Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies
Abstract
1. Introduction
2. Results and Discussion
2.1. Localization of RLs in Simplified Phospholipidic Biomimetic Plant Membranes
2.2. Impact of RL Insertion on Plant Phospholipidic Plasma Membrane Model: Dynamic Insight
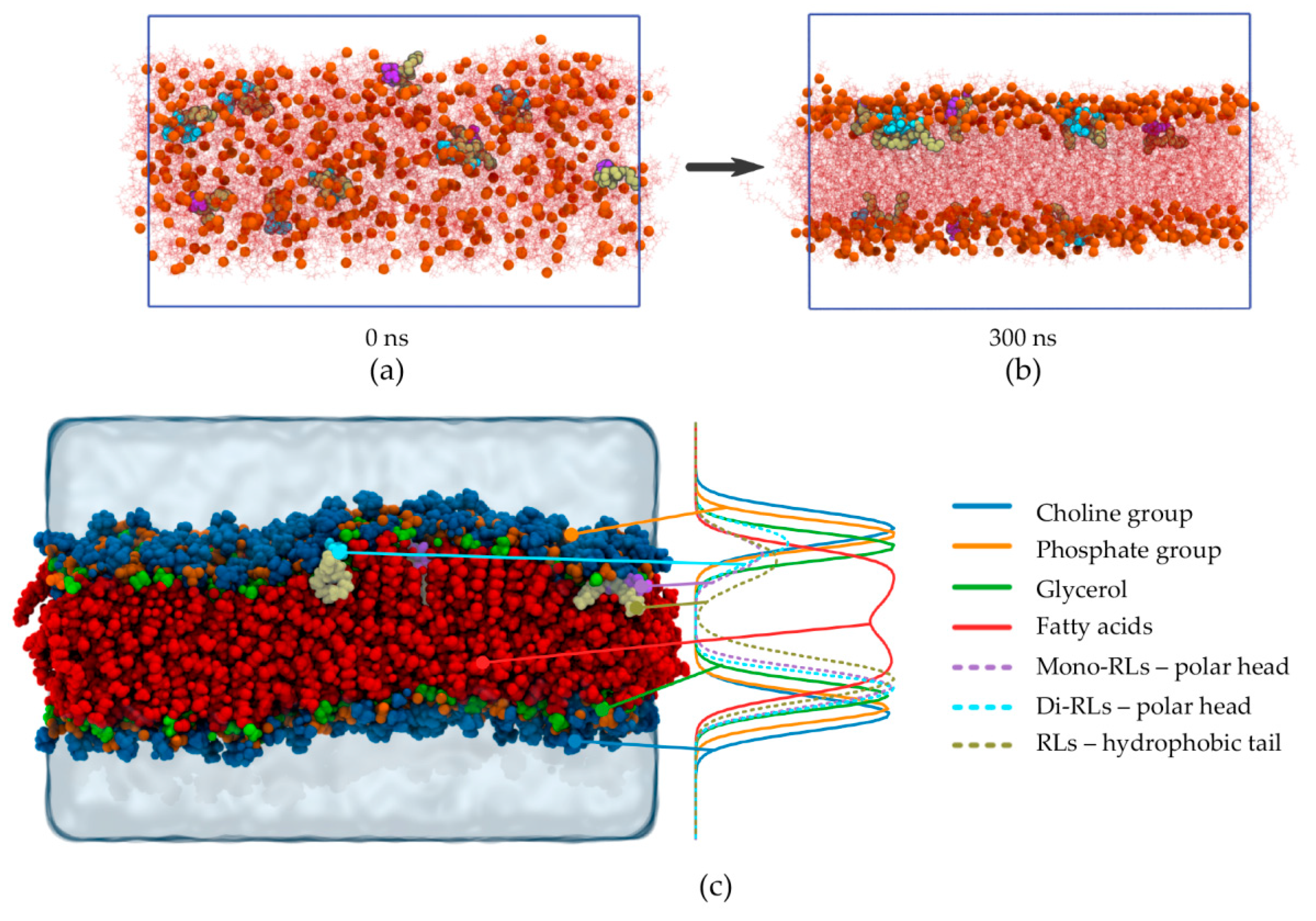
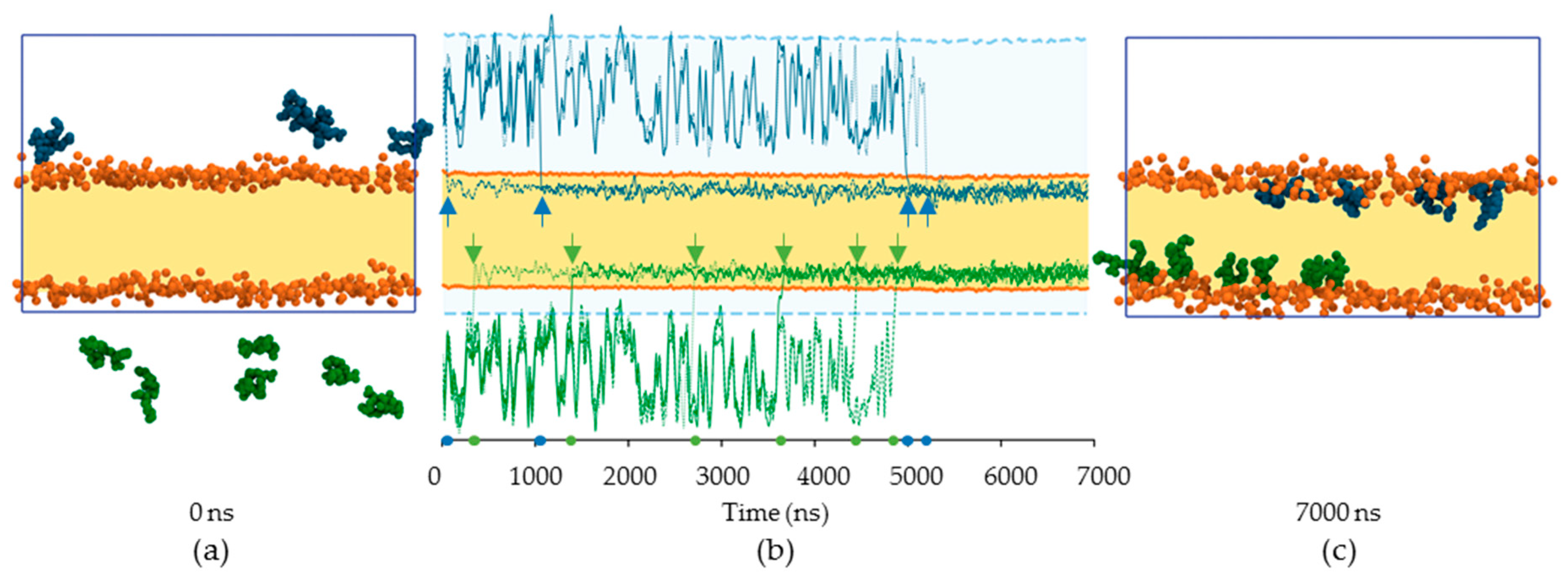
2.2.1. Kinematics of RL Insertion into Lipid Bilayers
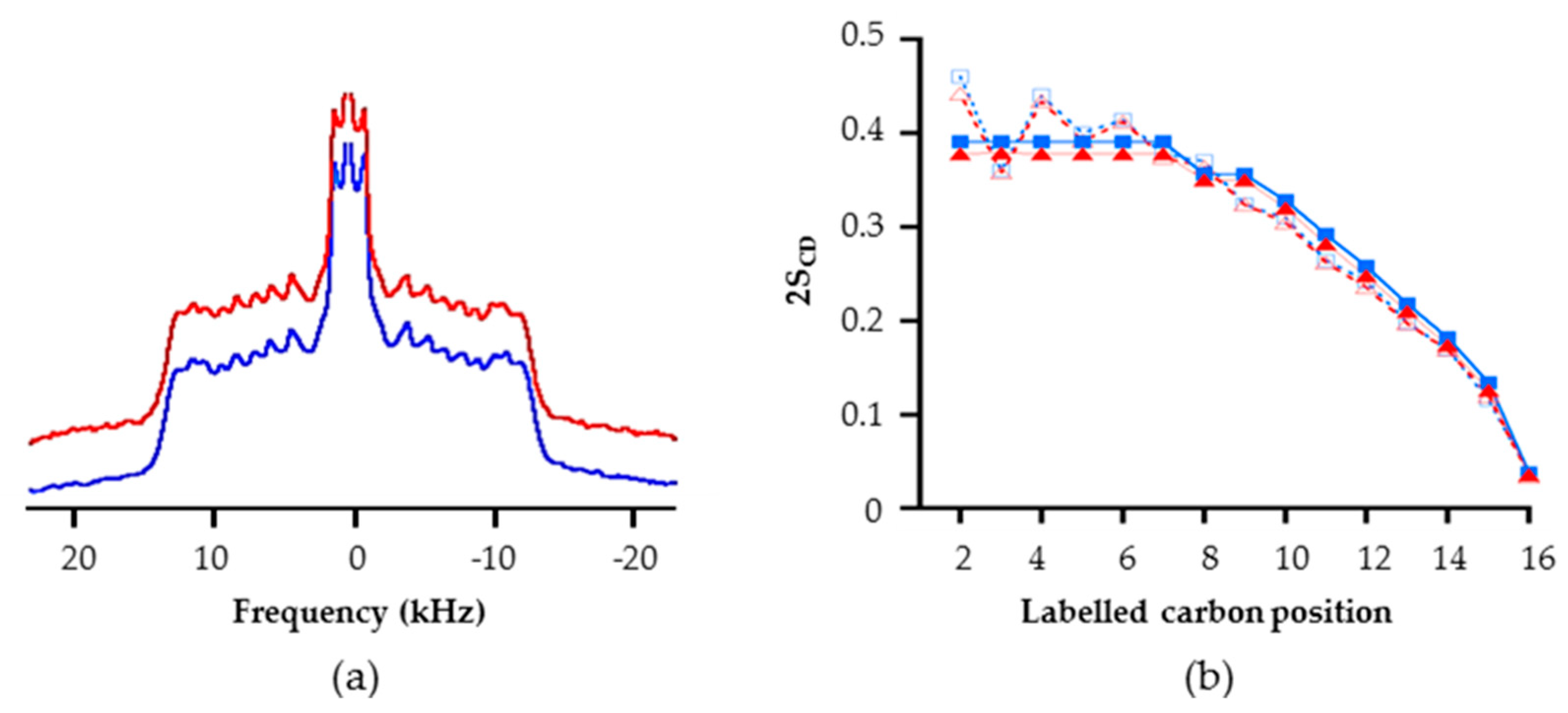
2.2.2. Impact of RL Presence on Lipid Chain Dynamic
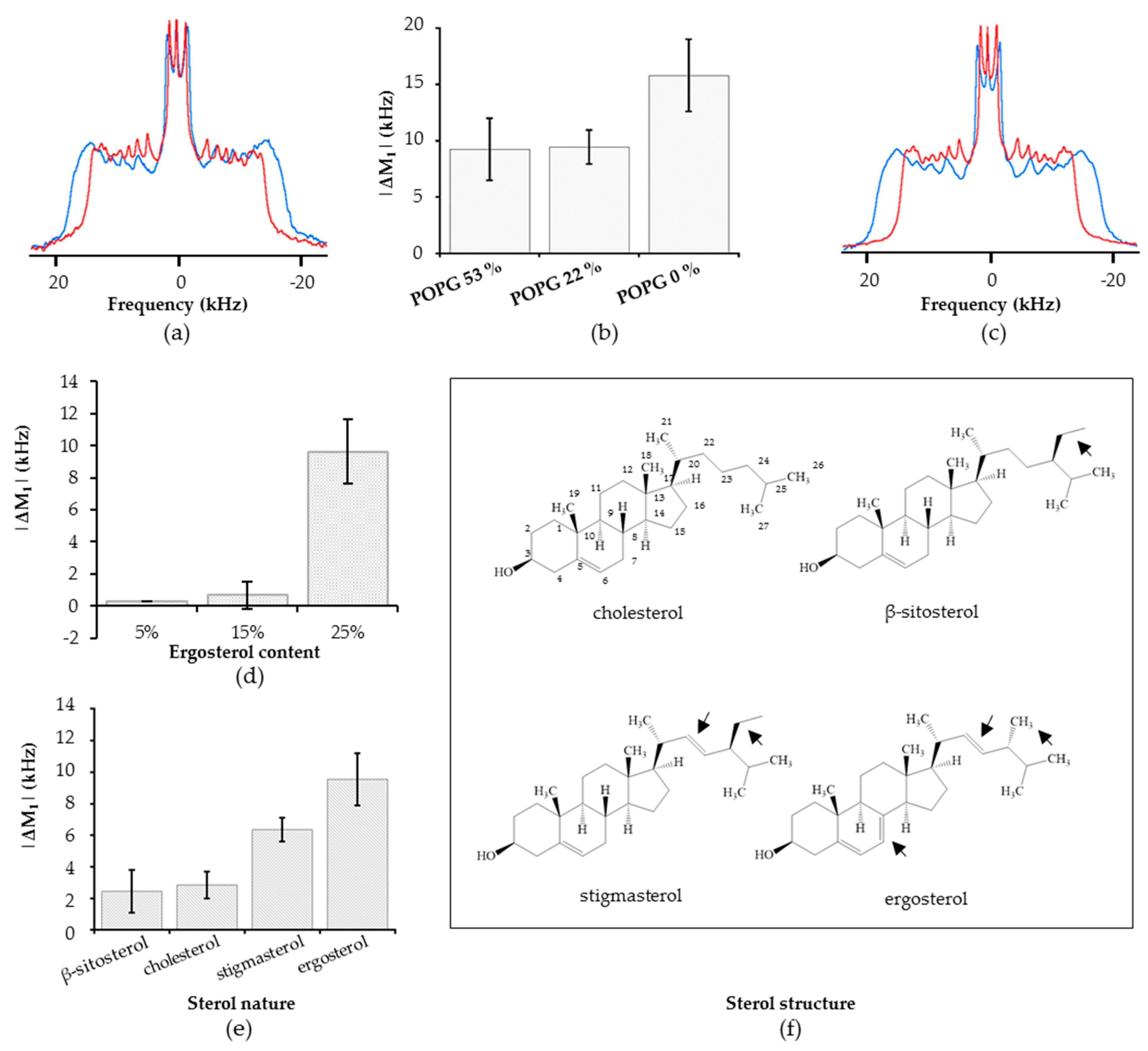
2.3. Getting Closer to Lipid Composition of Plant Plasma Membrane
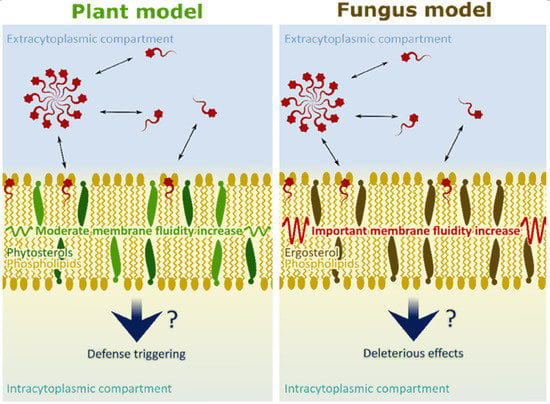
2.4. RL Effect on a Fungi Membrane Model Dynamic
3. Materials and Methods
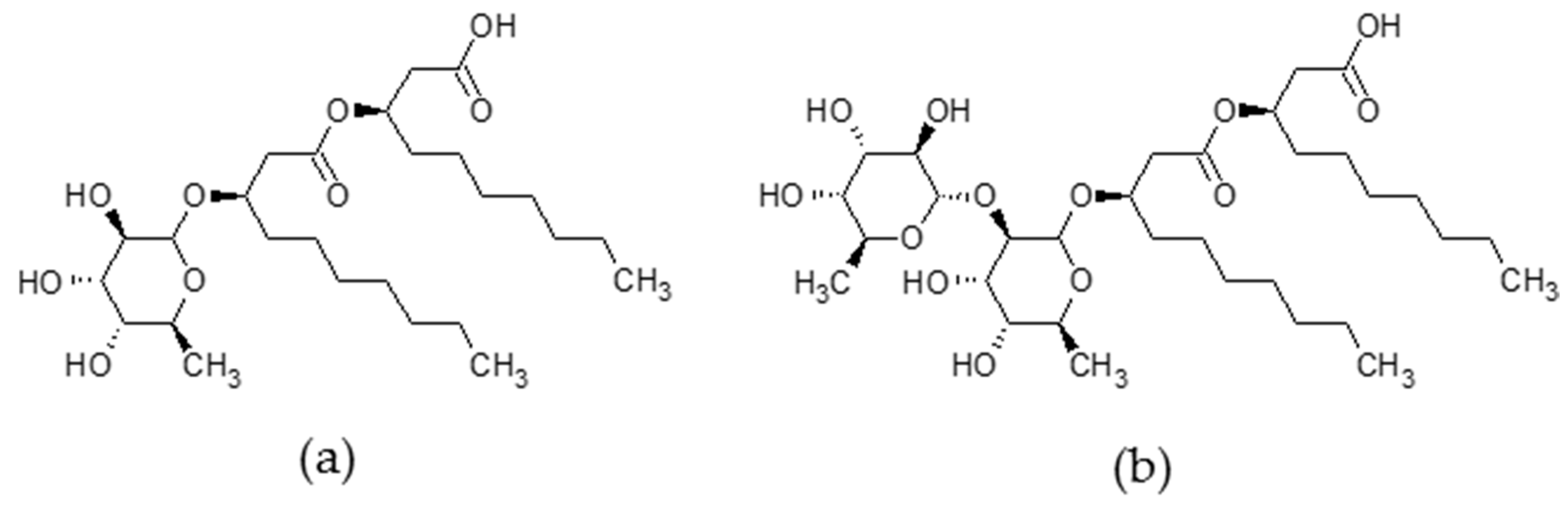
3.1. Materials
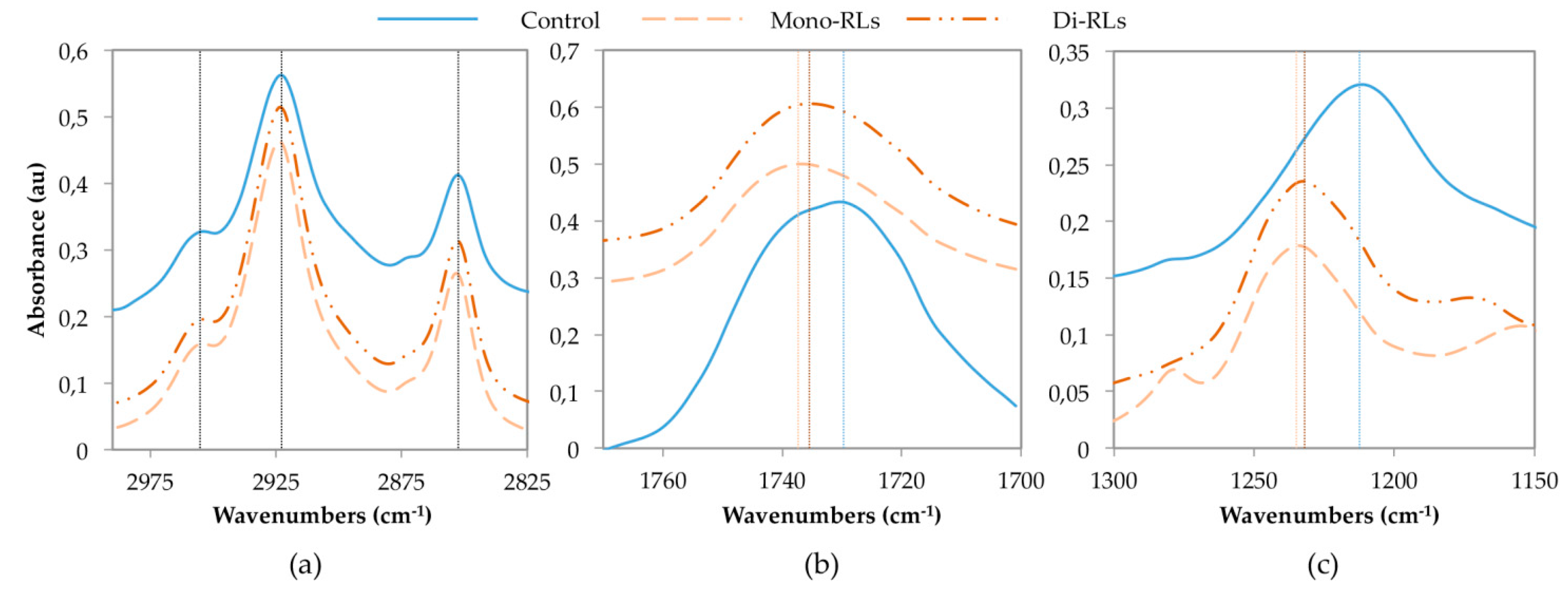
3.2. FTIR Experiments
3.2.1. Sample Preparation
3.2.2. Data Acquisition and Analysis
3.3. Molecular Dynamics Simulations
3.4. NMR Experiments
3.4.1. Sample Preparation
3.4.2. Data Acquisition and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACN | acetonitrile |
| DMPC | 1,2-dimyristoyl-sn-glycero-3-phosphocholine |
| DPPC | 1,2-dipalmitoyl-sn-glycero-3-phosphocholine |
| DTGS | deuterated tri glycine sulfate |
| ELSD | evaporative light scattering detector |
| ERDF | European regional development fund |
| FTIR | Fourier transform infrared spectroscopy |
| HdF | Hauts de France |
| HPLC | high-performance liquid chromatography |
| LINCS | linear constraint solver |
| M1 | first spectral moment |
| MD | molecular dynamics |
| NMR | nuclear magnetic resonance |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PG | phosphatidylglycerol |
| PI | phosphatidylinositol |
| PLPC | 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine |
| POPC | 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine |
| POPG | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) |
| PTFE | polytetrafluoroethylene |
| RL | rhamnolipid |
| SCD | notation for the first order parameter |
| SPMESCD | smooth particle mesh Ewaldfirst order parameter |
References
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, K.K.S.; Rahman, P.K.S.M. Rhamnolipid biosurfactants-past, present, and future scenario of global market. Front. Microbiol. 2014, 5, 454. [Google Scholar] [CrossRef]
- Santos, D.K.F.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional Biomolecules of the 21st Century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, Y.; Gama, Y.; Nagahora, H.; Yamaguchi, M.; Nakahara, H.; Kamata, T. The pH-sensitive conversion of molecular aggregates of rhamnolipid biosurfactant. Chem. Lett. 1987, 16, 763–766. [Google Scholar] [CrossRef]
- Kłosowska-Chomiczewska, I.E.; Mędrzycka, K.; Hallmann, E.; Karpenko, E.; Pokynbroda, T.; Macierzanka, A.; Jungnickel, C. Rhamnolipid CMC prediction. J. Colloid Interface Sci. 2017, 488, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Cieśla, J.; Koczańska, M.; Bieganowski, A. An interaction of rhamnolipids with Cu2+ ions. Molecules 2018, 23, 488. [Google Scholar] [CrossRef] [PubMed]
