From Human Cytogenetics to Human Chromosomics
Abstract
1. Introduction
2. Human Chromosomes
3. Contributions of Prof. Uwe Claussen to Human Cytogenetics
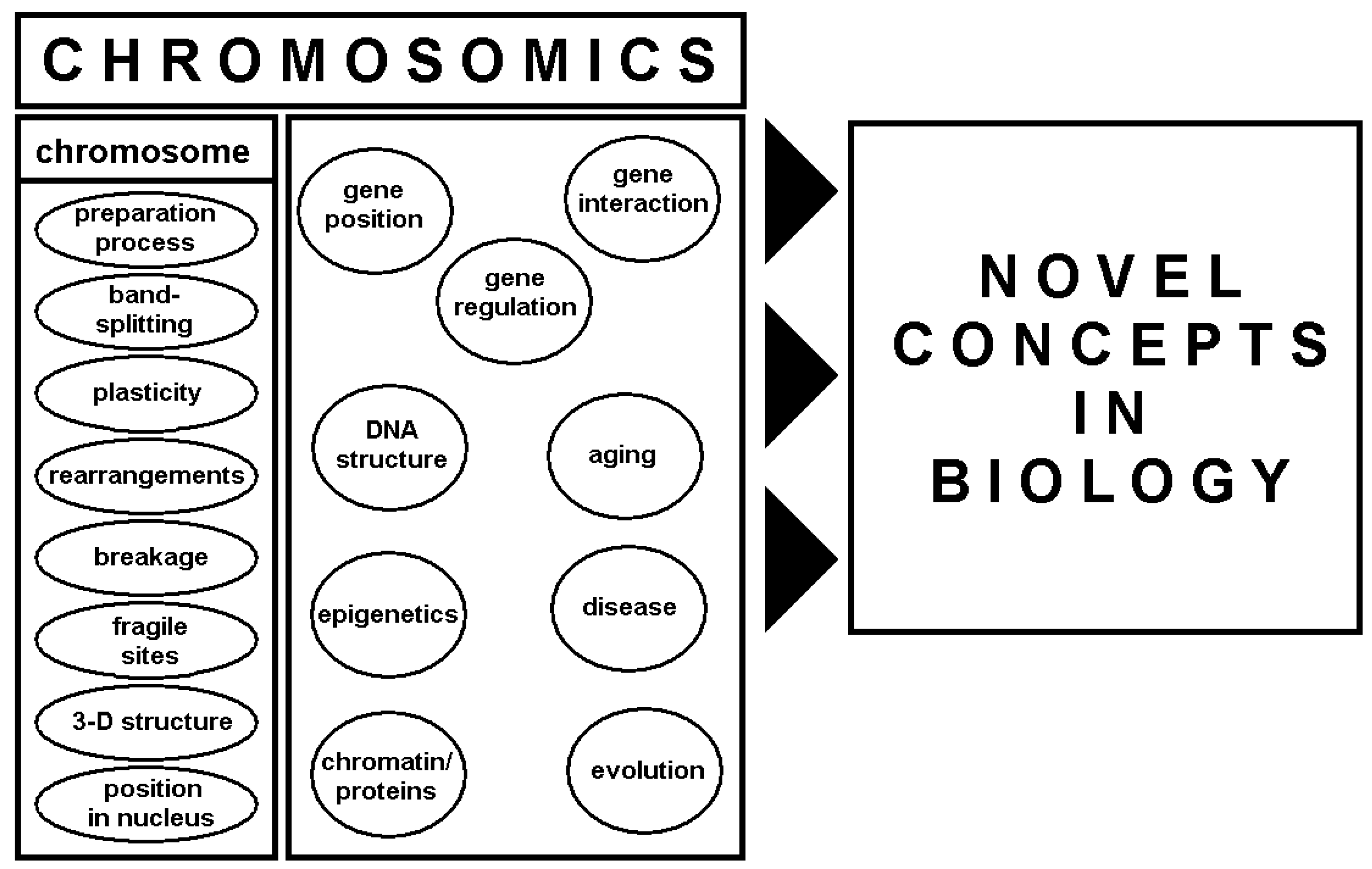
4. Chromosomics
- -
- “on plasticity of chromosomes in relation to the three-dimensional positions of genes, which affect cell function in a developmental and tissue-specific manner during the cell cycle” [3]. This included studies on chromosome structure in meta- and inter-phase [2,73,74,75,76,77,78,79,80,81], as well as studies by Thomas Cremer [82,83,84,85,86,87] and others [92,93,94,95,96,97,98] that used three-dimensional-FISH and HiC-analyses [42,99,100,101,102,103,104,105,106]. Today, it is theorized that gene expression is dependent upon and regulated by chromosome structure in interphase. Thus, new concepts are already being integrated into transcriptomic research, with chromatin modifications largely considered major epigenetic factors influencing gene expression [99].
