1. Introduction
ANO1 is a Ca
2+ activated Cl
− channel (CaCC), which is widely expressed in tissues such as sensory neurons, smooth muscle cells, and salivary glands. In salivary glands, ANO1 is one of the most important pathways for the apical Cl
− ion efflux required for fluid secretion [
1]. Therefore, ANO1 is thought to be a potential target for the treatment of salivary gland dysfunction and/or the generation of fully functional artificial salivary gland tissue. In adult salivary glands, ANO1 is mainly expressed in the apical membrane of the secretory acinar cells, but not in the epithelial cells that line the ducts of the gland [
2]. The mechanisms that determine this differential expression of ANO1 in the acinar and duct cells are still unknown. A previous study reported that ANO1 expression is regulated by epigenetic mechanisms involving DNA methylation of the ANO1 gene promoter region in head and neck squamous cell carcinomas, but similar studies have not been conducted on normal salivary gland tissue [
3].
Epigenetic mechanisms, such as DNA methylation and histone modification, are known to be involved in the regulation of gene expression without requiring any changes in the DNA sequence [
4,
5]. The hypo- and hypermethylation pattern of CpG islands modulates global and site-specific gene expression by altering the degree of chromatin condensation. This subsequently determines the binding of transcription factors to the target genes [
6]. The hypermethylation process is usually carried out by DNA methyltransferases (DNMTs) such as Dnmt1, Dnmt3a, and Dnmt3b [
5,
6,
7,
8]. Epigenetic patterning is crucial to the development and regeneration of various epithelial organs. For example, the differentiation of both liver and pancreas from the early endoderm is determined by epigenetic regulation of liver specific genes (
Alb1,
Ttr, and
Afp), and an early pancreatic gene (
PDX1), respectively [
9]. Furthermore, during pancreatic islet development, the fates of alpha and beta cells are determined by epigenetic regulation, so that even after full differentiation both cell types can be trans-differentiated in response to epigenetic modulations [
9]. The prostate gland is another example showing the importance of spatiotemporal methylation dynamics during organogenesis. Basal epithelium-specific DNA methylation of E-cadherin is required for proper budding and further branching morphogenesis of the prostate [
10]. In salivary glands, however, there have been no studies of such developmental epigenetic dynamics. In this paper, we address this omission, by using isolated embryonic submandibular glands (eSMG) to investigate the potential epigenetic determinants of ANO1 expression during salivary gland organogenesis.
Since tissue regeneration generally recapitulates the developmental process [
11,
12,
13], revealing the mechanisms behind cell type-dependent expression of ANO1 during development may be helpful in in vitro efforts to generate salivary glands expressing functional ANO1. It is known that both salivary gland acinar and duct cells arise from primitive progenitor cell populations during salivary gland organogenesis, and that salivary gland ductal progenitor cells remaining in the adult intercalated ducts can differentiate into acinar cells during regeneration of damaged salivary glands [
14]. Thus, it is possible that epigenetic modulation of gene expression is involved in the cell-type dependent expression of specific proteins associated with acinar and duct cells [
15,
16]. Our previous study revealed that epigenetic regulation is involved in the differential expression of Cystic fibrosis transmembrane conductance regulator (CFTR) in salivary gland acini and duct [
17]. However, this work was limited to artificial epigenetic modulation in different kinds of immortalized cell lines by 5-aza-2’-deoxycytidine (5-Aza-CdR), a DNMT inhibitor consequently inducing DNA demethylation. In this study, the epigenetic regulation of ANO1 in salivary gland duct and acinar cells is examined under more physiologic conditions by using Matrigel-induced trans-differentiation of the Human Salivary Gland (HSG) cell line as model of development. The HSG cell line is derived from intercalated ducts of irradiated human submandibular glands, and is known to trans-differentiate into acinar cells when cultured on basement membrane extracts such as Matrigel or laminin-1 [
18,
19,
20]. By using this and other experimental models, we demonstrate that the differential expression of ANO1 in acini and duct is regulated by epigenetic mechanisms.