- Vatsa, P.; Sanchez, L.; Clément, C.; Baillieul, F.; Dorey, S. Rhamnolipid biosurfactants as new players in animal and plant defense against microbes. Int. J. Mol. Sci. 2010, 11, 5095–5108. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Q.; Hua, Y.; Chen, J.; Zhang, H.; Wang, H. Potential applications of biosurfactant rhamnolipids in agriculture and biomedicine. Appl. Microbiol. Biotechnol. 2017, 101, 8309–8319. [Google Scholar] [CrossRef] [PubMed]
- Deleu, M.; Crowet, J.M.; Nasir, M.N.; Lins, L. Complementary biophysical tools to investigate lipid specificity in the interaction between bioactive molecules and the plasma membrane: A review. Biochim. Biophys. Acta Biomembr. 2014, 1838, 3171–3190. [Google Scholar] [CrossRef] [PubMed]
- Pashynska, V.A. Mass spectrometric study of rhamnolipid biosurfactants and their interactions with cell membrane phospholipids. Biopolym. Cell 2009, 25, 504–508. [Google Scholar] [CrossRef]
- Aranda, F.J.; Espuny, M.J.; Marque, A.; Teruel, J.A.; Manresa, A.; Ortiz, A. Thermodynamics of the Interaction of a Dirhamnolipid Biosurfactant Secreted by Pseudomonas aeruginosa with Phospholipid Membranes. Langmuir 2007, 23, 2700–2705. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Aranda, F.J.; Teruel, J.A.; Espuny, M.J.; Marqués, A.; Manresa, A.; Ortiz, A. Permeabilization of biological and artificial membranes by a bacterial dirhamnolipid produced by Pseudomonas aeruginosa. J. Colloid Interface Sci. 2010, 341, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, A.; Teruel, J.A.; Espuny, M.J.; Marqués, A.; Manresa, A.; Aranda, F.J. Effects of dirhamnolipid on the structural properties of phosphatidylcholine membranes. Int. J. Pharm. 2006, 325, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Aranda, F.J.; Teruel, J.A.; Ortiz, A. Interaction of a bacterial dirhamnolipid with phosphatidylcholine membranes: A biophysical study. Chem. Phys. Lipids 2009, 161, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, H.; Noghabi, K.A.; Ortiz, A. Interaction of a bacterial monorhamnolipid secreted by Pseudomonas aeruginosa MA01 with phosphatidylcholine model membranes. Chem. Phys. Lipids 2012, 165, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Aranda, F.J.; Teruel, J.A.; Ortiz, A. New pH-sensitive liposomes containing phosphatidylethanolamine and a bacterial dirhamnolipid. Chem. Phys. Lipids 2011, 164, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Haba, E.; Pinazo, A.; Pons, R.; Pérez, L.; Manresa, A. Complex rhamnolipid mixture characterization and its influence on DPPC bilayer organization. Biochim. Biophys. Acta 2014, 1838, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Moussa, Z.; Chebl, M.; Patra, D. Interaction of