- -
- “into chromatin-modification-mediated changes in the architecture of chromosomes, which may influence the functions and lifespans of cells, tissues, organs and individuals.” Insights into the flexible three-dimensional structures of metaphase chromosomes may also help to understand the influence of aforementioned “positional effects” on cells at different stages of their development. One important consideration is the recently demonstrated fact that each cell of the human body remembers which of the homologous chromosome sets derives from the mother and father of the individual [81]. In addition, effects of copy number alterations that appear during aging and their effects on nuclear architecture have yet to be established and further studies are warranted [107].
- -
- on “species-specific differences in the architecture of chromosomes, which has been overlooked in the past” [3]. In that sense, the use of the word chromosomics was correct as used the aforementioned Russian colleagues [88]. Chromosomics studies with evolution focus on the construction of the interphase stage and the effects of this architecture were already performed e.g. in different mammals, reptiles and other species [75,108,109,110,111]. It is still unknown if conserved genes in mammalians keep their position in the same kind of chromosomal band (Giemsa-dark or –light) during evolution. Further studies are needed to elucidate whether changes in position lead to differential expression.
- -
- on “the occurrence and prevalence of chromosomal gaps and breaks and interchanges” [3]. The focus here includes fragile sites [112] and their putative role as seeding points of (i) evolutionary conserved breakpoints [112,113,114], (ii) breakpoints observed in inherited [112,115], and (iii) acquired chromosomal aberrations in tumors [112,116,117]. Recently, as originally suggested by Prof. Claussen [3], fragile site related breaks were attributed to chromosome three-dimensional structure and function rather than to DNA-sequence [118,119,120].
5. Conclusions: Chromosomes and Their Appreciation in Nowadays Human Genetics
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aCGH | array comparative genomic hybridization |
| CNVs | copy number variants |
| DNA | deoxyribonucleic acid |
| FISH | fluorescence in situ hybridization |
| GTG | G-banding by Trypsin and Giemsa |
| Hi-C | high-throughput sequencing based chromosome conformation capture techniques |
| ISCN | International System for Human Cytogenomic Nomenclature |
| Mb | megabasepair |
| MLPA | multiplex-ligation dependent probe amplification |
| PCR | polymerase chain reaction |
| Prof. | professor |
| Q-banding | Quinacrine based banding of chromosomes |
References
- Liehr, T. In memoriam Prof. Dr. med. Uwe Claussen (* 30.04.1945 † 20.07.2008). ECA-Newsletter 2009, 23, 33. [Google Scholar]
- Lemke, J.; Claussen, J.; Michel, S.; Chudoba, I.; Mühlig, P.; Westermann, M.; Sperling, K.; Rubtsov, N.; Grummt, U.W.; Ullmann, P.; et al. The DNA-based structure of human chromosome 5 in interphase. Am. J. Hum. Genet. 2002, 71, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Claussen, U. Chromosomics. Cytogenet. Genome Res. 2005, 111, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Aardema, M.J.; MacGregor, J.T. Toxicology and genetic toxicology in the new era of “toxicogenomics”: Impact of “-omics” technologies. Mutat. Res. 2002, 499, 13–25. [Google Scholar] [CrossRef]
- van Ommen, B.; Stierum, R. Nutrigenomics: Exploiting systems biology in the nutrition and health arena. Curr. Opin. Biotechnol. 2002, 13, 517–521. [Google Scholar] [CrossRef]
- Robosky, L.C.; Robertson, D.G.; Baker, J.D.; Rane, S.; Reily, M.D. In vivo toxicity screening programs using metabonomics. Comb. Chem. High Throughput Screen. 2002, 5, 651–662. [Google Scholar] [CrossRef]
- Gong, J.P. From genomics, proteomics to cytomics, or from cytometry to cytomics. Ai Zheng 2003, 22, 449–451. [Google Scholar]
- Kiechle, F.L.; Zhang, X.; Holland-Staley, C.A. The -omics era and its impact. Arch. Pathol. Lab. Med. 2004, 128, 1337–1345. [Google Scholar]
- Gagna, C.E.; Winokur, D.; Clark Lambert, W. Cell biology, chemogenomics and chemoproteomics. Cell. Biol. Int. 2004, 28, 755–764. [Google Scholar] [CrossRef]
