Primary Neurons and Differentiated NSC-34 Cells Are More Susceptible to Arginine-Rich ALS Dipeptide Repeat Protein-Associated Toxicity than Non-Differentiated NSC-34 and CHO Cells
Abstract
1. Introduction
2. Results
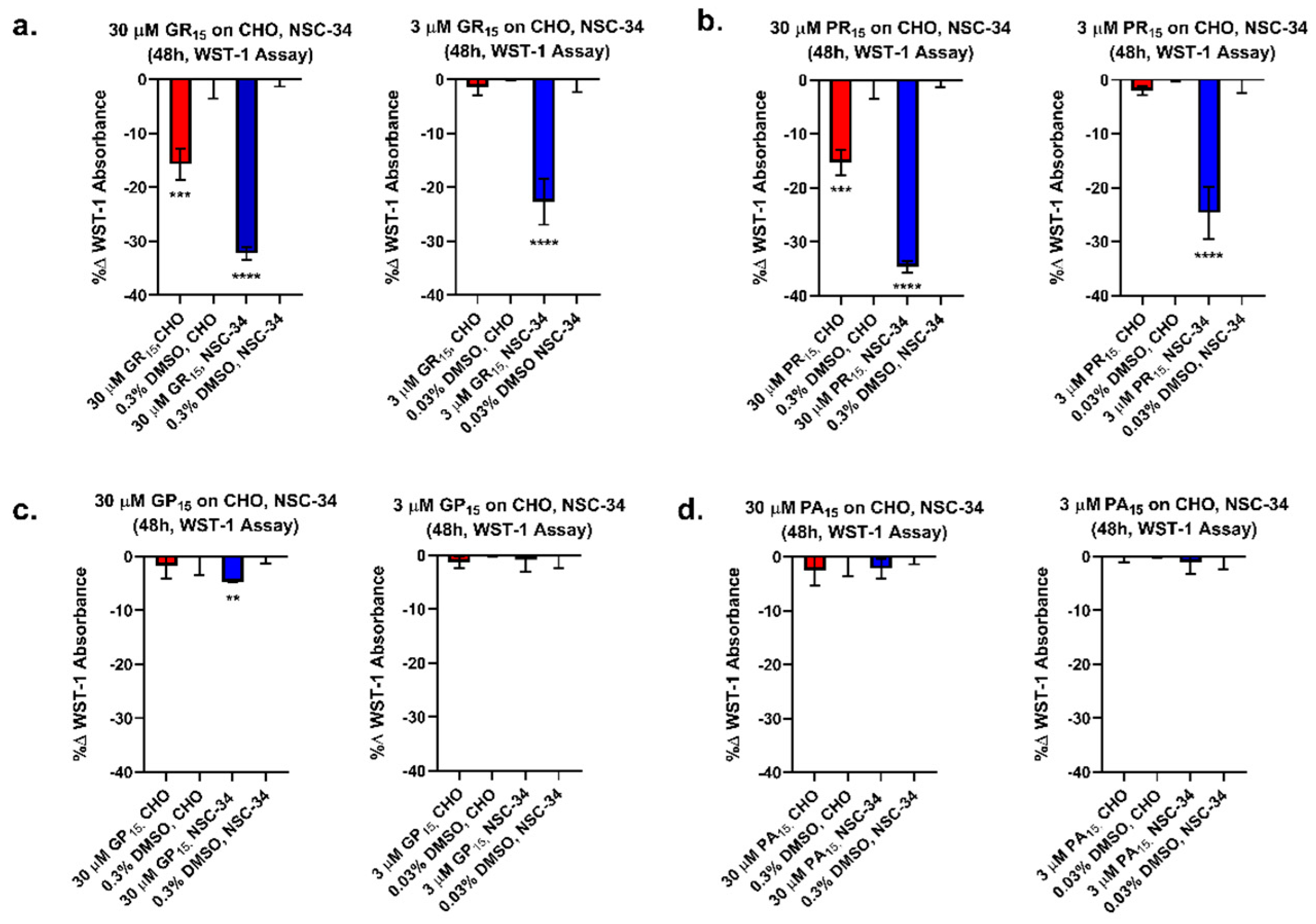
2.1. Arginine-Rich DRPs Exhibit Greater Cytotoxicity in a Neuron-Like Cell Line
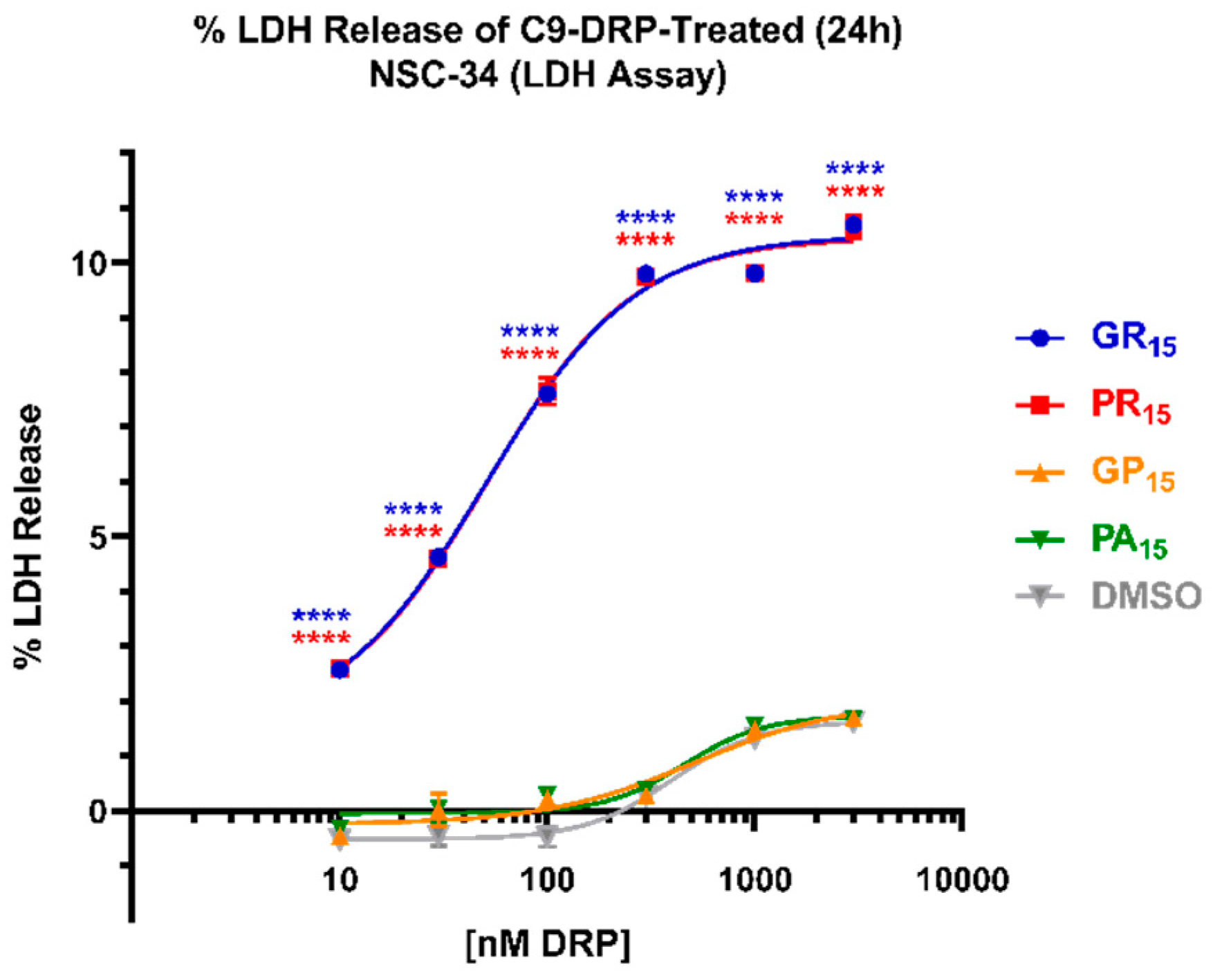
2.2. Arginine-Rich DRPs Induce Cytotoxicity in NSC-34 in a Significant, Dose-Dependent Manner