curcumin with 1,2-dioctadecanoyl-sn-glycero-3-phosphocholine liposomes: Intercalation of rhamnolipids enhances membrane fluidity, permeability and stability of drug molecule. Colloids Surf. B Biointerfaces 2017, 149, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.; Teruel, J.A.; Espuny, M.J.; Marqués, A.; Aranda, F.J.; Manresa, A.; Ortiz, A. Modulation of the physical properties of dielaidoylphosphatidylethanolamine membranes by a dirhamnolipid biosurfactant produced by Pseudomonas aeruginosa. Chem. Phys. Lipids 2006, 142, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, H.; Aranda, F.J.; Noghabi, K.A.; Ortiz, A. A bacterial monorhamnolipid alters the biophysical properties of phosphatidylethanolamine model membranes. Biochim. Biophys. Acta 2013, 1828, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- Stanghellini, M.E.; Miller, R.M. Biosurfactants: Their Identity and Potential Efficacy in the Biological Control of Zoosporic Plant Pathogens. Plant Dis. 1997, 81, 4–12. [Google Scholar] [CrossRef]
- Kim, B.S.; Lee, J.Y.; Hwang, B.K. In vivo control and in vitro antifungal activity of rhamnolipid B, a glycolipid antibiotic, against Phytophthora capsici and Colletotrichum orbiculare. Pest Manag. Sci. 2000, 56, 1029–1035. [Google Scholar] [CrossRef]
- Varnier, A.-L.; Sanchez, L.; Vatsa, P.; Boudesocque, L.; Garcia-Brugger, A.; Rabenoelina, F.; Sorokin, A.; Renault, J.-H.; Kauffmann, S.; Pugin, A.; et al. Bacterial rhamnolipids are novel MAMPs conferring resistance to Botrytis cinerea in grapevine. Plant Cell Environ. 2009, 32, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Monnier, N.; Furlan, A.; Botcazon, C.; Dahi, A.; Mongelard, G.; Cordelier, S.; Clément, C.; Dorey, S.; Sarazin, C.; Rippa, S. Rhamnolipids From Pseudomonas aeruginosa Are Elicitors Triggering Brassica napus Protection Against Botrytis cinerea Without Physiological Disorders. Front. Plant Sci. 2018, 9, 1170. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.; Courteaux, B.; Hubert, J.; Kauffmann, S.; Renault, J.-H.; Clément, C.; Baillieul, F.; Dorey, S. Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid. Plant Physiol. 2012, 160, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Hu, H.; Lu, L.; Zheng, X. Rhamnolipids induce oxidative stress responses in cherry tomato fruit to Alternaria alternata. Pest Manag. Sci. 2015, 72, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S. Sensing of molecular patterns through cell surface immune receptors. Curr. Opin. Plant Biol. 2017, 38, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Brandenburg, K.; Zähringer, U.; Rademann, J. Chemical Synthesis of a Glycolipid Library by a Solid-Phase Strategy Allows Elucidation of the Structural Specificity of Immunostimulation by Rhamnolipids. Chem. Eur. J. 2006, 12, 7116–7124. [Google Scholar] [CrossRef] [PubMed]