- Dettmer, K.; Hammock, B.D. Metabolomics—A new exciting field within the “omics” sciences. Environ. Health Perspect. 2004, 112, A396–A397. [Google Scholar] [CrossRef]
- Thongboonkerd, V. Genomics, proteomics and integrative “omics” in hypertension research. Curr. Opin. Nephrol. Hypertens. 2005, 14, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Banfield, J.F.; Verberkmoes, N.C.; Hettich, R.L.; Thelen, M.P. Proteogenomic approaches for the molecular characterization of natural microbial communities. OMICS 2005, 9, 301–333. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J. Beobachtungen über Kerntheilungen in den Zellen der Geschwülste. Virchow’s Arch. 1879, 78, 279–301. [Google Scholar] [CrossRef]
- Flemming, W. Zur Kenntniss der Zelle und ihrer Theilungs-Erscheinungen. Schriften des Naturwissenschaftlichen Vereins für Schleswig-Holstein 1878, 3, 23–27. [Google Scholar]
- Waldeyer, W. Uber Karyokinese und ihre Beziehungen zu den Befruchtungsvorgangen. Arch. Mikros Anat. 1888, 32, 1–122. [Google Scholar] [CrossRef]
- Mendel, G. Versuche über Plflanzenhybriden. Verhandlungen des naturforschenden Vereines in Brünn 1866, IV, 3–47. [Google Scholar]
- Ferguson-Smith, M.A. History and evolution of cytogenetics. Mol. Cytogenet. 2015, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Claussen, U. Current developments in human molecular cytogenetic techniques. Curr. Mol. Med. 2002, 2, 283–297. [Google Scholar] [CrossRef]
- Flemming, W. Beiträge zur Kenntniss der Zelle und ihrer Lebenserscheinungen. Archiv für mikroskopische Anatomie 1879, 16, 302–436. [Google Scholar] [CrossRef]
- Sutton, W.S. On the morphology of the chromosome group in Brachystola magna. Biol. Bull 1902, 4, 24–39. [Google Scholar] [CrossRef]
- Baltzer, F. Theodor Boveri. Science 1964, 144, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Crow, E.W.; Crow, J.F. 100 Years Ago: Walter Sutton and the chromosome theory of heredity. Genetics 2001, 160, 1–4. [Google Scholar]
- Boveri, Τ. Über die Konstitution der chromatischen Kernsubstanz. In Verhandlungen der Deutschen Zoologischen Gesellschaft, 13. Jahresversammlung zu Würzburg; Wilhelm Engelmann: Leipzig, Germany, 1903; pp. 10–33. [Google Scholar]
- Sutton, W.S. The chromosomes in heredity. Biol. Bull 1903, 4, 231–251. [Google Scholar] [CrossRef]
- Boveri, T. Zur Frage der Entstehung maligner Tumoren. J. Cell. Sci. 2008, 121, 1–84. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.H. Sex limited inheritance in Drosophila. Science 1910, 32, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.H. Die stoffliche Grundlage der Vererbung; Gebrüder Borntraeger: Berlin, Germany, 1921; pp. 1–291. [Google Scholar]
- Watson, J.D.; Crick, F.H.C. A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Genetical implication for the structure of deoxyribonucleic acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Painter, T.S. The Y-chromosome in mammals. Science 1921, 53, 503–504. [Google Scholar] [CrossRef]
- Hsu, T.C. Mammalian chromosomes in vitro. I. The karyotype of man. J. Hered. 1952, 43, 167–172. [Google Scholar] [CrossRef]
- Tjio, J.H.; Levan, A. The chromosome number of man. Hereditas 1956, 42, 1–6. [Google Scholar] [CrossRef]
- Schlegelberger, B. In memoriam: Prof. Dr. rer. nat. Dr. med. h.c. Lore Zech; 24.9.1923–13.3.2013: Honorary member of the European Society of Human Genetics, Honorary member of the German Society of Human Genetics, Doctor laureate, the University of Kiel, Germany. Mol. Cytogenet. 2013, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Caspersson, T.; Zech, L.; Johansson, C. Analysis of human metaphase chromosomes set by aid of DNAbinding fluorescent agents. Expl. Cell. Res. 1970, 62, 490–492. [Google Scholar] [CrossRef]
- Liehr, T.; Othman, M.A.; Rittscher, K.; Alhourani, E. The current state of molecular cytogenetics in cancer diagnosis. Expert Rev. Mol. Diagn. 2015, 15, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Carreira, I.M.; Aktas, D.; Bakker, E.; Rodríguez de Alba, M.; Coviello, D.A.; Florentin, L.; Scheffer, H.; Rincic, M. European registration process for Clinical Laboratory Geneticists in genetic healthcare. Eur. J. Hum. Genet. 2017, 25, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Manolakos, E.; Vetro, A.; Kefalas, K.; Rapti, S.M.; Louizou, E.; Garas, A.; Kitsos, G.; Vasileiadis, L.; Tsoplou, P.; Eleftheriades, M.; et al. The use of array-CGH in a cohort of Greek children with developmental delay. Mol. Cytogenet. 