3. Discussion
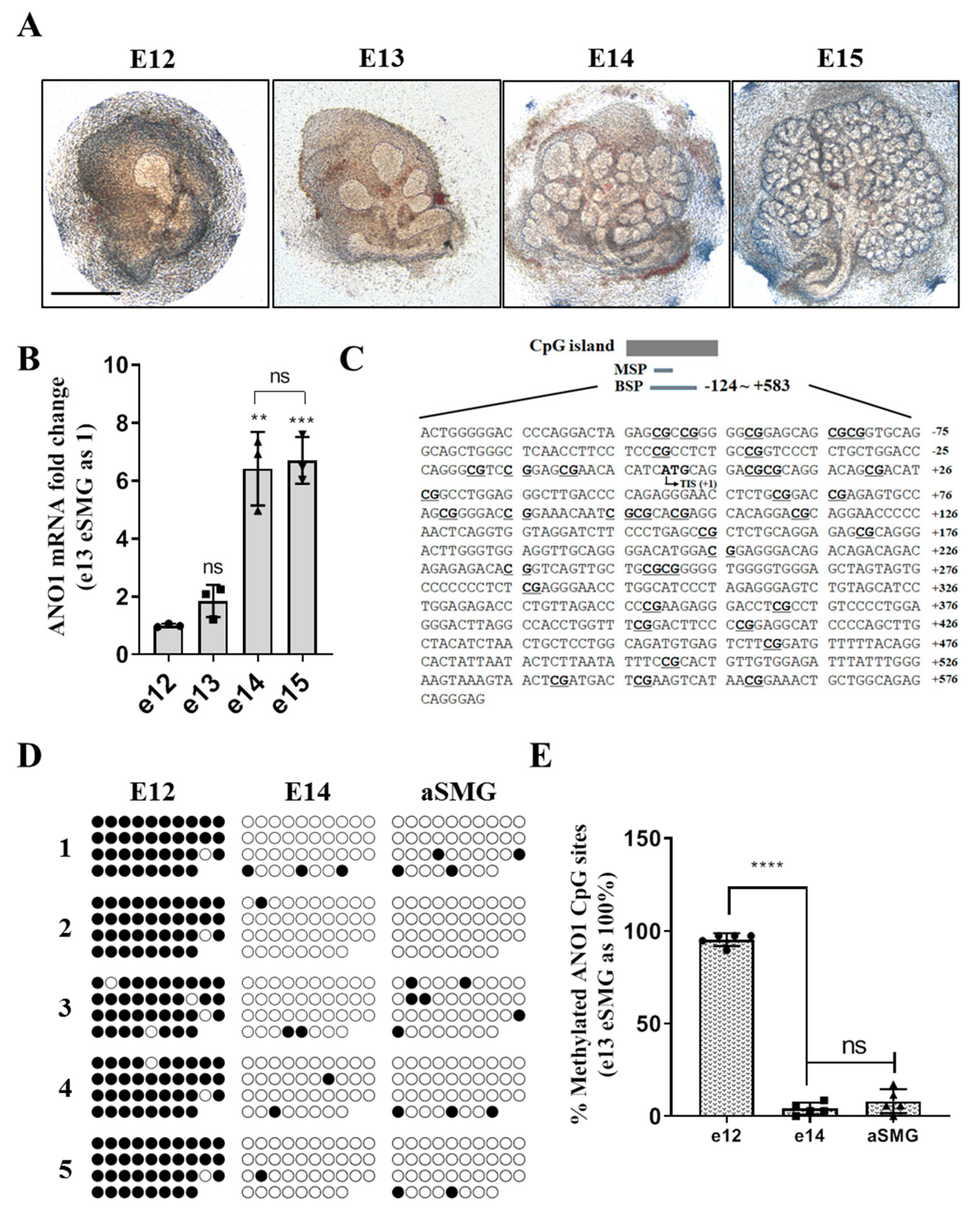
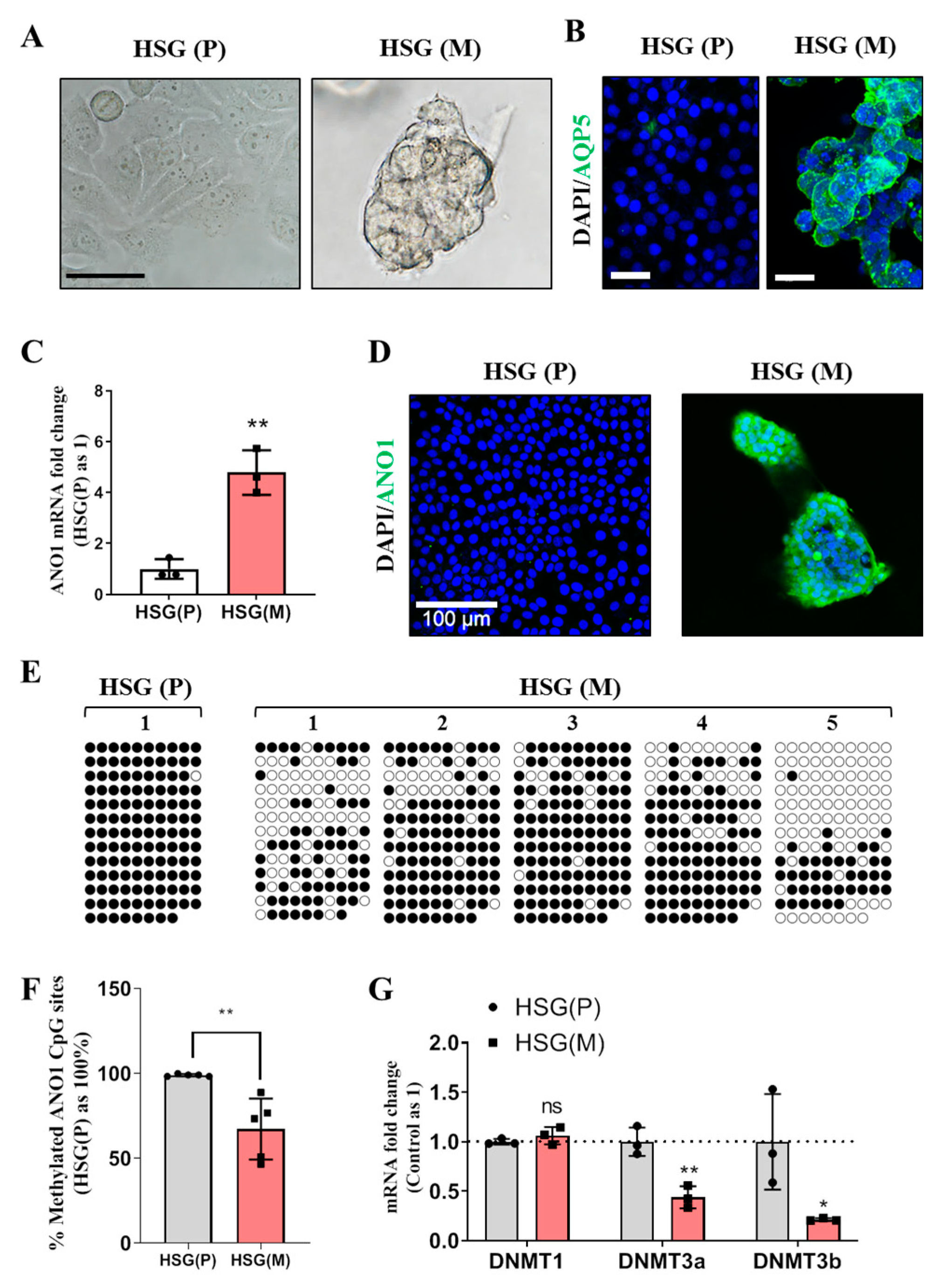
In this study, various experimental models including: eSMG, mouse SMG, and in vitro trans-differentiation HSG cell line have been used to obtain evidence for epigenetic regulation of ANO1 expression in the development of SMGs. ANO1 CpG islands in e12 eSMG are highly methylated, but those in e14 eSMG or adult SMG are significantly unmethylated. The differential expression pattern of ANO1 in duct and acini is defined at e14. When artificial demethylation is induced by treatment of 5-Aza-CdR, both duct-origin HSG cell line and adult mouse SMG duct cells exhibited the expression of ANO1. During the trans-differentiation of duct-origin HSG cells into acinar cells within Matrigel, significant demethylation of ANO1 CpG islands is observed, confirming that the expression of ANO1 in salivary glands is mainly regulated by epigenetic modulation.
DNA methylation is involved in various cellular processes including embryonic development, regulation of gene expression, and determination of chromosomal structure and activity [
8]. During early developmental stages, the degree of DNA methylation is also highly elevated to map the specific regulatory patterns of gene promoter regions required for further developmental processes [
8]. Therefore, precise DNA methylation patterning is essential for maintaining normal organogenesis. Although there has been no study on global epigenetic patterning during organogenesis of salivary glands, unpublished data from this laboratory show that the treatment of 5-Aza-CdR to ex vivo cultured e13 eSMG completely abolished branching morphogenesis and induced rapid degeneration of epithelium (data not shown). This is because demethylation by 