- Kouzayha, A.; Wattraint, O.; Sarazin, C. Interactions of two transmembrane peptides in supported lipid bilayers studied by a 31P and 15N MAOSS NMR strategy. Biochimie 2009, 91, 774–778. [Google Scholar] [CrossRef] [PubMed]
- Rippa, S.; Eid, M.; Formaggio, F.; Toniolo, C.; Béven, L. Hypersensitive-like response to the pore-former peptaibol alamethicin in Arabidopsis thaliana. Chembiochem 2010, 11, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Haapalainen, M.; Engelhardt, S.; Küfner, I.; Li, C.M.; Nürnberger, T.; Lee, J.; Romantschuk, M.; Taira, S. Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol. Plant Pathol. 2011, 12, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Buchholz, G.; Nick, P. The cytoskeleton is disrupted by the bacterial effector HrpZ, but not by the bacterial PAMP flg22, in tobacco BY-2 cells. J. Exp. Bot. 2013, 64, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Henry, G.; Deleu, M.; Jourdan, E.; Thonart, P.; Ongena, M. The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell. Microbiol. 2011, 13, 1824–1837. [Google Scholar] [CrossRef] [PubMed]
- Nasir, M.N.; Lins, L.; Crowet, J.M.; Ongena, M.; Dorey, S.; Dhondt-Cordelier, S.; Clément, C.; Bouquillon, S.; Haudrechy, A.; Sarazin, C.; et al. Differential Interaction of Synthetic Glycolipids with Biomimetic Plasma Membrane Lipids Correlates with the Plant Biological Response. Langmuir 2017, 33, 9979–9987. [Google Scholar] [CrossRef] [PubMed]
- Luzuriaga-Loaiza, W.P.; Schellenberger, R.; De Gaetano, Y.; Obounou Akong, F.; Villaume, S.; Crouzet, J.; Haudrechy, A.; Baillieul, F.; Clément, C.; Lins, L.; et al. Synthetic Rhamnolipid Bolaforms trigger an innate immune response in Arabidopsis thaliana. Sci. Rep. 2018, 8, 8534. [Google Scholar] [CrossRef] [PubMed]
- Buchoux, S.; Lai-Kee-Him, J.; Garnier, M.; Tsan, P.; Besson, F.; Brisson, A.; Dufourc, E.J. Surfactin-triggered small vesicle formation of negatively charged membranes: A novel membrane-lysis mechanism. Biophys. J. 2008, 95, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Heerklotz, H.; Wieprecht, T.; Seelig, J. Membrane Perturbation by the Lipopeptide Surfactin and Detergents as Studied by Deuterium NMR. J. Phys. Chem. B 2004, 108, 4909–4915. [Google Scholar] [CrossRef]
- Kell, H.; Holzwarth, J.F.; Boettcher, C.; Heenan, R.K.; Vater, J. Physicochemical studies of the interaction of the lipoheptapeptide surfactin with lipid bilayers of L-alpha-dimyristoyl phosphatidylcholine. Biophys. Chem. 2007, 128, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Grau, A.; Fernández, J.G.; Peypoux, F.; Ortiz, A. A study on the interactions of surfactin with phospholipid vesicles. Biochim. Biophys. Acta 1999, 1418, 307–319. [Google Scholar] [CrossRef]