2010, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Alhourani, E.; Rincic, M.; Othman, M.A.; Pohle, B.; Schlie, C.; Glaser, A.; Liehr, T. Comprehensive chronic lymphocytic leukemia diagnostics by combined multiplex ligation dependent probe amplification (MLPA) and interphase fluorescence in situ hybridization (iFISH). Mol. Cytogenet. 2014, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Cytogenetic contribution to uniparental disomy (UPD). Mol. Cytogenet. 2010, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Nazaryan, L.; Stefanou, E.G.; Hansen, C.; Kosyakova, N.; Bak, M.; Sharkey, F.H.; Mantziou, T.; Papanastasiou, A.D.; Velissariou, V.; Liehr, T.; et al. The strength of combined cytogenetic and mate-pair sequencing techniques illustrated by a germline chromothripsis rearrangement involving FOXP2. Eur. J. Hum. Genet. 2014, 22, 338–343. [Google Scholar] [CrossRef]
- Liehr, T. Expert knowledge on human genetic counselling and chromosomics are necessary for sound genetic laboratory diagnostics. Mol. Exp. Biol. Med. 2017, 1, 1–3. [Google Scholar]
- Maass, P.G.; Weise, A.; Rittscher, K.; Lichtenwald, J.; Barutcu, A.R.; Liehr, T.; Aydin, A.; Wefeld-Neuenfeld, Y.; Pölsler, L.; Tinschert, S.; et al. Reorganization of inter-chromosomal interactions in the 2q37-deletion syndrome. EMBO J. 2018, 37, e96257. [Google Scholar] [CrossRef]
- Meridies, R.; Maar, K.; Claussen, U. Clinical and genetic aspects of the horseshoe kidney (author’s transl). Urol. Int. 1976, 31, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.H.; Claussen, U.; Lüdecke, H.J.; Horsthemke, B.; Ellis, D.; Oakey, H.; Wilson, D.; Burn, J.; Williamson, R.; Scambler, P.J. Interstitial deletions in DiGeorge syndrome detected with microclones from 22q11. Mamm. Genome 1992, 3, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Voigt, H.J.; Beinder, E.; Claussen, U. [Ultrasound detection of suspected chromosome abnormalities in the 1st and 2nd trimester. Results of a prospective study]. Geburtshilfe Frauenheilkd 1994, 54, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Koscielny, S.; Dahse, R.; Sonntag, J.; Riese, U.; Theuer, C.; Hofmann, M.E.; von Eggeling, F.; Claussen, U.; Beleites, E.; Ernst, G.; et al. Clinical implications of telomerase activity and inactivation of the tumor suppressor gene p16 (CDKN2A) in head and neck cancer. Otolaryngol. Pol. 2000, 54, 291–295. [Google Scholar] [PubMed]
- Horsthemke, B.; Nazlican, H.; Hüsing, J.; Klein-Hitpass, L.; Claussen, U.; Michel, S.; Lich, C.; Gillessen-Kaesbach, G.; Buiting, K. Somatic mosaicism for maternal uniparental disomy 15 in a girl with Prader-Willi syndrome: Confirmation by cell cloning and identification of candidate downstream genes. Hum. Mol. Genet. 2003, 12, 2723–2732. [Google Scholar] [CrossRef]
- Nazlican, H.; Zeschnigk, M.; Claussen, U.; Michel, S.; Boehringer, S.; Gillessen-Kaesbach, G.; Buiting, K.; Horsthemke, B. Somatic mosaicism in patients with Angelman syndrome and an imprinting defect. Hum. Mol. Genet. 2004, 13, 2547–2555. [Google Scholar] [CrossRef] [PubMed]
- Schreyer, I.; Neumann, A.; Beensen, V.; Eichhorn, K.H.; Heller, A.; Claussen, U.; Liehr, T. Dup(13)(q14.2-q14.3): Yet another new differential diagnostic aspect for short stature-like phenotype. J. Histochem. Cytochem. 2005, 53, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Kuechler, A.; Eller, C.D.; Sahoo, T.; Baldermann, C.; Lieser, U.; Hesse, M.; Gläser, C.; Hagemann, M.; Yatsenko, S.A.; et al. Minimal phenotype in a girl with trisomy 15q due to t(X;15)(q22.3;q11.2) translocation. Am. J. Med. Genet. A 2006, 140, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Hering, A.; Guratowska, M.; Bucsky, P.; Claussen, U.; Decker, J.; Ernst, G.; Hoeppner, W.; Michel, S.; Neumann, H.; Parlowsky, T.; et al. Characteristic genomic imbalances in pediatric pheochromocytoma. Genes Chromosomes Cancer 2006, 4, 602–607. [Google Scholar] [CrossRef]
- Claussen, U. The pipette method: A new rapid technique for chromosome analysis in prenatal diagnosis. Hum. Genet. 1980, 54, 277–278. [Google Scholar] [CrossRef]
- Bullerdiek, J.; Heyat, M.; Bartnitzke, S.; Claussen, U.; Schloot, W. The pipette-method: Its application to cytogenetic studies of tumor cells cloned in semisolid media. Anticancer Res. 1985, 5, 411–413. [Google Scholar] [PubMed]