5-Aza-CdR is non-specific, thus, we are only able to observe epigenetic regulation of ANO1 during organogenesis, rather than modulating it. In addition, since the current study is limited to the negative modulation of ANO1′s methylation status by using 5-Aza-CdR, it will be necessary to confirm our hypothesis with more advanced epigenetic modulation system such as dCas9-DNA methyltransferase MQ1 fusion protein which targets specific gene to produce methylation [
23]. Furthermore, the molecular cues which initiate methylation or demethylation of ANO1 CpG islands during salivary gland organogenesis or trans-differentiation of HSG cells also remain to be elucidated.
Based on our results from trans-differentiating HSG cells into acinar cells, it can be speculated that downregulation of DNMTs by basement matrix may be responsible for demethylation of ANO1 during organogenesis. This hypothesis can be supported by a classic methodology, the epithelial rudiment culture technique, which has been routinely used for studying salivary gland branching morphogenesis. Epithelial rudiments of eSMG without mesenchyme can branch and differentiate within Matrigel [
24]. However, since Matrigel is heterogeneous ECM mixtures isolated from Engelbreth-Holm-Swarm mouse sarcoma [
25], key ECM substances triggering the demethylation of ANO1 CpG islands in HSG cells should be further specified. One study reports that the expansion of c-kit+ epithelial progenitor cells during salivary gland organogenesis is epigenetically regulated by a microRNA secreted from mesenchyme [
26]. Therefore, it is also possible that the demethylation of ANO1 CpG islands can be triggered by single or multiple mesenchymal factors including miRNAs, soluble growth factors, and extracellular matrix substances.