- Liu, J.; Hagberg, I.; Novitsky, L.; Hadj-Moussa, H.; Avis, T.J. Interaction of antimicrobial cyclic lipopeptides from Bacillus subtilis influences their effect on spore germination and membrane permeability in fungal plant pathogens. Fungal Biol. 2014, 118, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Lebrón-Paler, A.; Pemberton, J.E.; Becker, B.A.; Otto, W.H.; Larive, C.K.; Maier, R.M. Determination of the acid dissociation constant of the biosurfactant monorhamnolipid in aqueous solution by potentiometric and spectroscopic methods. Anal. Chem. 2006, 78, 7649–7658. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mawgoud, A.M.; Aboulwafa, M.M.; Hassouna, N.A.-H. Characterization of Rhamnolipid Produced by Pseudomonas aeruginosa Isolate Bs20. Appl. Biochem. Biotechnol. 2009, 157, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Coss, C.S.; Mudalige, A.; Polt, R.L.; Pemberton, J.E. A PM-IRRAS investigation of monorhamnolipid orientation at the air-water interface. Langmuir 2013, 29, 4441–4450. [Google Scholar] [CrossRef] [PubMed]
- Salnikov, E.S.; Mason, A.J.; Bechinger, B. Membrane order perturbation in the presence of antimicrobial peptides by 2H solid-state NMR spectroscopy. Biochimie 2009, 91, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Joseph, R.A. Cold Acclimation of Arabidopsis Thaliana. Effect on Plasma Membrane Lipid Composition and Freeze-lnduced Lesions. Plant Physiol. 1995, 109, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Goni, F.M.; Arrondo, J.L.R. A study of phospholipid phosphate groups in model membranes by Fourier transform IR spectroscopy. Faraday Discuss. Chem. Soc. 1986, 81, 117–126. [Google Scholar] [CrossRef]
- Mason, A.J.; Marquette, A.; Bechinger, B. Zwitterionic Phospholipids and Sterols Modulate Antimicrobial Peptide-Induced Membrane Destabilization. Biophys. J. 2007, 93, 4289–4299. [Google Scholar] [CrossRef] [PubMed]
- Svetlovics, J.A.; Wheaten, S.A.; Almeida, P.F. Phase separation and fluctuations in mixtures of a saturated and an unsaturated phospholipid. Biophys. J. 2012, 102, 2526–2535. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Bhunia, A.; Kotler, S.A.; Ramamoorthy, A. Detergent-Type Membrane Fragmentation by MSI-78, MSI-367, MSI-594, and MSI-843 Antimicrobial Peptides and Inhibition by Cholesterol: A Solid-State Nuclear Magnetic Resonance Study. Biochemistry 2015, 54, 1897–1907. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Penfold, J.; Thomas, R.K.; Smyth, T.J.P.; Perfumo, A.; Marchant, R.; Banat, I.M.; Stevenson, P.; Parry, A.; Tucker, I.; et al. Solution Self-Assembly and Adsorption at the Air−Water Interface of the Monorhamnose and Dirhamnose Rhamnolipids and Their Mixtures. Langmuir 2010, 26, 18281–18292. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta Rev. Biomembr. 1983, 737, 117–171. [Google Scholar] [CrossRef]
- Brown, M.; Lope-Piedrafita, S. Solid-State Deuterium NMR Spectroscopy of Membranes; Webb, G.A., Ed.; Springer: New York, NY, USA, 2006; ISBN 978-1-4020-3894-5. [Google Scholar]