- Lüdecke, H.J.; Senger, G.; Claussen, U.; Horsthemke, B. Cloning defined regions of the human genome by microdissection of banded chromosomes and enzymatic amplification. Nature 1989, 338, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Lüdecke, H.J.; Senger, G.; Claussen, U.; Horsthemke, B. Construction and characterization of band-specific DNA libraries. Hum. Genet. 1990, 84, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Senger, G.; Lüdecke, H.J.; Horsthemke, B.; Claussen, U. Microdissection of banded human chromosomes. Hum. Genet. 1990, 84, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, W.; Claussen, U.; Lüdecke, H.J.; Senger, G.; Horsthemke, B.; Geurts Van Kessel, A.; Goertzen, W.; Fahsold, R. New markers for the neurofibromatosis-2 region generated by microdissection of chromosome 22. Genomics 1991, 10, 786–791. [Google Scholar] [CrossRef]
- Claussen, U.; Mazur, A.; Rubtsov, N. Chromosomes are highly elastic and can be stretched. Cytogenet. Cell. Genet. 1994, 66, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Hliscs, R.; Mühlig, P.; Claussen, U. The spreading of metaphases is a slow process which leads to a stretching of chromosomes. Cytogenet. Cell. Genet. 1997, 76, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Claussen, U.; Kleider, W.; Müller, H.G.; Wille, N.; Baumann, H.A. Quality control in routine chromosome analysis: Prediction of total number of bands for the individual case analyzed. Clin. Genet. 1992, 41, 100–104. [Google Scholar] [CrossRef] [PubMed]
- McGowan-Jordan, J.; Simons, A.; Schmid, M. International System Of Cytogenomic Nomenclature (ISCN); S. Karger: Basel, Switzerland, 2016. [Google Scholar]
- Hliscs, R.; Mühlig, P.; Claussen, U. The nature of G-bands analyzed by chromosome stretching. Cytogenet. Cell Genet. 1997, 79, 162–166. [Google Scholar] [CrossRef]
- Kuechler, A.; Mueller, C.R.; Liehr, T.; Claussen, U. Detection of microdeletions in the short arm of the X chromosome by chromosome stretching. Cytogenet. Cell. Genet. 2001, 95, 12–16. [Google Scholar] [CrossRef]
- Lehrer, H.; Weise, A.; Michel, S.; Starke, H.; Mrasek, K.; Heller, A.; Kuechler, A.; Claussen, U.; Liehr, T. The hierarchically organized splitting of chromosome bands into sub-bands analyzed by multicolor banding (MCB). Cytogenet. Genome. Res. 2004, 105, 25–28. [Google Scholar] [CrossRef]
- Kosyakova, N.; Weise, A.; Mrasek, K.; Claussen, U.; Liehr, T.; Nelle, H. The hierarchically organized splitting of chromosomal bands for all human chromosomes. Mol. Cytogenet. 2009, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Claussen, U.; Michel, S.; Mühlig, P.; Westermann, M.; Grummt, U.W.; Kromeyer-Hauschild, K.; Liehr, T. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet. Genome Res. 2002, 98, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Chudoba, I.; Plesch, A.; Lörch, T.; Lemke, J.; Claussen, U.; Senger, G. High resolution multicolor-banding: A new technique for refined FISH analysis of human chromosomes. Cytogenet. Cell. Genet. 1999, 84, 156–160. [Google Scholar] [CrossRef]
- Liehr, T.; Heller, A.; Starke, H.; Rubtsov, N.; Trifonov, V.; Mrasek, K.; Weise, A.; Kuechler, A.; Claussen, U. Microdissection based high resolution multicolor banding for all 24 human chromosomes. Int. J. Mol. Med. 2002, 9, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Weise, A.; Heller, A.; Starke, H.; Mrasek, K.; Kuechler, A.; Weier, H.U.; Claussen, U. Multicolor chromosome banding (MCB) with YAC/BAC-based probes and region-specific microdissection DNA libraries. Cytogenet. Genome Res. 2002, 97, 43–50. [Google Scholar] [CrossRef]
- Nietzel, A.; Rocchi, M.; Starke, H.; Heller, A.; Fiedler, W.; Wlodarska, I.; Loncarevic, I.F.; Beensen, V.; Claussen, U.; Liehr, T. A new multicolor-FISH approach for the characterization of marker chromosomes: Centromere-specific multicolor-FISH (cenM-FISH). Hum. Genet. 2001, 108, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Weise, A.; Gross, M.; Mrasek, K.; Mkrtchyan, H.; Horsthemke, B.; Jonsrud, C.; von Eggeling, F.; Hinreiner, S.; Witthuhn, V.; Claussen, U.; et al. Parental-origin-determination fluorescence in situ hybridization distinguishes homologous human chromosomes on a single-cell level. Int. J. Mol. Med. 2008, 21, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Steinhaeuser, U.; Starke, H.; Nietzel, A.; Lindenau, J.; Ullmann, P.; Claussen, U.; Liehr, T. Suspension (S)-FISH, a new technique for interphase nuclei. J. Histochem. Cytochem. 2002, 50, 1697–1698. [Google Scholar] [CrossRef] [PubMed]
