DNA Repair Gene ZmRAD51A Improves Rice and Arabidopsis Resistance to Disease
Abstract
1. Introduction
2. Results
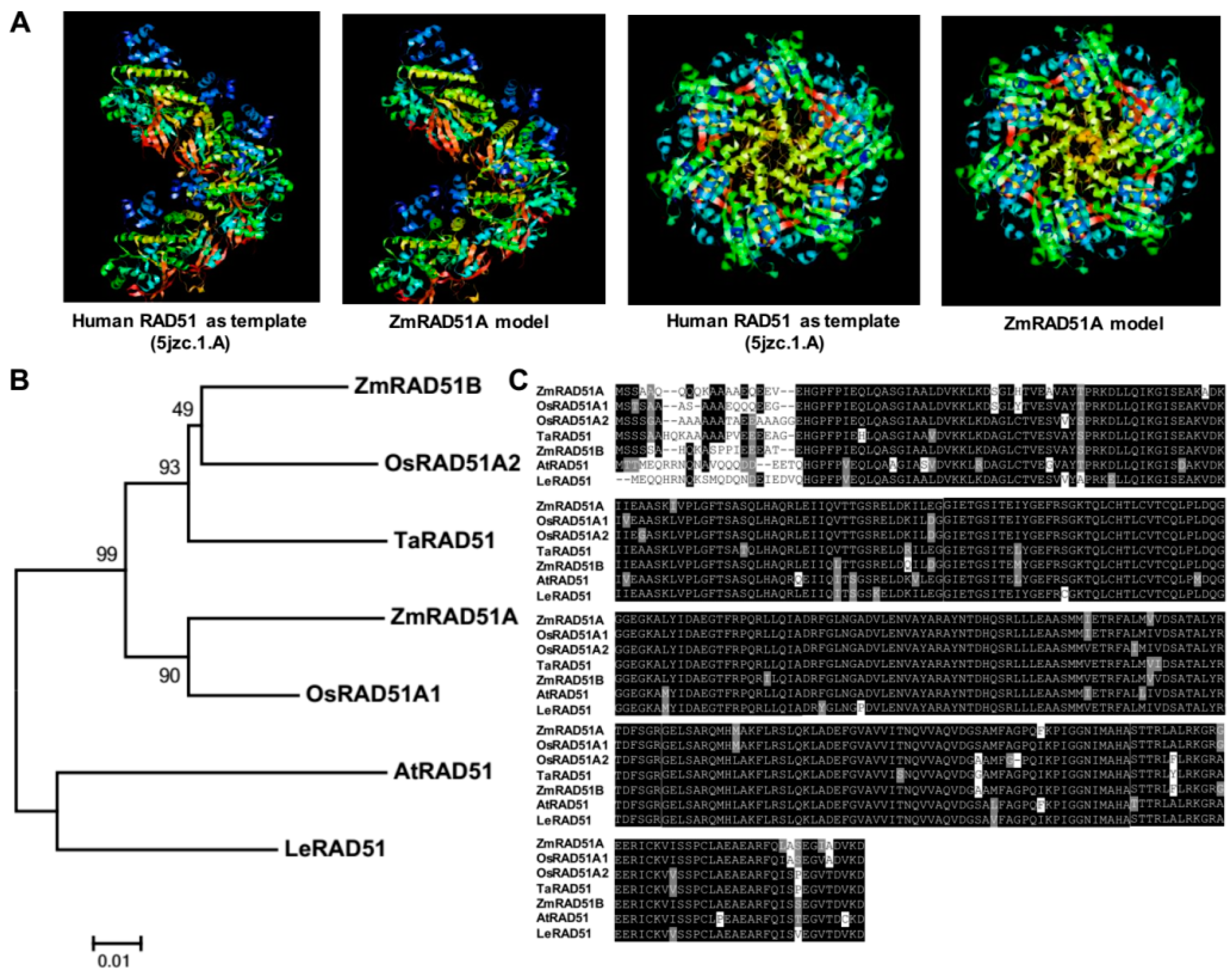
2.1. Cloning and Characterization of ZmRAD51A
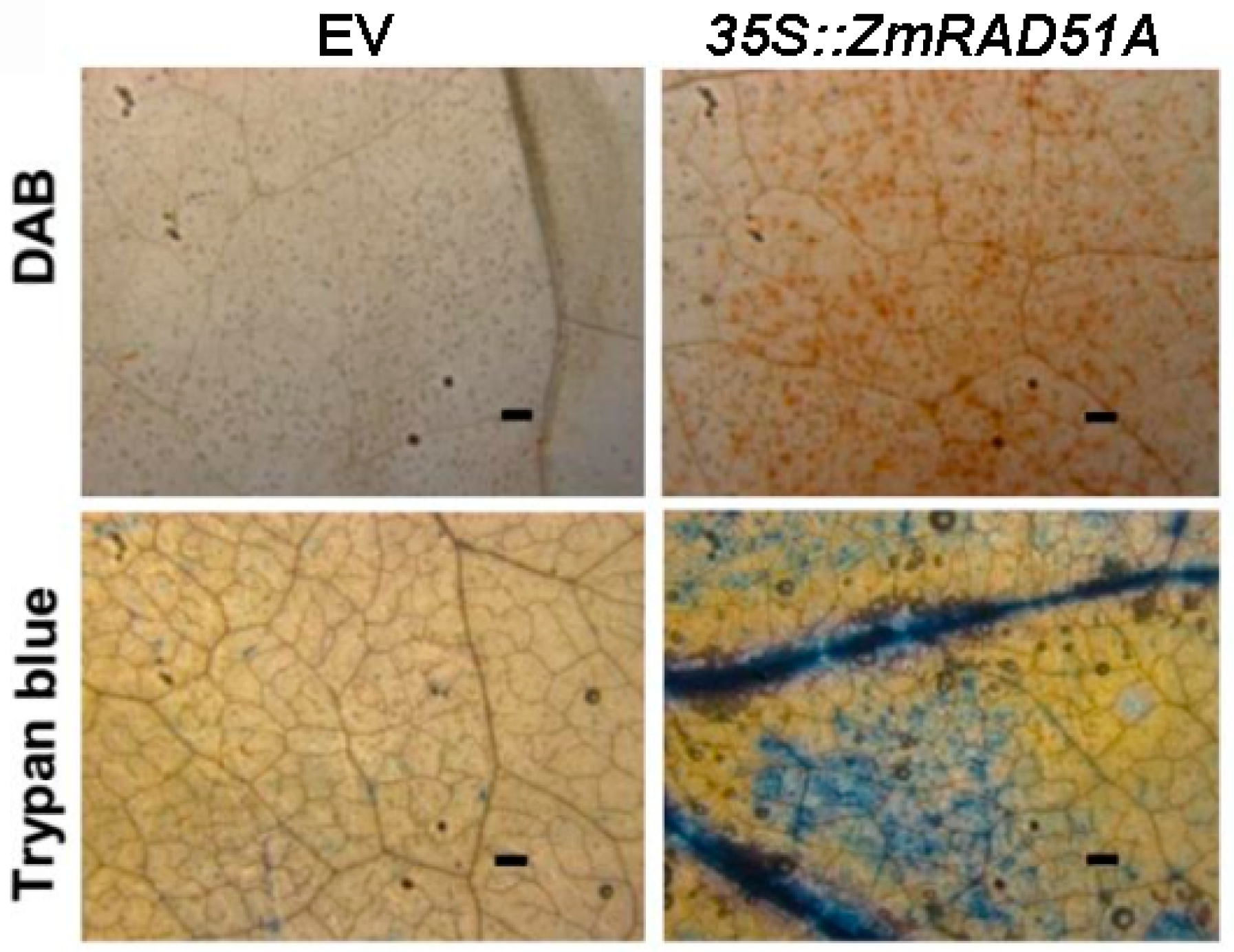
2.2. Transient Overexpression of ZmRAD51A in Nicotiana Benthamiana Leaves Induced a Hypersensitive Response
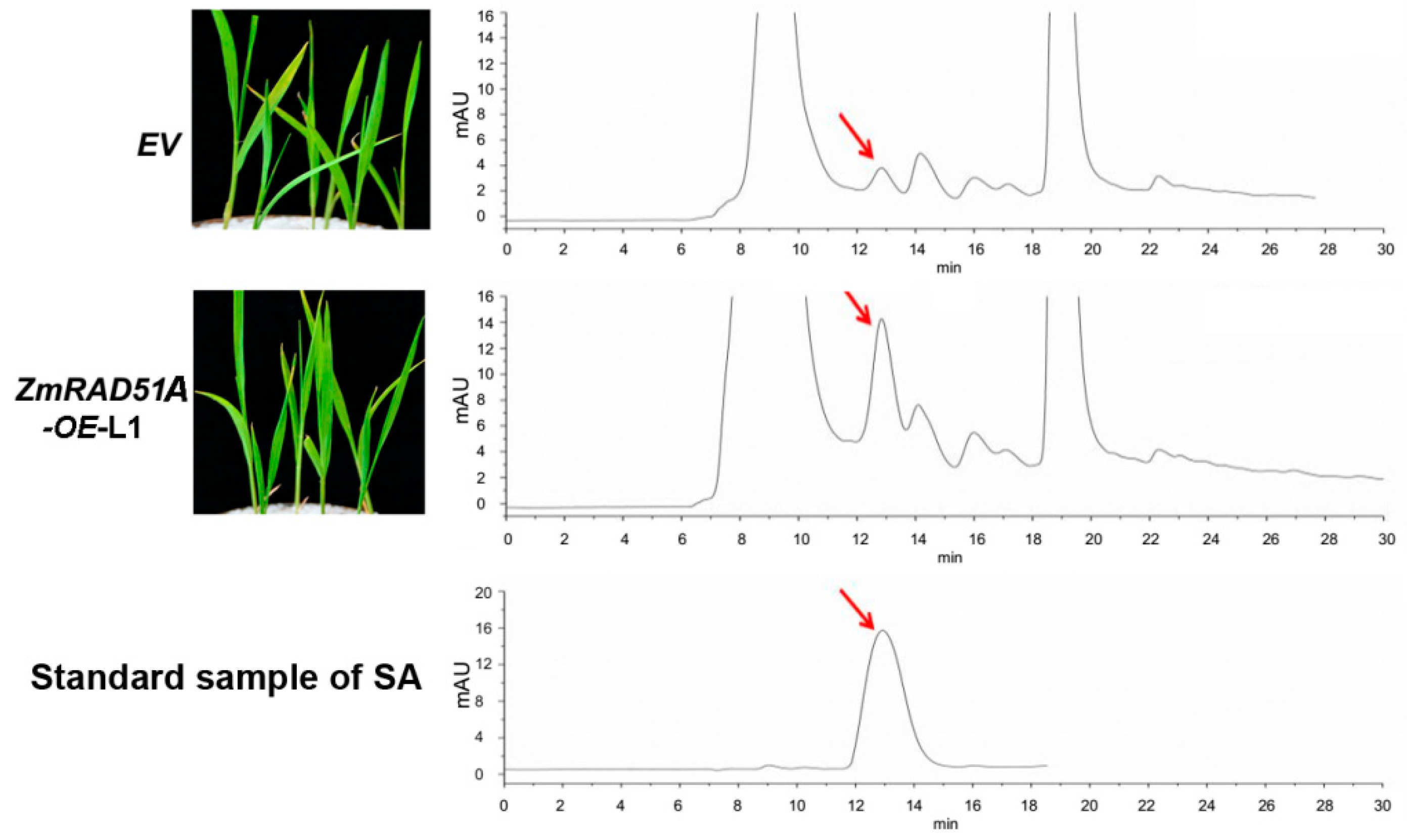
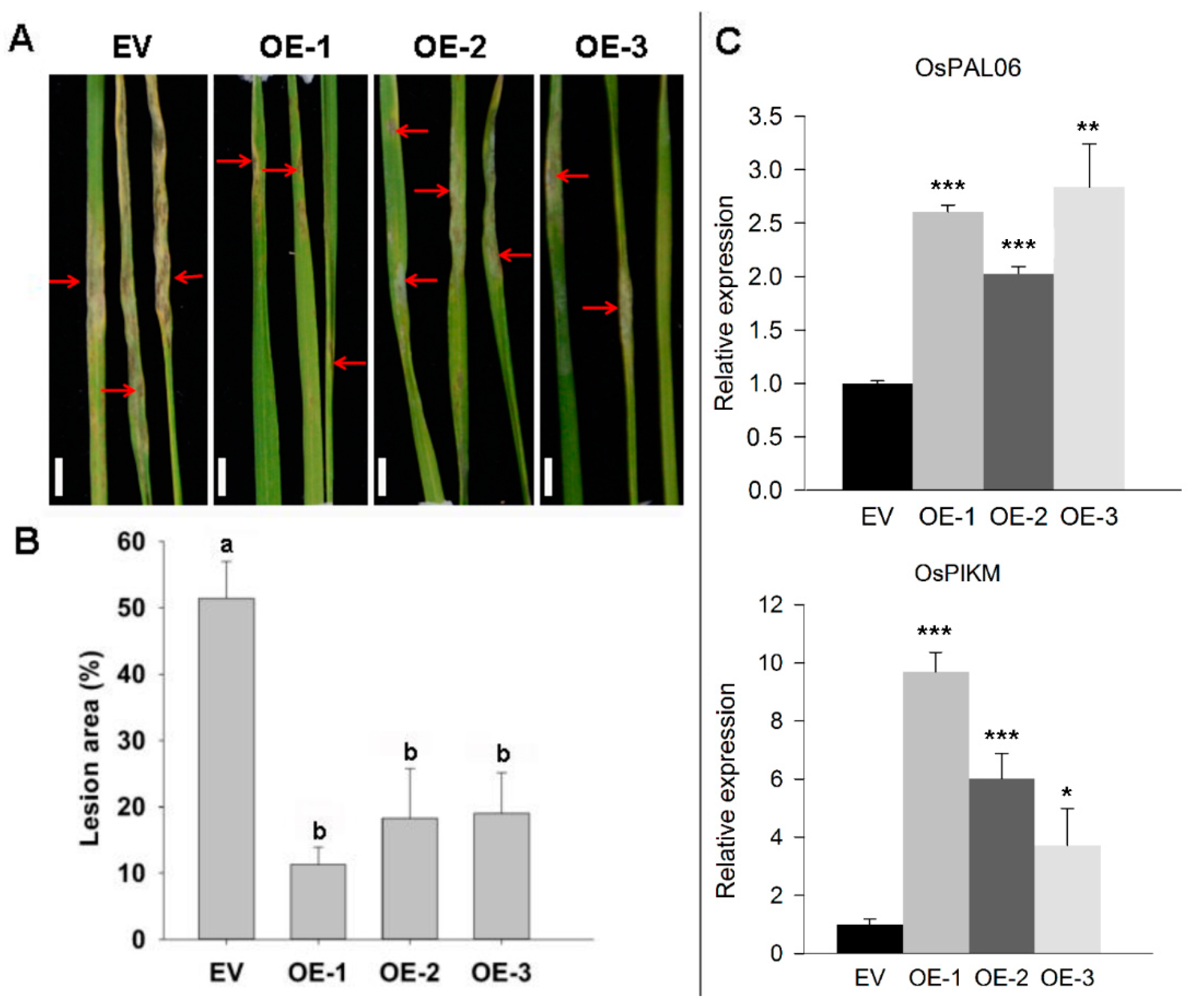
2.3. Transgenic Overexpression of ZmRAD51A Increased SA Synthesis and Conferred Rice Resistance to M. oryzae
2.4. ZmRAD51A Overexpression in Arabidopsis Enhanced Resistance to Pst DC3000 Triggering by Increased SA-Related Genes Expression
3. Discussion
3.1. ZmRAD51A Is a Conserved DNA Repair Protein in Maize
3.2. ZmRAD51A Is Involved in SA-Signal Defense Responses
3.3. ZmRAD51A Confers Disease Resistance in Transgenic Plants
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
4.3. Bioinformatics Analysis
4.4. Full-Length cDNA Cloning and Vector Construction
4.5. Cell Death Assays in Nicotiana Benthamiana
4.6. Rice Transformation
4.7. Measurement of SA
4.8. Rice Pathogens Inoculation
4.9. Arabidopsis Transformation
4.10. Arabidopsis Pathogen Inoculation
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| SA | Salicylic acid |
| HPLC | High-performance liquid chromatography |
| PTI | PAMP-triggered immunity |
| ETI | Effector-triggered immunity |
| NPR1 | NONEXPRESSOR OF PR1 GENES |
| DSBs | DNA double strand breaks |
| SAR | systemic acquired resistance |
References
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007, 50, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Loon, L.C.V.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 2003, 113, 935. [Google Scholar] [CrossRef]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-Mediated Turnover of the Transcription Co-Activator NPR1 Plays Dual Roles in Regulating Plant Immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Plant Physiol. 