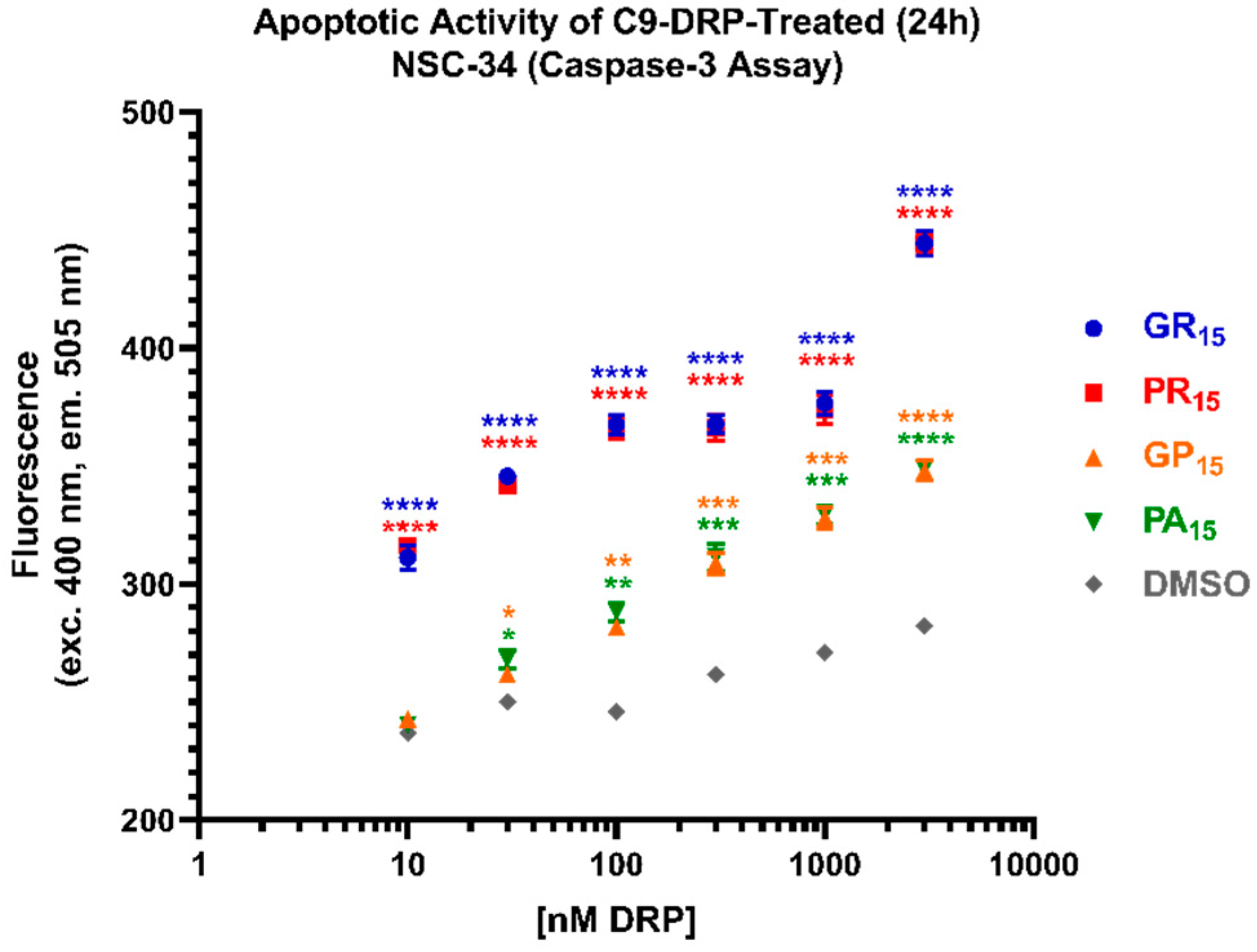
2.3. All DRPs Induce Apoptosis in a Significant, Dose-Dependent Manner
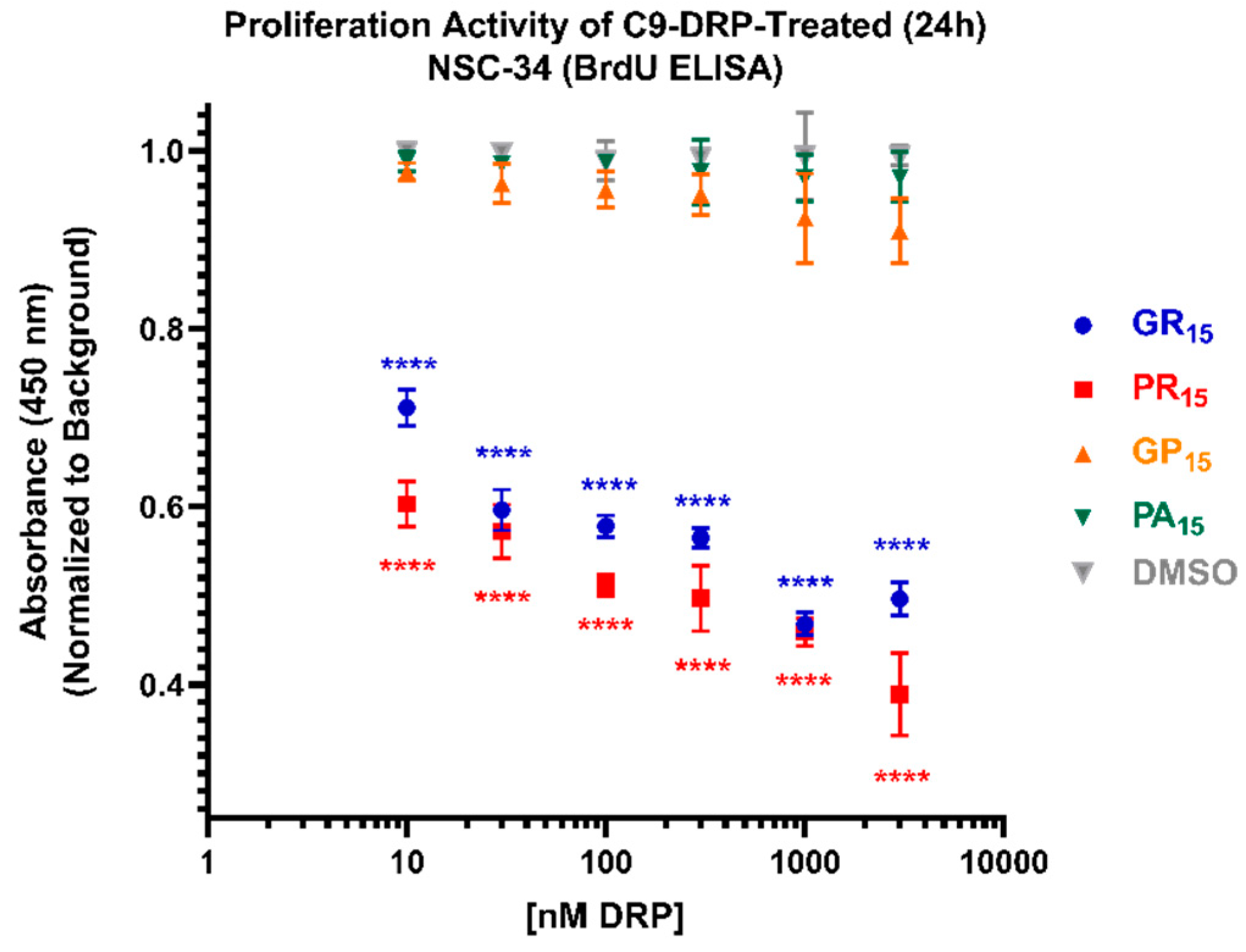
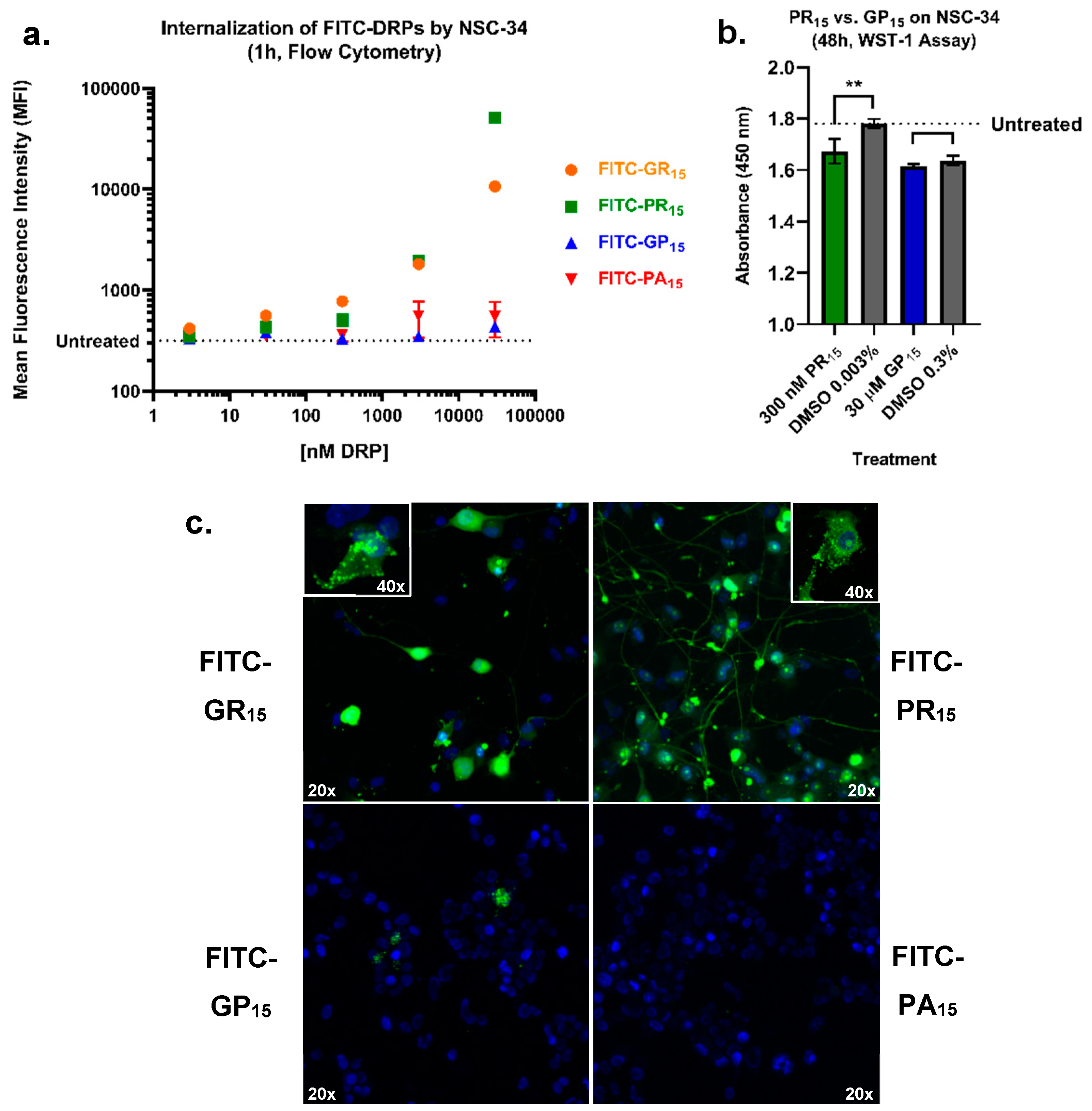
2.4. Arginine-Rich DRPs Interfere with NSC-34 Proliferation Activity
2.5. Arginine-Rich FITC-Labeled DRP Incubation Results in Increased Labeling of NSC-34 Cells Compared to Non-Arginine-Rich DRPs
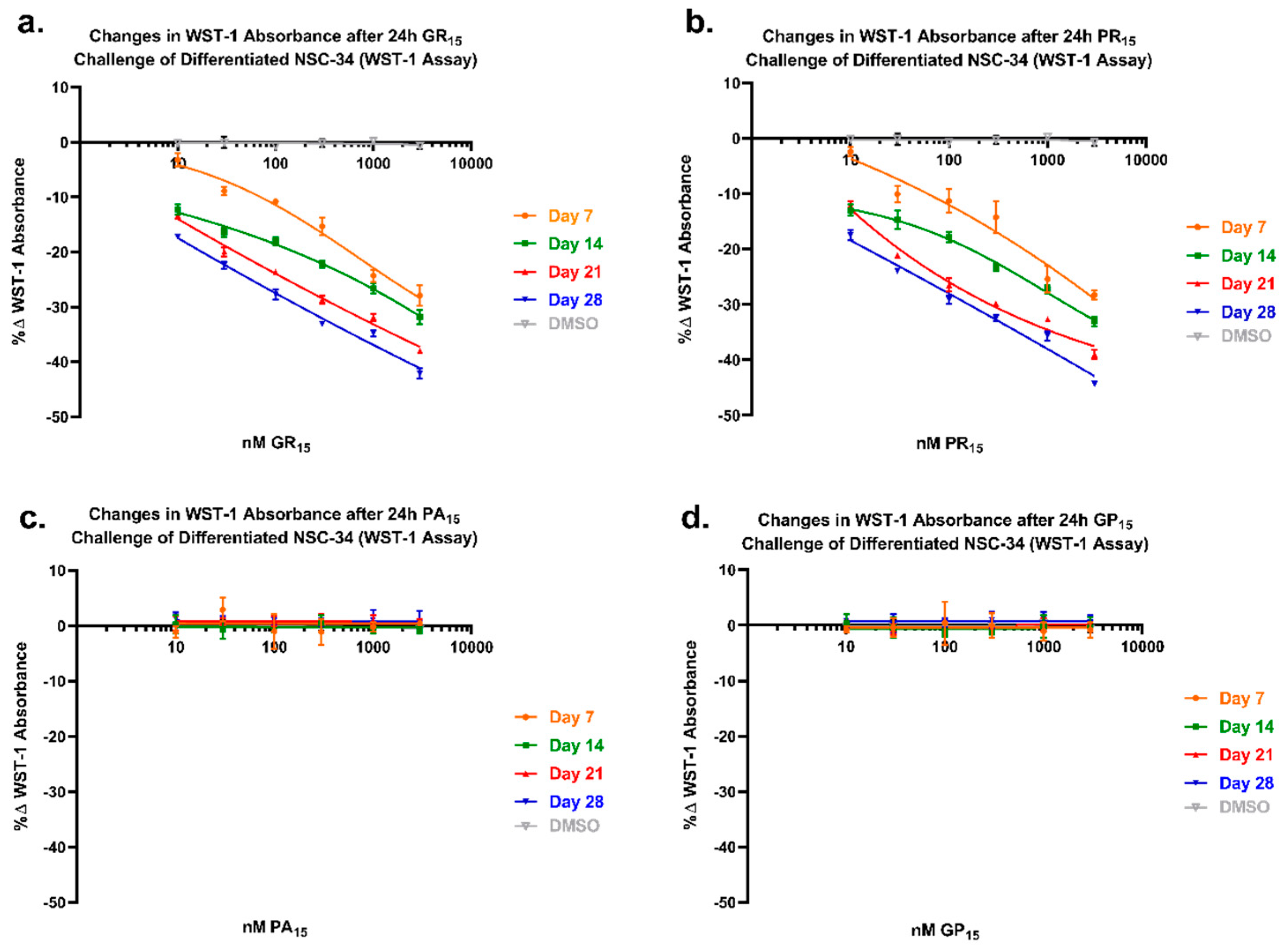