- Grélard, A.; Loudet, C.; Diller, A.; Dufourc, E.J. NMR Spectroscopy of Lipid Bilayers. In Membrane Protein Structure Determination: Methods and Protocols; Lacapère, J.-J., Ed.; Springer: New York, NY, USA, 2010; Volume 654, p. 459. ISBN 9781607617617. [Google Scholar]
- Davis, J.H.; Maraviglia, B.; Weeks, G.; Godin, D.V. Bilayer rigidity of the erythrocyte membrane 2H-NMR of a perdeuterated palmitic acid probe. Biochim. Biophys. Acta 1979, 550, 362–366. [Google Scholar] [CrossRef]
- Leung, S.S.W.; Thewalt, J. Deuterium NMR of Mixed Lipid Membranes; Separovic, F., Naito, A., Eds.; Royal Society of Chemistry: London, UK, 2014; ISBN 978-1-84973-910-8. [Google Scholar]
- Felle, H.H. pH: Signal and Messenger in Plant Cells. Plant Biol. 2001, 3, 577–591. [Google Scholar] [CrossRef]
- Minami, A.; Fujiwara, M.; Furuto, A.; Fukao, Y.; Yamashita, T.; Kamo, M.; Kawamura, Y.; Uemura, M. Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 2009, 50, 341–359. [Google Scholar] [CrossRef] [PubMed]
- Funnekotter, B.; Kaczmarczyk, A.; Turner, S.R.; Bunn, E.; Zhou, W.; Smith, S.; Flematti, G.; Mancera, R.L. Acclimation-induced changes in cell membrane composition and influence on cryotolerance of in vitro shoots of native plant species. Plant Cell Tissue Organ Cult. 2013, 114, 83–96. [Google Scholar] [CrossRef]
- Ipsen, J.H.; Mouritsen, O.G.; Zuckermann, M.J. Theory of thermal anomalies in the specific heat of lipid bilayers containing cholesterol. Biophys. J. 1989, 56, 661–667. [Google Scholar] [CrossRef]
- Vist, M.R.; Davis, J.H. Phase Equilibria of Cholesterol/Dipalmitoylphosphatidylcholine Mixtures: 2H Nuclear Magnetic Resonance and Differential Scanning Calorimetry. Biochemistry 1990, 29, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Veatch, S.L.; Keller, S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003, 85, 3074–3083. [Google Scholar] [CrossRef]
- Beck, J.G.; Mathieu, D.; Loudet, C.; Buchoux, S.; Dufourc, E.J. Plant sterols in “rafts”: A better way to regulate membrane thermal shocks. FASEB J. 2007, 21, 1714–1723. [Google Scholar] [CrossRef] [PubMed]
- Bartels, T.; Lankalapalli, R.S.; Bittman, R.; Beyer, K.; Brown, M.F. Raftlike mixtures of sphingomyelin and cholesterol investigated by solid-state 2H NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 14521–14532. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, K.; Mongrand, S.; Beney, L.; Simon-Plas, F.; Gerbeau-Pissot, P. Differential Effect of Plant Lipids on Membrane Organization. J. Biol. Chem. 2015, 290, 5810–5825. [Google Scholar] [CrossRef] [PubMed]