- Weise, A.; Starke, H.; Heller, A.; Claussen, U.; Liehr, T. Evidence for interphase DNA decondensation transverse to the chromosome axis: A multicolor banding analysis. Int. J. Mol. Med. 2002, 9, 359–361. [Google Scholar] [CrossRef]
- Felka, T.; Lemke, J.; Lemke, C.; Michel, S.; Liehr, T.; Claussen, U. DNA degradation during maturation of erythrocytes—Molecular cytogenetic characterization of Howell-Jolly bodies. Cytogenet. Genome Res. 2007, 119, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Manvelyan, M.; Hunstig, F.; Mrasek, K.; Bhatt, S.; Pellestor, F.; Weise, A.; Liehr, T. Position of chromosomes 18, 19, 21 and 22 in 3D-preserved interphase nuclei of human and gorilla and white hand gibbon. Mol. Cytogenet. 2008, 1, 9. [Google Scholar] [CrossRef] [PubMed]
- Manvelyan, M.; Hunstig, F.; Bhatt, S.; Mrasek, K.; Pellestor, F.; Weise, A.; Simonyan, I.; Aroutiounian, R.; Liehr, T. Chromosome distribution in human sperm—A 3D multicolor banding-study. Mol. Cytogenet. 2008, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Manvelyan, M.; Kempf, P.; Weise, A.; Mrasek, K.; Heller, A.; Lier, A.; Höffken, K.; Fricke, H.J.; Sayer, H.G.; Liehr, T.; et al. Preferred co-localization of chromosome 8 and 21 in myeloid bone marrow cells detected by three dimensional molecular cytogenetics. Int. J. Mol. Med. 2009, 24, 335–341. [Google Scholar] [PubMed]
- Klein, E.; Manvelyan, M.; Simonyan, I.; Hamid, A.B.; Guilherme, R.S.; Liehr, T.; Karamysheva, T. Centromeric association of small supernumerary marker chromosomes with their sister-chromosomes detected by three dimensional molecular cytogenetics. Mol. Cytogenet. 2012, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Roediger, J.; Hessenkemper, W.; Bartsch, S.; Manvelyan, M.; Huettner, S.S.; Liehr, T.; Esmaeili, M.; Foller, S.; Petersen, I.; Grimm, M.O.; et al. Supraphysiological androgen levels induce cellular senescence in human prostate cancer cells through the Src-Akt pathway. Mol. Cancer 2014, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Karamysheva, T.; Kosyakova, N.; Guediche, N.; Liehr, T. Small supernumerary marker chromosomes and the nuclear architecture of sperm—A study in a fertile and an infertile brother. Syst. Biol. Reprod. Med. 2015, 61, 32–36. [Google Scholar] [CrossRef]
- Weise, A.; Bhatt, S.; Piaszinski, K.; Kosyakova, N.; Fan, X.; Altendorf-Hofmann, A.; Tanomtong, A.; Chaveerach, A.; de Cioffi, M.B.; de Oliveira, E.; et al. Chromosomes in a genome-wise order: Evidence for metaphase architecture. Mol. Cytogenet. 2016, 9, 36. [Google Scholar] [CrossRef]
- Cremer, C.; Münkel, C.; Granzow, M.; Jauch, A.; Dietzel, S.; Eils, R.; Guan, X.Y.; Meltzer, P.S.; Trent, J.M.; Langowski, J.; et al. Nuclear architecture and the induction of chromosomal aberrations. Mutat. Res. 1996, 366, 97–116. [Google Scholar] [CrossRef]
- Zink, D.; Cremer, T. Cell nucleus: Chromosome dynamics in nuclei of living cells. Curr. Biol. 1998, 8, R321–R324. [Google Scholar] [CrossRef]
- Mateos-Langerak, J.; Goetze, S.; Leonhardt, H.; Cremer, T.; van Driel, R.; Lanctôt, C. Nuclear architecture: Is it important for genome function and can we prove it? J. Cell. Biochem. 2007, 102, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Schmid, VJ.; Cremer, M.; Cremer, T. Quantitative analyses of the 3D nuclear landscape recorded with super-resolved fluorescence microscopy. Methods 2017, 123, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M.; Cremer, C. The 4D nucleome: Genome compartmentalization in an evolutionary context. Biochemistry 2018, 83, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Grafodatskiĭ, A.S. Comparative chromosomics. Mol. Biol. 2007, 41, 408–422. [Google Scholar] [CrossRef]
- Ji, Z.; Zhang, L. Chromosomics: Detection of numerical and structural alterations in all 24 human chromosomes simultaneously using a novel OctoChrome FISH assay. J. Vis. Exp. 2012, 60, 3619. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://patents.justia.com/assignee/trans-chromosomics-inc (accessed on 9 January 2019).
- Available online: https://wikis.nyu.edu/display/Vogel/03+Chromosomics (accessed on 9 January 2019).