2.6. NSC-34 Sensitivity to Arginine-Rich DRPs Increases with Neuronal Differentiation
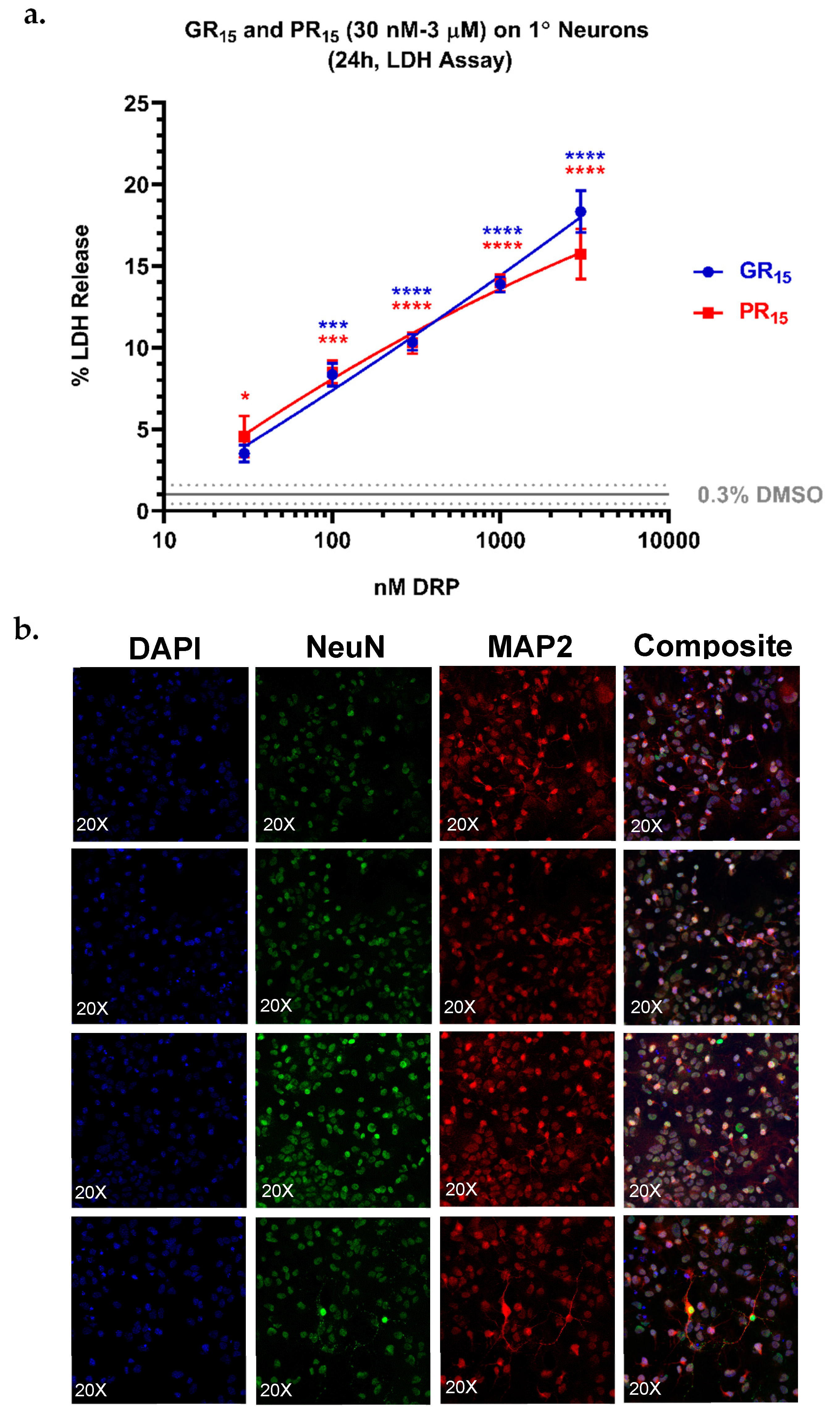
2.7. Neonatal Mouse Derived Spinal Neurons are Most Sensitive to Arginine-Rich DRPs
3. Discussion
4. Materials and Methods
4.1. CHO and NSC-34 Cell Culture and NSC-34 Differentiation
4.2. Dipeptide Repeat Proteins for Testing
4.3. WST-1 Assay
4.4. LDH Assay
4.5. Caspase-3 Assay
4.6. BrdU ELISA
4.7. Flow Cytometry