2013, 42, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Wang, S.; Dong, X. Arabidopsis SNI1 and RAD51D Regulate Both Gene Transcription and DNA Recombination during the Defense Response. Proc. Natl. Acad. Sci. USA 2007, 104, 4223. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Durrant, W.E.; Wang, S.; Yan, S.; Tan, E.H.; Dong, X. DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 2011, 9, 115–124. [Google Scholar] [CrossRef]
- Shinohara, A.; Ogawa, H.; Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 1992, 69, 457–470. [Google Scholar] [CrossRef]
- Kou, Y.; Chang, Y.; Li, X.; Xiao, J.; Wang, S. The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J. Exp. Bot. 2012, 63, 5323. [Google Scholar] [CrossRef]
- Symington, L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002, 66, 630. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Fedoroff, N. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 2004, 101, 10596–10601. [Google Scholar] [CrossRef] [PubMed]
- Doutriaux, M.P.; Couteau, F.; Bergounioux, C.; White, C. Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 1998, 257, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Osakabe, K.; Nakayama, S.; Endo, M.; Tagiri, A.; Todoriki, S.; Ichikawa, H.; Toki, S. Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 2005, 139, 896. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Abe, K.; Yamanouchi, H.; Takyuu, T.; Yoshioka, T.; Ito, Y.; Kato, T.; Tabata, S.; Kurei, S.; Yoshioka, Y. Arabidopsis Rad51B is important for double-strand DNA breaks repair in somatic cells. Plant Mol. Biol. 2005, 57, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Rajanikant, C.; Melzer, M.; Rao, B.J.; Sainis, J.K. Homologous recombination properties of OsRad51, a recombinase from rice. Plant Mol. Biol. 2008, 68, 479. [Google Scholar] [CrossRef]