- Heng, H.H.; Krawetz, S.A.; Lu, W.; Bremer, S.; Liu, G.; Ye, C.J. Re-defining the chromatin loop domain. Cytogenet. Cell. Genet. 2001, 93, 155–161. [Google Scholar] [CrossRef]
- Baumgartner, M.; Dutrillaux, B.; Lemieux, N.; Lilienbaum, A.; Paulin, D.; Viegas-Péquignot, E. Genes occupy a fixed and symmetrical position on sister chromatids. Cell 1991, 64, 761–766. [Google Scholar] [CrossRef]
- Volpi, E.V.; Chevret, E.; Jones, T.; Vatcheva, R.; Williamson, J.; Beck, S.; Campbell, R.D.; Goldsworthy, M.; Powis, S.H.; Ragoussis, J.; et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell. Sci. 2000, 113, 1565–1566. [Google Scholar]
- Borden, J.; Manuelidis, L. Movement of the X chromosome in epilepsy. Science 1988, 242, 1687–1691. [Google Scholar] [CrossRef]
- Bridger, J.M.; Boyle, S.; Kill, I.R.; Bickmore, W.A. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 2000, 10, 149–152. [Google Scholar] [CrossRef]
- Chevret, E.; Volpi, E.V.; Sheer, D. Mini review: Form and function in the human interphase chromosome. Cytogenet. Cell. Genet. 2000, 90, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L. The dynamics of chromosome organization and gene regulation. Ann. Rev. Biol. 2003, 72, 573–608. [Google Scholar] [CrossRef] [PubMed]
- Ranade, D.; Koul, S.; Thompson, J.; Prasad, K.B.; Sengupta, K. Chromosomal aneuploidies induced upon Lamin B2 depletion are mislocalized in the interphase nucleus. Chromosoma 2017, 126, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, Y.; Zhao, F.; Liu, J.; Yin, J.; Chen, D.; Ma, W.; Ke, X. A new classification of interphase nuclei based on spatial organizations of chromosome 8 and 21 for t(8;21)(q22;q22) acute myeloid leukemia by three-dimensional fluorescence in situ hybridization. Leuk. Res. 2015, 39, 1414–1420. [Google Scholar] [CrossRef] [PubMed]
- Bártová, E.; Jirsová, P.; Fojtová, M.; Soucek, K.; Kozubek, S. Chromosomal territory segmentation in apoptotic cells. Cell. Mol. Life Sci. 2003, 60, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Bonora, G.; Disteche, C.M. Structural aspects of the inactive X chromosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160357. [Google Scholar] [CrossRef]
- Grob, S.; Cavalli, G. Technical Review: A Hitchhiker’s Guide to chromosome conformation capture. Methods Mol. Biol. 2018, 1675, 233–246. [Google Scholar] [PubMed]
- Schmitt, A.D.; Hu, M.; Ren, B. Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell. Biol. 2016, 17, 743–755. [Google Scholar] [CrossRef]
- Corces, M.R.; Corces, V.G. The three-dimensional cancer genome. Curr. Opin. Genet. Dev. 2016, 36, 1–7. [Google Scholar] [CrossRef]
- Bernardi, G. Genome organization and chromosome architecture. Cold Spring Harb. Symp. Quant. Biol. 2015, 80, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Mkrtchyan, H.; Gross, M.; Hinreiner, S.; Polytiko, A.; Manvelyan, M.; Mrasek, K.; Kosyakova, N.; Ewers, E.; Nelle, H.; Liehr, T.; et al. The human genome puzzle—The role of copy number variation in somatic mosaicism. Curr. Genomics 2010, 11, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Kreysing, M.; Lanctôt, C.; Kösem, S.; Peichl, L.; Cremer, T.; Guck, J.; Joffe, B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 2009, 137, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Joffe, B. Inverted nuclear architecture and its development during differentiation of mouse rod photoreceptor cells: A new model to study nuclear architecture. Genetika 2010, 46, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, H.; Habermann, F.A.; Solovei, I.; Cremer, M.; Cremer, T. Non-random radial arrangements of interphase chromosome territories: Evolutionary considerations and functional implications. Mutat. Res. 2002, 504, 37–45. [Google Scholar] [CrossRef]
- Lomiento, M.; Grasser, F.; Rocchi, M.; Müller, S. The interplay between genome organization and nuclear architecture of primate evolutionary neo-centromeres. Genomics 2013, 102, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Mrasek, K.; Schoder, C.; Teichmann, A.C.; Behr, K.; Franze, B.; Wilhelm, K.; Blaurock, N.; Claussen, U.; Liehr, T.; Weise, A. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int. J. Oncol. 2010, 36, 929–940. [Google Scholar]
- Weise, A.; Kosyakova, N.; Voigt, M.; Aust, N.; Mrasek, K.; Löhmer, S.; Rubtsov, N.; Karamysheva, T.V.; Trifonov, V.A.; Hardekopf, D.; et al. Comprehensive analyses of white-handed Gibbon chromosomes enables access to 92 evolutionary conserved breakpoints compared to the human genome. Cytogenet. Genome Res. 2015, 145, 42–49. [Google Scholar] [CrossRef]
- Fan, X.; Supiwong, W.; Weise, A.; Mrasek, K.; Kosyakova, N.; Tanomtong, A.; Pinthong, K.; Trifonov, V.A.; Cioffi de B., M.; Grothmann, P.; et al. Comprehensive characterization of evolutionary conserved breakpoints in four New World Monkey karyotypes compared to Chlorocebus aethiops and Homo sapiens. Heliyon 2015, 1, e00042. [Google Scholar] [CrossRef]
- Liehr, T.; Kosyakova, N.; Schröder, J.; Ziegler, M.; Kreskowski, K.; Pohle, B.; Bhatt, S.; Theuss, L.; Wilhelm, K.; Weise, A.; et al. Evidence for correlation of fragile sites and chromosomal breakpoints in carriers of constitutional balanced chromosomal rearrangements. Balkan J. Med. Genet. 2011, 14, 13–16. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Q.; Mao, J.H.; Weise, A.; Mrasek, K.; Fan, X.; Zhang, X.; Liehr, T.; Lu, K.H.; Balmain, A.; et al. An HDAC1-binding domain within FATS bridges p21 turnover to radiation-induced tumorigenesis. Oncogene 2010, 29, 2659–2671. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Qiu, L.; Mrasek, K.; Zhang, J.; Liehr, T.; Quintana, L.G.; Li, Z. Common fragile sites: Genomic hotspots of DNA damage and carcinogenesis. Int. J. Mol. Sci. 2012, 13, 11974–11999. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Das, I.; Arlt, M.; Patel, N.; Kraftson, S.; Glover, T.W.; Sekiguchi, J.M. The SNM1B/APOLLO DNA nuclease functions in resolution of replication stress and maintenance of common fragile site stability. Hum. Mol. Genet. 2013, 22, 4901–4913. [Google Scholar] [CrossRef] [PubMed]
- Lukusa, T.; Fryns, J.P. Human chromosome fragility. Biochem. Biophys. Acta 2008, 1779, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Minocherhomji, S.; Hickson, I.D. Structure-specific endonucleases: Guardians of fragile site stability. Trends Cell. Biol. 2014, 24, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Available online: https//www.eshg.org/index.php?id=95 (accessed on 9 January 2019).