- Sha, R.; Meng, Q. Antifungal activity of rhamnolipids against dimorphic fungi. J. Gen. Appl. Microbiol. 2016, 62, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Avis, T.J.; Bélanger, R.R. Specificity and mode of action of the antifungal fatty acid cis-9-heptadecenoic acid produced by Pseudozyma flocculosa. Appl. Environ. Microbiol. 2001, 67, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Wise, C.; Falardeau, J.; Hagberg, I.; Avis, T.J. Cellular Lipid Composition Affects Sensitivity of Plant Pathogens to Fengycin, an Antifungal Compound Produced by Bacillus subtilis Strain CU12. Phytopathology 2014, 104, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Rella, A.; Farnoud, A.M.; Del Poeta, M. Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 2016, 61, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Gronnier, J.; Gerbeau-Pissot, P.; Germain, V.; Mongrand, S.; Simon-Plas, F. Divide and Rule: Plant Plasma Membrane Organization. Trends Plant Sci. 2018, 23, 899–917. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Mizutani, M.; Aoki, N.; Watanabe, B.; Saga, H.; Saito, S.; Oikawa, A.; Suzuki, H.; Sakurai, N.; Shibata, D.; et al. Cytochrome P450 CYP710A Encodes the Sterol C-22 Desaturase in Arabidopsis and Tomato. Plant Cell 2006, 18, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Targeting pathogen sterols: Defence and counterdefence? PLoS Pathog. 2017, 13, e1006297. [Google Scholar] [CrossRef] [PubMed]
- Gamir, J.; Darwiche, R.; Van’t Hof, P.; Choudhary, V.; Stumpe, M.; Schneiter, R.; Mauch, F. The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 2017, 89, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Mas, A.; Navarro-Pedreño, J.; Cooke, D.T.; James, C.S. Characterization and lipid composition of the plasma membrane in grape leaves. Phytochemistry 1994, 35, 1249–1253. [Google Scholar] [CrossRef]
- Borner, G.H.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; Macaskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of Detergent-Resistant Membranes in Arabidopsis. Evidence for Plasma Membrane Lipid Rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Laloi, M.; Perret, A.-M.; Chatre, L.; Melser, S.; Cantrel, C.; Vaultier, M.-N.; Zachowski, A.; Bathany, K.; Schmitter, J.-M.; Vallet, M.; et al. Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 2007, 143, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Chalbi, N.; Martínez-Ballesta, M.C.; Youssef, N.B.; Carvajal, M. Intrinsic stability of Brassicaceae plasma membrane in relation to changes in proteins and lipids as a response to salinity. J. Plant Physiol. 2015, 175, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Keinath, N.F.; Kierszniowska, S.; Lorek, J.; Bourdais, G.; Kessler, S.A.; Shimosato-Asano, H.; Grossniklaus, U.; Schulze, W.X.; Robatzek, S.; Panstruga, R. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 2010, 285, 39140–39149. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Fan, L.; Chen, T.; Li, R.; Li, X.; He, Q.; Botella, M.A.; Lin, J. Clathrin and Membrane Microdomains Cooperatively Regulate RbohD Dynamics and Activity in Arabidopsis. Plant Cell 2014, 26, 1729–1745. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Van Der Spoel, D.; Van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softw. X 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- ATB|Topology Converter. Available online: https://atb.uq.edu.au/index.py?tab=topology_converter (accessed on 3 December 2018).