- Byun, M.Y.; Kim, W.T. Suppression of OsRAD51D results in defects in reproductive development in rice (Oryza sativa L.). Plant J. 2014, 79, 256–269. [Google Scholar] [CrossRef]
- Franklin, A.E.; Mcelver, J.; Sunjevaric, I.; Rothstein, R.; Bowen, B.; Cande, W.Z. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 1999, 11, 809–824. [Google Scholar] [CrossRef]
- Franklin, A.E.; Golubovskaya, I.N.; Bass, H.W.; Cande, W.Z. Improper chromosome synapsis is associated with elongated RAD51 structures in the maize desynaptic2 mutant. Chromosoma 2003, 112, 17–25. [Google Scholar]
- Pawlowski, W.P.; Golubovskaya, I.N.; Cande, W.Z. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 2003, 15, 1807. [Google Scholar] [CrossRef]
- Devisetty, U.K.; Mayes, K.; Mayes, S. The RAD51 and DMC1 homoeologous genes of bread wheat: Cloning, molecular characterization and expression analysis. BMC Res. Notes 2010, 3, 245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, X.; Zuo, K.; Zhang, J.; Sun, X.; Tang, K. Molecular Cloning and Characterization of a Novel Gossypium barbadense L. RAD-Like Gene. Plant Mol. Biol. Report. 2011, 29, 324–333. [Google Scholar] [CrossRef]
- Wang, S.; Schulze-Lefert, P. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. USA 2010, 107, 22716. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lin, X.; Poland, J.; Trick, H.; Leach, J.; Hulbert, S. A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. USA 2005, 102, 15383–15388. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.; Drake, J.; Ayliffe, M.; Sun, Q.; Ellis, J.; Hulbert, S.; Pryor, T. Molecular Characterization of the Maize Rp1-D Rust Resistance Haplotype and Its Mutants. Plant Cell 1999, 11, 1365–1376. [Google Scholar] [CrossRef]