In salivary glands, the Cl
− channels CFTR and ANO1 are known to play a major role in salivary secretion [
2,
27]. CFTR is a cyclic AMP-activated Cl
− channel, whereas ANO1 is a Ca2+-activated Cl
− channel [
2,
27,
28,
29]. CFTR is predominantly expressed in SMG ducts but not in acinar cells, and in a previous study, we demonstrated that this differential expression is due to the DNA methylation of CFTR CpG islands [
17]. By contrast, ANO1 is expressed in SMG acinar cells but not in the ducts [
2,
27]. Although differential expression patterns of ANO1 or CFTR in acini and duct are explained by epigenetic modification, not all acini or duct-specific proteins are regulated by epigenetic mechanisms. In the case of AQP5, a representative marker for the salivary gland acinar cell, their mRNA transcripts exist in HSG cells cultured on plastic surface, but because of an unknown mechanism, the AQP5 mRNA cannot be translated into functional proteins [
20]. However, when they are cultured in Matrigel, the AQP5 mRNAs are translated into functional proteins [
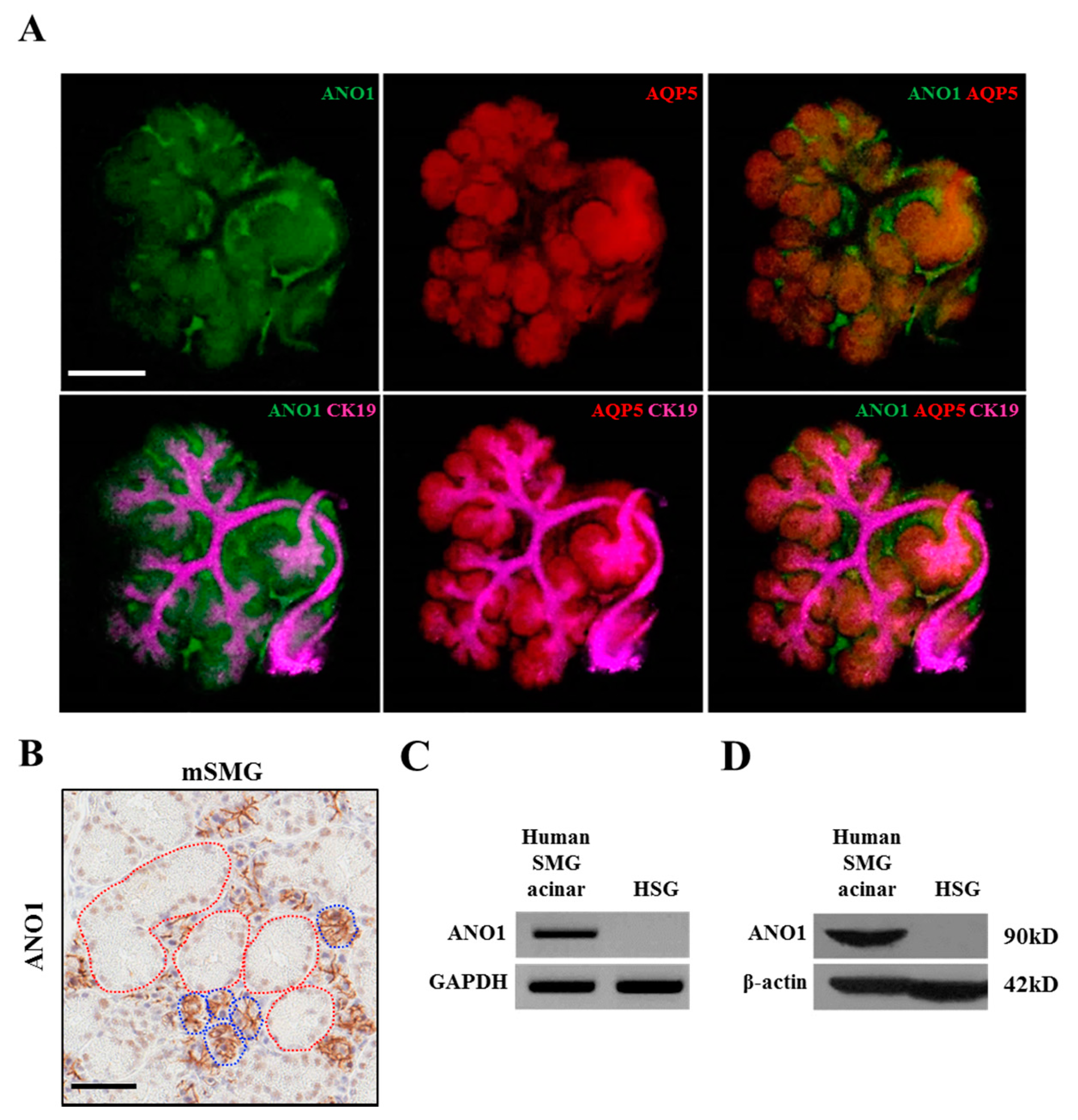
20]. Therefore, although AQP5 is exclusively expressed in acini like ANO1, it is not regulated by epigenetic mechanisms.