- Jämbeck, J.P.M.; Lyubartsev, A.P. An extension and further validation of an all-atomistic force field for biological membranes. J. Chem. Theory Comput. 2012, 8, 2938–2948. [Google Scholar] [CrossRef] [PubMed]
- Jämbeck, J.P.M.; Lyubartsev, A.P. Derivation and systematic validation of a refined all-atom force field for phosphatidylcholine lipids. J. Phys. Chem. B 2012, 116, 3164–3179. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. ISBN 978-90-481-8368-5. [Google Scholar]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.J.E.; Melo, M.N.; Seyler, S.L.; Domański, J.; Dotson, D.L.; Buchoux, S.; Kenney, I.M.; et al. MDAnalysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations. In Proceedings of the 15th Python in Science Conference, Austin, TX, USA, 11–17 July 2016; pp. 98–105. [Google Scholar]
- Visual Molecular Dynamic. Available online: https://www.ks.uiuc.edu/Research/vmd/ (accessed on 30 January 2019).
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Hoffman, R.E.; Becker, E.D. Temperature dependence of the 1H chemical shift of tetramethylsilane in chloroform, methanol, and dimethylsulfoxide. J. Magn. Reson. 2005, 176, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H.; Jeffrey, K.R.; Bloom, M.; Valic, M.I.; Higgs, T.P. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem. Phys. Lett. 1976, 42, 390–394. [Google Scholar] [CrossRef]
- Buchoux, S. Nmrdepaker in Launchpad. Available online: https://launchpad.net/nmrdepaker (accessed on 30 November 2018).
- Burnett, L.J.; Muller, B.H. Deuteron Quadrupole Coupling Constants in Three Solid Deuterated Paraffin Hydrocarbons: C2D6, C4D10, C6D14. J. Chem. Phys. 1971, 55, 5829–5831. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monnier, N.; Furlan, A.L.; Buchoux, S.; Deleu, M.; Dauchez, M.; Rippa, S.; Sarazin, C. Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies. Int. J. Mol. Sci. 2019, 20, 1009. https://doi.org/10.3390/ijms20051009
Monnier N, Furlan AL, Buchoux S, Deleu M, Dauchez M, Rippa S, Sarazin C. Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies. International Journal of Molecular Sciences. 2019; 20(5):1009. https://doi.org/10.3390/ijms20051009
Chicago/Turabian StyleMonnier, Noadya, Aurélien L. Furlan, Sébastien Buchoux, Magali Deleu, Manuel Dauchez, Sonia Rippa, and Catherine Sarazin. 2019. "Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies" International Journal of Molecular Sciences 20, no. 5: 1009. https://doi.org/10.3390/ijms20051009
APA StyleMonnier, N., Furlan, A. L., Buchoux, S., Deleu, M., Dauchez, M., Rippa, S., & Sarazin, C. (2019). Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies. International Journal of Molecular Sciences, 20(5), 1009. https://doi.org/10.3390/ijms20051009