- Hurni, S.; Scheuermann, D.; Krattinger, S.G.; Kessel, B.; Wicker, T.; Herren, G.; Fitze, M.N.; Breen, J.; Presterl, T.; Ouzunova, M. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. USA 2015, 112, 8780. [Google Scholar] [CrossRef]
- Li, J.; Wen, T.J.; Schnable, P.S. Role of RAD51 in the Repair of MuDR-Induced Double-Strand Breaks in Maize (Zea mays L.). Genetics 2008, 178, 57. [Google Scholar] [CrossRef]
- Yang, C.; Deng, W.; Tang, N.; Wang, X.; Fang, Y.; Lin, D.; Li, Z. Overexpression of ZmAFB2, the maize homologue of AFB2 gene, enhances salt tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. 2013, 112, 171–179. [Google Scholar] [CrossRef]
- Zhang, H.; Teng, W.; Liang, J.; Liu, X.; Zhang, H.; Zhang, Z.; Zheng, X. MADS1, a novel MADS-box protein, is involved in the response of Nicotiana benthamiana to bacterial harpin (Xoo). J. Exp. Bot. 2016, 67, 131–141. [Google Scholar] [CrossRef]
- Li, L.Y.; Wang, L.; Jing, J.X.; Li, Z.Q.; Lin, F.; Huang, L.F.; Pan, Q.H. The Pik m gene, conferring stable resistance to isolates of Magnaporthe oryzae, was finely mapped in a crossover-cold region on rice chromosome 11. Mol. Breed. 2007, 20, 179–188. [Google Scholar] [CrossRef]
- Qiu, D.; Xiao, J.; Ding, X.; Xiong, M.; Cai, M.; Cao, Y.; Li, X.; Xu, C.; Wang, S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant-Microbe Interact. 2007, 20, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.; Daniels, M.J.; Parker, J.E. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292. [Google Scholar] [CrossRef] [PubMed]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Chung, E.H.; Eitas, T.K.; Dangl, J.L. Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 2011, 108, 7619–7624. [Google Scholar] [PubMed]
- Eitas, T.K.; Nimchuk, Z.L.; Dangl, J.L. Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 2008, 105, 6475–6480. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, L.; Liu, R.; Jiao, B.; Zhao, X.; Ling, Z.; Luo, K. Overexpression of Poplar PtrWRKY89 in Transgenic Arabidopsis Leads to a Reduction of Disease Resistance by Regulating Defense-Related Genes in Salicylate- and Jasmonate-Dependent Signaling. PLoS ONE 2016, 11, e0149137. [Google Scholar] [CrossRef]