From a tissue-engineering perspective, restoring expression of acini-specific functional proteins in ductal cells by using the clinically available demethylating agent, 5-Aza-CdR, has enormous therapeutic potential for salivary gland dysfunction. Radiation-induced xerostomia is one of the most common complications caused by head and neck cancer radiotherapy [
30,
31]. Although the mechanisms are not yet fully understood, radiation significantly destroys acini while not harming ducts [
32]. Therefore, one study suggests delivering the Aquaporin-1 gene to duct cells using a viral vector for the restoration of salivary flow [
33,
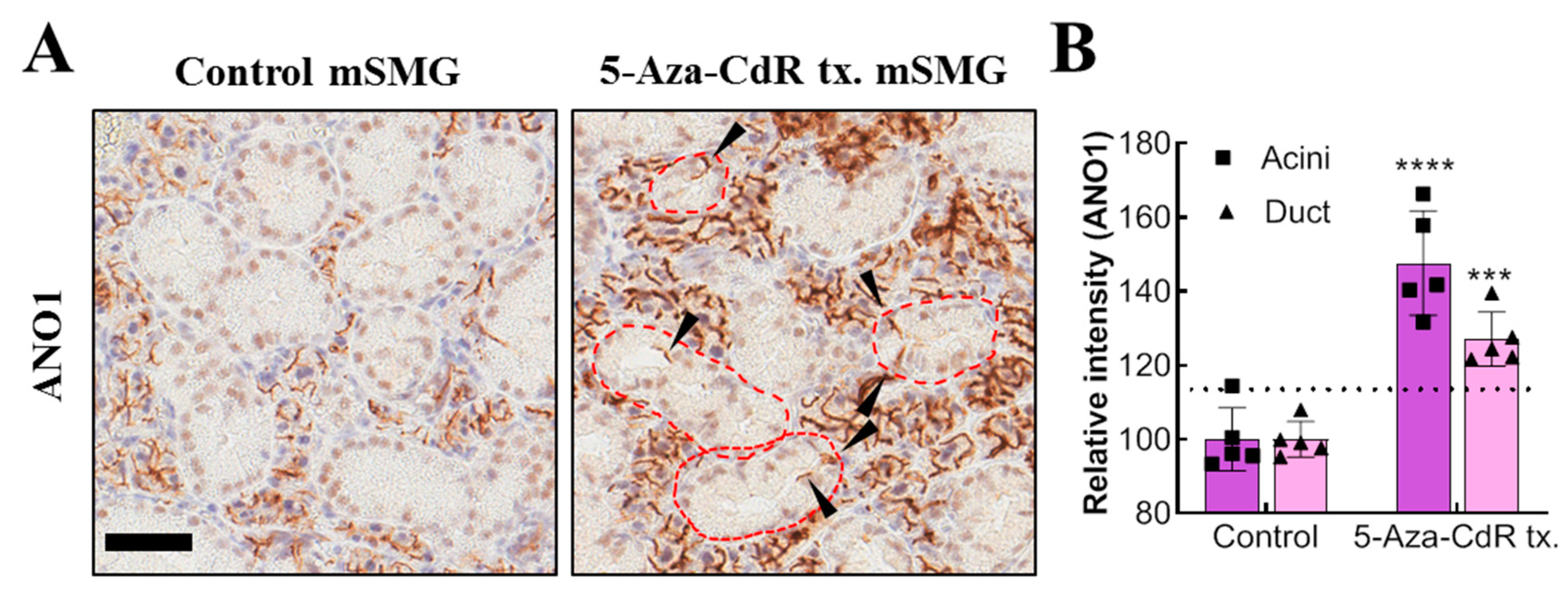
34]. Likewise, if it is possible to induce expression of acini-specific functional proteins in the remaining duct cells by using 5-Aza-CdR; they may replace the functions of acinar cells. In this study, continuous injection of 5-Aza-CdR successfully induces ANO1 in duct cells, showing the potential application of demethylating agents for the treatment of salivary gland dysfunction. It is notable that treatment of 5-Aza-CdR also increases ANO1 expression in acinar cells, which may lead to enhancement of salivation, but this requires further examination.
The mechanism for the significantly different degree of expressional increase in duct and acinar ANO1 by in vivo injections of high dose 5-Aza-CdR also still needs to be explained. This might be due to the difference in physiological methylation levels in duct and acinar cells and insufficient 5-Aza-CdR delivery to submandibular glands. In fact, mice treated with low dose 5-Aza-CdR showed no such increased expression of ANO1 in acini or duct, suggesting that there is a threshold concentration for 5-Aza-cdR to achieve sufficient demethylation for the significant expressional increment of ANO1. Since high dose 5-Aza-CdR has potential risk of leukopenia or bone marrow hypoplasia [
35], targeted delivery of concentrated 5-Aza-CdR via advanced drug delivery system may more efficiently recover ductal ANO1 expression with minimal systemic toxicity. There are several studies used continuous 5-Aza-CdR injections in vivo to correct non-cancerous diseases. For examples, altered methylation status of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase 1, 2 in injured kidney was corrected by series in vivo injection of 5-Aza-CdR, resulting in amelioration of renal fibrosis in mice [
36]. Another possible application is cell therapy using in vitro-cultured immortalized salivary gland cell lines. Although HSG cell line is derived from neoplastic cells, it is a well-established candidate for the generation of artificial salivary glands [
37]. First, HSG cells can differentiate into acinar cells when cultured in Matrigel [
20,
38]. Second, they functionally express muscarinic and purinergic receptors to increase intracellular calcium like primary salivary gland cells [
39,
40]. If expression of other acini-specific proteins can be induced by treatment of 5-Aza-CdR in HSG cultured without Matrigel, less immunogenic and more economic materials will be able to replace Matrigel. Therefore, global expressional changes of such genes should be examined via Next Generation Sequencing or proteome array after an artificial epigenetic modulation. It is also noted that the activity of ANO1 restored by 5-Aza-CdR in HSG cells seems lower than that of primary mouse SMG acinar cells, indicating that other channels or proteins expressed on primary acinar cells may have synergistic effect on ANO1 activity.
