trypsin | 1BJU | |
| Vascular Endothelial Growth Factor Receptor 2 (VEGFr2) | 1VR2 | |
| Cathepsin G | 1KYN | MUV |
| Coagulation Factor XI (FXI) | 4D76 | |
| Focal Adhesion Kinase (FAK) | 4Q9S | |
| Muscarinic Acetylcholine Receptor M1 | 5CXV | |
| cAMP-dependent Protein Kinase (PRKACA) | 5BX6 | |
| Sphingosine 1-Phosphate Receptor 1 (S1PR1) | 3V2W | |
| Aurora kinase A | 4ZS0 | ChEMBL |
| Beta-2 Adrenergic Receptor | 3P0G | |
| Beta-secretase 1 | 4RCD | |
| Cathepsin B | 3AI8 | |
| Cathepsin L | 5F02 | |
| Cathepsin S | 4PE6 | |
| Dopamine Receptor D3 | 3PBL | |
| Glycogen Phosphorylase | 3DD1 | |
| Insulin-like Growth Factor 1 Receptor (IGF1R) | 5HZN | |
| Leukotriene A-4 Hydrolase | 5BPP | |
| Macrophage Colony-Stimulating Factor 1 Receptor | 4RTH | |
| Muscarinic Acetylcholine Receptor M3 | 4DAJ | |
| OX2 orexin receptor | 4S0V | |
| Receptor-type tyrosine-protein kinase FLT3 | 4RT7 | |
| Renin | 4RYC | |
| Serine/threonine-protein kinase B-Raf | 5JRQ | |
| Thymidylate synthase | 5IOQ | |
| Tyrosine-protein Kinase ABL1 | 4ZOG | |
| Vasopressin Receptor V1R | 1YTV |
| Docking Procedures | TPR (%) | FDR (%) |
|---|---|---|
| Autodock | 27% | 76% |
| Fred | 36% | 69% |
| Dock6 | 25% | 79% |
| Glamdock | 31% | 73% |
| GlideSP | 36% | 67% |
| GlideXP | 30% | 73% |
| GoldASP | 28% | 73% |
| GoldCSCORE | 29% | 74% |
| GoldGSCORE | 25% | 77% |
| GoldPLP | 31% | 72% |
| Plants | 33% | 70% |
| rDock | 28% | 76% |
| Vina | 22% | 81% |
| Consensus Docking | 36% | 67% |
| Target | Consensus Level | Target | Consensus Level |
|---|---|---|---|
| ACHE | 2 | SRC | 2 |
| ALR2 | 1.5 | THROMBIN | 6 |
| AMPC | 2 | TK | 11 |
| AR | 12 | TRYPSIN | 10 |
| CDK2 | 2 | VEGFR2 | 1.5 |
| COX1 | 9 | ABL1 | 2.5 |
| COX2 | 6 | B-Raf | 4.5 |
| DHFR | 4 | Cathepsin B | 1 |
| EGFR | 1.5 | Cathepsin L | 2 |
| ER_ago | 12 | Renin | 6.5 |
| ER_ant | 10.5 | Glycogen Phosphorylase | 2 |
| FGFR1 | 1 | Cathepsin S | 1 |
| FXA | 7.5 | Beta-secretase 1 | 3 |
| GART | 11.5 | FLT3 | 5 |
| GPB | 4 | McsFR-1 | 1.5 |
| GR | 11 | OX2 | 4 |
| HIVPR | 7 | D3 | 1 |
| HIVRT | 9 | V1R | 1.5 |
| HMGA | 5 | Cathepsin G | 1.5 |
| HSP90 | 2.5 | Aurora kinase A | 2 |
| INHA | 0 | M3 | 1.5 |
| MR | 12 | IGF1R | 2 |
| NA | 11.5 | Leukotriene A-4 Hydrolase | 5.5 |
| P38 MAP | 2.5 | Thymidylate synthase | 6.5 |
| PARP | 3 | S1PR1 | 3 |
| PNP | 3 | β2 Receptor | 1.5 |
| PPARG | 6 | FXI | 0 |
| PR | 11 | FAK | 1 |
| RXR | 12 | PRKACA | 2.5 |
| SAHH | 10 | M1 | 1.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lapillo, M.; Tuccinardi, T.; Martinelli, A.; Macchia, M.; Giordano, A.; Poli, G. Extensive Reliability Evaluation of Docking-Based Target-Fishing Strategies. Int. J. Mol. Sci. 2019, 20, 1023. https://doi.org/10.3390/ijms20051023
Lapillo M, Tuccinardi T, Martinelli A, Macchia M, Giordano A, Poli G. Extensive Reliability Evaluation of Docking-Based Target-Fishing Strategies. International Journal of Molecular Sciences. 2019; 20(5):1023. https://doi.org/10.3390/ijms20051023
Chicago/Turabian StyleLapillo, Margherita, Tiziano Tuccinardi, Adriano Martinelli, Marco Macchia, Antonio Giordano, and Giulio Poli. 2019. "Extensive Reliability Evaluation of Docking-Based Target-Fishing Strategies" International Journal of Molecular Sciences 20, no. 5: 1023. https://doi.org/10.3390/ijms20051023
APA StyleLapillo, M., Tuccinardi, T., Martinelli, A., Macchia, M., Giordano, A., & Poli, G. (2019). Extensive Reliability Evaluation of Docking-Based Target-Fishing Strategies. International Journal of Molecular Sciences, 20(5), 1023. https://doi.org/10.3390/ijms20051023