- Datta, K.; Koukolikova-Nicola, Z.; Baisakh, N.; Oliva, N.; Datta, S.K. Agrobacterium-mediated engineering for sheath blight resistance of indica rice cultivars from different ecosystems. Theor. Appl. Genet. 2000, 100, 832–839. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, H.; Cai, T.; Deng, Y.; Zhuang, R.; Zhang, N.; Zeng, Y.; Zheng, Y.; Tang, R.; Pan, R.; et al. Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 2017, 15, 39–55. [Google Scholar] [CrossRef]
- Divi, U.K.; Rahman, T.; Krishna, P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 2010, 10, 151. [Google Scholar] [CrossRef]
- Yang, D.L.; Yang, Y.N.; He, Z.H. Roles of Plant Hormones and Their Interplay in Rice Immunity. Mol. Plant 2013, 6, 675–685. [Google Scholar] [CrossRef]
- Novakova, M.; Sasek, V.; Dobrev, P.I.; Valentova, O.; Burketova, L. Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum—Reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 2014, 80, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef]
- Grant, J.J.; Chini, A.; Basu DLoake, G.J. Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant-Microbe Interact. 2003, 16, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Olivier, J.; Theulieres, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Liu, H.; Li, X.; Xiao, J.; Wang, S. Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol. Plant. 2014, 152, 486. [Google Scholar] [CrossRef] [PubMed]
- Coppinger, P.; Repetti, P.P.; Day, B.; Dahlbeck, D.; Mehlert, A.; Staskawicz, B.J. Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 2004, 40, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Bent, A.F. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLOS Pathog. 2014, 10, e1004030. [Google Scholar] [CrossRef]