In summary, this study indicates that epigenetic modification by DNA methylation is a key regulatory mechanism for silencing and upregulation, respectively, of ANO1 expression in salivary gland acini and duct. Considering the importance of ANO1 in salivary secretion, an epigenetic approach to enhance or restore ANO1 expression has therapeutic potentials for the treatment of salivary gland dysfunction.
4. Material and Methods
4.1. Cell Culture
Human submandibular gland cells, HSG, were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Media was changed every 48 h, and the cells were passaged every 72 h. The cells were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
4.2. Matrigel Culture of HSG
Growth factor-reduced Matrigel (Corning, Corning city, NY, USA) was thawed on ice. 10,000 HSG cells were mixed with 500 μL of the Matrigel on ice, and then, 20 μL Matrigel-HSG mixture were pipetted on to cell culture plates. The plates were incubated in 37 °C in a humidified atmosphere of 95% air and 5% CO2 for 15 min to solidify the Matrigel/HSG mixture. DMEM supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin was slowly added on the Matrigel dome. The cells were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
4.3. 5-Aza-CdR Treatment and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
HSG cells were treated with 10 μM 5-Aza-CdR for 1, 2, 3, and 4 days, and the medium containing 5-Aza-CdR was changed daily. Total RNA was isolated from untreated and 5-Aza-CdR treated HSG cells using the Trizol reagent according to the manufacturer’s instructions (Ambion, Life Technologies, Carlsbad, CA, USA). The purity and concentration of the total RNA were measured using a NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was also isolated by the same method from: human submandibular gland acinar (human SMG acinar) cells (see below), A253, and SGT cells.
cDNA was synthesized from 1 μg of total RNA with reverse transcriptase. Primer sequences for ANO1 amplification and GAPDH used as an endogenous control were as follows: ANO1: forward 5′-AACCACACCCTCTCCTCCTT-3′ and reverse 5′-CTTTGGTGTTGTGGTGGTTG-3′; GAPDH: forward 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′. cDNA was applied as a template for ANO1 amplification, and the RT-PCR conditions were as follows: 95 °C for 5 min; followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s; and a final step at 72 °C for 10 min. The PCR products were separated by 1% agarose gel electrophoresis.
4.4. Western Blot Analysis
Cells were washed with phosphate buffered saline (PBS), harvested, and dissolved in a lysis buffer. The protein samples were separated using 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. After blocking with 10% non-fat milk (Seoul Milk, Seoul, Korea), the membrane was incubated at 4 °C with a primary rabbit antibody ANO1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:1000), and then washed with Tris buffered saline + Tween 20 (TBST). The membrane was then incubated with an HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Dallas, TX, USA, 1:5000) for 1 h at room temperature, which was visualized using an Enhanced chemiluminescence (ECL) reagent (Thermo Fisher Scientific, Waltham, MA, USA).
4.5. Intracellular Cl− Ion Flux Monitoring
Intracellular Cl
− ion was monitored by modified version of the method of Choi et al. [
21]. Briefly, HSG cells were harvested from culture dish and loaded with 10mM MQAE dissolved in DMEM for 30 min at 37 °C. The cells were washed three times with HBSS, and attached to a cover-glass coated with Cell-Tak (Corning, NY, USA). The cover-glass formed the bottom of the experimental chamber. Fluorescence was monitored using a LSM700 Confocal Laser Scanning Microscope, fitted with a filter cube No. 49 (Ex/Em = 365/445, Zeiss, Oberkochen, Germany). Baseline intensity calibration was measured for 50 s. After that, cells were treated with 10 μM Eact, and fluorescence changes measured for a further 300 s at 5 s intervals.
4.6. Ex vivo SMG Organ Harvesting and Immunostaining
Embryonic SMGs (eSMGs) from Institute of Cancer Research (ICR) mice were harvested and stained according to previously described methods [
41]. Fetal mice were collected from e12 and 13 embryonic sacs and the eSMGs isolated. The isolated eSMGs were placed on Nucleopore filters (0.1 µm, Little Chalfont, Buckinghamshire, UK) floating on phenol red-free DMEM/F12 (1:1) containing 1% Pen/Strep, 0.15 µg/µL transferrin, and 5 ng/µL L-ascrobic acid. e12 and e13 eSMGs were stabilized for two hours or further cultured to obtain e14 and 15 eSMGs. After the incubation, bright field images were obtained by Nikon Ti digital inverted fluorescence microscope (Shinagawa, Tokyo, Japan). eSMGs at e14 stage were fixed with 4% paraformaldehyde and washed with PBS tween 0.5%. The fixed eSMGs were permeabilized with 0.1% Triton X-100 and blocked in PBS Tween 0.5% containing 10% donkey serum and 3% bovine serum albumin (BSA). The eSMGs were incubated overnight with primary antibodies against: ANO1 (1:200; Santa Cruz Biotechnology), Aquaporin 5 (1:200; Alomone Labs, Israel), or Cytokeratin 19 (1:200; DSHB, Iowa city, IA, USA). After a further overnight incubation with appropriate secondary antibodies conjugated with either: Cy2, Cy3, or Cy5, the eSMGs were mounted on slides. Immunocytochemistry staining was analyzed using an LSM700 Laser Confocal Scanning Microscope (Carl Zeiss, Oberkochen, Germany). The 3 × 3 image stitching mode was used to obtain images of whole eSMGs.