- Houge, G.; Liehr, T.; Schoumans, J.; Ness, G.O.; Solland, K.; Starke, H.; Claussen, U.; Strømme, P.; Akre, B.; Vermeulen, S. Ten years follow up of a boy with a complex chromosomal rearrangement: Going from a > 5 to 15-breakpoint CCR. Am. J. Med. Genet. A 2003, 118A, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Weise, A.; Rittinger, O.; Starke, H.; Ziegler, M.; Claussen, U.; Liehr, T. De novo 9-break-event in one chromosome 21 combined with a microdeletion in 21q22.11 in a mentally retarded boy with short stature. Cytogenet. Genome Res. 2003, 103, 14–16. [Google Scholar] [CrossRef]
- Norrby, E.; Levan, A.; Nichols, W.W. The correlation between the chromosome pulverization effect and other biological activities of measles virus preparations. Exp. Cell. Res. 1965, 41, 483–491. [Google Scholar] [CrossRef]
- Taylor, J.H.; Haut, W.F.; Tung, J. Effects of fluorodeoxyuridine on DNA replication, chromosome breakage, and reunion. Proc. Natl. Acad. Sci. USA 1962, 48, 190–198. [Google Scholar] [CrossRef]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Mark, J. Double-minutes—A chromosomal aberration in Rous sarcomas in mice. Hereditas 1967, 57, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Biedler, J.L.; Spengler, B.A. Metaphase chromosome anomaly: Association with drug resistance and cell-specific products. Science 1976, 191, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Pellman, D. Cancer genomes evolve by pulverizing single chromosomes. Cell 2011, 144, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Quigley, D.A.; Dang, H.X.; Zhao, S.G.; Lloyd, P.; Aggarwal, R.; Alumkal, J.J.; Foye, A.; Kothari, V.; Perry, M.D.; Bailey, A.M.; et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018, 174, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Iafrate, A.J.; Feuk, L.; Rivera, M.N.; Listewnik, M.L.; Donahoe, P.K.; Qi, Y.; Scherer, S.W.; Lee, C. Detection of large-scale variation in the human genome. Nat. Genet. 2004, 36, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Sebat, J.; Lakshmi, B.; Troge, J.; Alexander, J.; Young, J.; Lundin, P.; Månér, S.; Massa, H.; Walker, M.; Chi, M.; et al. Large-scale copy number polymorphism in the human genome. Science 2004, 305, 525–528. [Google Scholar] [CrossRef]
- Liehr, T. Cytogenetically visible copy number variations (CG-CNVs) in banding and molecular cytogenetics of human; about heteromorphisms and euchromatic variants. Mol. Cytogenet. 2016, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Gosden, J.R.; Lawrie, S.S.; Gosden, C.M. Satellite DNA sequences in the human acrocentric chromosomes: Information from translocations and heteromorphisms. Am. J. Hum. Genet. 1981, 33, 243–251. [Google Scholar]
- Hasegawa, T.; Asamura, S.; Nagai, T.; Tsuchiya, Y. An unusual variant of chromosome 16 in three generations. Acta Paediatr. Jpn. 1992, 34, 166–168. [Google Scholar] [CrossRef]
- Chiang, J.C.; Jiang, J.; Newburger, P.E.; Lawrence, J.B. Trisomy silencing by XIST normalizes Down syndrome cell pathogenesis demonstrated for hematopoietic defects in vitro. Nat. Commun. 2018, 9, 5180. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liehr, T. From Human Cytogenetics to Human Chromosomics. Int. J. Mol. Sci. 2019, 20, 826. https://doi.org/10.3390/ijms20040826
Liehr T. From Human Cytogenetics to Human Chromosomics. International Journal of Molecular Sciences. 2019; 20(4):826. https://doi.org/10.3390/ijms20040826
Chicago/Turabian StyleLiehr, Thomas. 2019. "From Human Cytogenetics to Human Chromosomics" International Journal of Molecular Sciences 20, no. 4: 826. https://doi.org/10.3390/ijms20040826
APA StyleLiehr, T. (2019). From Human Cytogenetics to Human Chromosomics. International Journal of Molecular Sciences, 20(4), 826. https://doi.org/10.3390/ijms20040826