- Gasser, S.; Orsulic, S.; Brown, E.J.; Raulet, D.H. The DNA damage pathway regulates innate immune system ligands for the NKG2D receptor. Nature 2005, 436, 1186–1190. [Google Scholar] [CrossRef]
- Yan, S.; Wang, W.; Marqués, J.; Mohan, R.; Saleh, A.; Durrant, W.E.; Song, J.; Dong, X. Salicylic Acid Activates DNA Damage Responses to Potentiate Plant Immunity. Mol. Cell 2013, 52, 602–610. [Google Scholar] [CrossRef]
- Tanaka, K.; Hadwiger, L.A. Nonhost resistance: Reactive oxygen species (ROS) signal causes DNA damage prior to the induction of PR genes and disease resistance in pea tissue. Physiol. Mol. Plant Pathol. 2017, 98, 18–24. [Google Scholar]
- Zhang, C.; Zhang, H.; Zhao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 2013, 32, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- Short, J.M.; Liu, Y.; Chen, S.; Soni, N.; Madhusudhan, M.S.; Shivji, M.K.K.; Venkitaraman, A.R. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res. 2016, 44, 9017–9030. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, Y.J.; Jiang, H.H.; Jiang, C.S.; Du, Y.B.; Gong, C.; Wang, W.; Zhu, S.W.; Han, G.M.; Cheng, B.J. Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Lin, H.N.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Medina-Rivera, A.; Defrance, M.; Sand, O.; Herrmann, C.; Castro-Mondragon, J.A.; Delerce, J.; Jaeger, S.; Blanchet, C.; Vincens, P.; Caron, C.; et al. RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 2015, 43, W50–W56. [Google Scholar] [CrossRef]
- Cesari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef]
- Hwang, I.S.; Hwang, B.K. The Pepper Mannose-Binding Lectin Gene CaMBL1 Is Required to Regulate Cell Death and Defense Responses to Microbial Pathogens. Plant Physiol. 2011, 155, 447–463. [Google Scholar] [CrossRef]