4.7. Immunofluorescence Staining of HSG Cells
HSG cells were grown on cell culture slides and treated with 10 μM 5-Aza-CdR (Sigma Aldrich, St. Louis, MO, USA) for 3 days. After treatment with 5-Aza-CdR, the HSG cells were fixed in 4% paraformaldehyde for 10 min and washed with PBS. For Matrigel-encapsulated HSG cells (3-day culture), they were fixed in ice cold 4% paraformaldehyde for 30 min and washed with PBS. Cells were permeabilized with 0.1% Triton X-100 for 10 min. Cells were then blocked with 10% donkey serum and 3% BSA for 60 min and washed with cold PBS/Glycine. After that, cells were incubated overnight with an anti-human ANO1 (1:200, Abcam, ab53212, Cambridge, UK) or anti-human AQP5 primary (1:200, Alomone Labs, #AQP-005, Hadassah Ein Kerem, Jerusalem, Israel) antibody at 4°C. After washing with PBS, the cells were incubated with an Alexa Fluor®® 488 donkey anti-mouse or anti-rabbit IgG secondary antibody (1:200) and DAPI (1:1000) for 2 h at room temperature. HSG cells treated with or without 5-Aza-CdR were mounted with FLUO-GEL WITH TRIS BUFFER (Electron Microscopy Science, Hatfield, PA, USA) and visualized by an LSM700 (Carl Zeiss, Oberkochen, Germany).
4.8. Immunohistochemistry of Adult Mouse SMG
Adult mouse SMGs in paraffin block sections were de-paraffinized and hydrated. For the de-paraffinization step, the sections were incubated in dry oven at 60C for 1 h and de-waxed in xylene five times for 4 min. The mouse SMG sections were rehydrated in 100%, 95%, and 75% ethanol for 3 min each and immersed in tap water for 5 min. For the antigen retrieval step, the sections were immersed into citrate buffer (0.01 M, pH 6.0) and microwaved at high, medium, and low power for 5 min each. After that, the sections were immersed in cold PBS. Next, to quench endogenous peroxidase, the sections were immersed in peroxidase-blocking solution (Agilent DAKO, S2023, Santa Clara, CA, USA) and washed three times with PBS for 5 min each. The sections were then incubated with anti-ANO1 primary antibody (100:1, Abcam, ab532132) for 30 min. The sections were washed in PBS three times, and incubated with EnVision +systems-HRP labelled polymer anti-rabbit (Agilent DAKO, K4003) for 30 min. The sections were washed in PBS three times for 5 min each. After that, the sections were incubated in DAB solution (Agilent DAKO, GV825) for 5 min, and the reaction was stopped by washing in tap water. For the counterstaining, the sections were immersed in Mayer’s hematoxylin solution for 1 min. After the dehydration step (immersing in 75%, 80%, 95%, and 100% ethanol for 1 min each), the slide sections were cleared in xylene 4 times for 5 min each. Finally, the slide sections were mounted with Permount Mounting Medium (Fisher chemical, SP15-100, Waltham, MA, USA) for further analysis.
4.9. Quantification of ANO1 Expression from Immunohistochemistry of Adult Mouse SMG
Immunohistochemistry images of adult mouse SMG stained with anti-ANO1 antibody (Abcam, ab532132) were digitally scanned by Aperio AT2 (Leica, Wetzlar, Germany). The scanned images were then captured by Aperio ImageScope version 12.3.2.8013 (Leica, Wetzlar, Germany). The ANO1 expression in duct and acini was quantified by SABIA ver 1.0.0.0 (Solution for Automatic Bio-Image Analysis, Seoul, Korea). In detail, areas of duct or acini were manually selected based on their morphological difference, and then, the relative ANO1-positive areas were automatically calculated within the previously selected area.
4.10. Animal Experiments
Eight-week old ICR mice were purchased from Daehan BioLink (Choong-buk, Korea). Mice were injected intraperitoneally with either PBS or low (2.5 mg/kg)/high dose (25 mg/kg) 5-Aza-CdR for 7 consecutive days. The mice were then sacrificed and the SMGs are harvested. The animal experiment protocol used in this study was approved by Seoul National University Institutional Animal Care and Use Committee (Approval number: SNU-190610-5-1, Approval date: 17 June 2019).