- Cho, K.; Han, O.; Tamogami, S.; Shibato, J.; Kubo, A.; Agrawal, G.K.; Rakwal, R. Quantification of jasmonic and salicylic acids in rice seedling leaves. Methods Mol. Biol. 2013, 956, 185–200. [Google Scholar] [CrossRef]
- Meuwly, P.; Metraux, J.P. Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal. Biochem. 1993, 214, 500–505. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Sucher, J.; Selter, L.L.; Chauhan, H.; Zhou, B.; Tang, M.Z.; Upadhyaya, N.M.; Mieulet, D.; Guiderdoni, E.; Weidenbach, D.; et al. The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol. J. 2016, 14, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. CSH Protoc. 2009, 2009, pdb-prot5177. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.D.; Liu, H.X.; Jiang, C.H.; Wang, Y.P.; Wang, Q.Y.; Jin, H.L.; Guo, J.H. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant-Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Xu, Y.; Zhou, L.; Ali, A.; Jiang, H.; Zhu, S.; Li, X. DNA Repair Gene ZmRAD51A Improves Rice and Arabidopsis Resistance to Disease. Int. J. Mol. Sci. 2019, 20, 807. https://doi.org/10.3390/ijms20040807
Liu F, Xu Y, Zhou L, Ali A, Jiang H, Zhu S, Li X. DNA Repair Gene ZmRAD51A Improves Rice and Arabidopsis Resistance to Disease. International Journal of Molecular Sciences. 2019; 20(4):807. https://doi.org/10.3390/ijms20040807
Chicago/Turabian StyleLiu, Fang, Yunjian Xu, Lingyan Zhou, Asif Ali, Haiyang Jiang, Suwen Zhu, and Xiaoyu Li. 2019. "DNA Repair Gene ZmRAD51A Improves Rice and Arabidopsis Resistance to Disease" International Journal of Molecular Sciences 20, no. 4: 807. https://doi.org/10.3390/ijms20040807
APA StyleLiu, F., Xu, Y., Zhou, L., Ali, A., Jiang, H., Zhu, S., & Li, X. (2019). DNA Repair Gene ZmRAD51A Improves Rice and Arabidopsis Resistance to Disease. International Journal of Molecular Sciences, 20(4), 807. https://doi.org/10.3390/ijms20040807