4.11. Methylation-Specific PCR
Genomic DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen, Venlo, Netherlands) from: HSG cells (before and after treatment with 5-Aza-CdR), human SMG cells, A253 cells, and SGT cells. For methylation-specific PCR (MSP), bisulfite modification of genomic DNA was performed with the EpiTect Bisulfite Kit (Qiagen) as described previously [
17]. MSP was performed with bisulfite-treated genomic DNA using specific primers for the methylated or unmethylated forms of the ANO1 CpG islands. The following methylation-specific primer sequences were used: M forward 5′-TTTTAAGGTAAAGGCGGGTC-3′ and M reverse 5′-CTCGATACGAAAAACGCCTA-3′; U forward 5′-TATTTTTAAGGTAAAGGTGGGTT-3′ and U reverse 5′-CTCAATACAAAAAACACCTAAAC-3′.
4.12. Bisulfite Sequencing
2 μg of genomic DNA was modified by a sodium bisulfite conversion reaction with the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s instructions. The modified genomic DNA was amplified with bisulfite primer sequences of the ANO1 CpG island locus (NCBI accession: NC_000011, region: 70078169 to 70079120, 952 bp) designed by the Methyl Primer Express
TM software v1.0 as follows: forward 5′-AAAAATAAAATTTGGAGGGGTT-3′ and reverse 5′-CCCTAACTACCCCAACAAATAC-3′. The PCR reactions were carried out as follows: 94 °C for 5 min; 35 cycles of 94 °C for 45 s and 55 °C for 45 s; and a final cycle of 72 °C for 45 s at the end. The PCR products were purified with a Gel Extraction Kit (Qiagen) and ligated into the pGEM-T easy vector (Promega, Madison, WI, USA). Five separate clones each from HSG cells, eSMGs, and adult SMGs were selected for bisulfite sequencing analysis according to the previously described method [
17].
4.13. Human and Mouse Submandibular Gland (SMG) Acianr Cell Isolation
Human SMG tissues were obtained from the patients visiting the Seoul National University Dental Hospital (SNUDH) from 2016 to 2018. Biopsied tissues were confirmed as normal SMG tissue by pathology department of SNUDH. Mouse SMGs were harvested from 8-week old male ICR mice. SMG tissues were immersed in dissociation buffer (RPMI supplemented with 1 mg/mL Collagenase IV and 1mg/mL DNase 1) and placed in the Gentle MACS machine (Miltenyi Biotech, North Rhine-Westphalia, Germany). By using a preprogrammed protocol (M_impTumor3) in Gentle MACS, the SMG tissues were mechanically dissociated and incubated in 37 °C water bath for 30 min. This step was repeated once more, and dissociated SMG cells were centrifuged at 1500 rpm for 5 min. After washing with HBSS three times, dissociated SMG cells were filtered through 70 μm strainer (Miltenyi Biotech, North Rhine-Westphalia, Germany). Acinar cells were isolated from the dissociated SMG cells by density gradient cell separation method using Accudenz described by Xin Xu et al. (Accurate Chemical and Scientific Corporation, Westbury, NY, USA) [
42]. All ethical guidelines and consent forms were approved by Institutional Review Board of SNUDH (Approval number: CRI11023G, approval date: 5 July 2011).
4.14. RT-PCR
mRNAs in Human SMG acinar cells, mouse eSMG, and HSG cells were extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was prepared from 2μg of total RNA using reverse transcriptase with oligo-dT primer. The RT-PCR was performed with specific primer for ANO1. The primer sequences are as follows: For human ANO1, forward 5′-AACCACACCCTCTCCTCCTT-3′ and reverse 5′-CTTTGGTGTTGTGGTGGTTG-3′. For mouse ANO1, forward 5′-GAGGCCAGTAGCCATCAGAG-3′ and reverse 5′-GAGAGCGTGTGATTGACGAA-3′. For GAPDH, forward 5’-GAAGGTGAAGGTCGGAGTC-3’ and reverse 5’-GAAGATGGTGATGGGATTTC-3′. The RT-PCR was performed with a specific condition: 35 cycles of denaturation at 94 °C for 40 sec, annealing at 55 °C for 40 sec, and extension at 72 °C for 40 sec, with a final extension at 72 °C for 10 min. cDNA from samples were amplified using PCR amplification machine (PTC-1148C, Bio-Rad Laboratories Inc., Hercules, CA, USA). The RT-PCR products were loaded in 1.2% agarose gels (Sigma Aldrich, St. Louis, MO, USA) supplemented with 0.1 μg/mL ethidium bromide (Sigma Aldrich, St. Louis, MO, USA). Gel-running was performed by an electrophoresis machine (Mupid -2 plus, OPTIMA, Tokyo, Japan). Finally, PCR bands in the gel were visualized by UV light of bioimaging system (TS-312R, Spectoline, Westbury, NY, USA).
4.15. Statistical Analysis
All experiments were repeated in triplicate. Statistical analysis was performed by unpaired t-test or one-way ANOVA with Tukey’s multiple comparisons test, using Graph Pad Prism 8 software (GraphPad Software, Inc., La Jolla, CA, USA); p-values less than 0.05 were considered as statistically significant.