4.8. Immunohistochemistry and Slide Preparation for Microscopy
4.9. Confocal Laser Scanning Microscopy
4.10. Isolation and Culture of Primary Neurons from Spinal Cords of Neonatal Mice
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DRP | Dipeptide Repeat Protein |
| HRE | Hexanucleotide Repeat Expansion |
| C9HRE | C9orf72 Hexanucleotide Repeat Expansion Mutation |
| RAN | Repeat-Associated non-ATG |
| ALS | Amyotrophic Lateral Sclerosis |
| FTD | Frontotemporal Dementia |
| C9ALS/FTD | C9orf72 Amyotrophic Lateral Sclerosis/Frontotemporal Dementia |
| CHO | Chinese Hamster Ovary cell line |
| NSC-34 | Mouse Spinal Cord x Neuroblastoma Hybrid cell line |
References
- Dejesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Sanchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Weng, S.-M.; Arzberger, T.; May, S.; Rentzsch, K.; Kremmer, E.; Schmid, B.; Kretzshmar, H.A.; Cruts, M.; Van Broeckhoven, C.; et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science 2013, 339, 1335–1338. [Google Scholar] [CrossRef] [PubMed]
- Van Mossevelde, S.; van der Zee, J.; Cruts, M.; Van Broeckhoven, C. Relationship between C9orf72 repeat size and clinical phenotype. Curr. Opin. Genet. Dev. 2017, 44, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Zu, T.; Gibbens, B.; Doty, N.S.; Gomes-Pereira, M.; Huguet, A.; Stone, M.D.; Margolis, J.; Peterson, M.; Markowski, T.W.; Ingram, M.A.C.; et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA 2011, 108, 260–265. [Google Scholar] [CrossRef]
- Zamiri, B.; Mirceta, M.; Bomsztyk, K.; Macgregor, R.B., Jr.; Pearson, C.E. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8•(GGCCCC)8 repeat: Effect of CpG methylation. Nucleic Acids Res. 2015, 43, 10055–10064. [Google Scholar]
- Fratta, P.; Mizielinska, S.; Nicoll, A.J.; Zloh, M.; Fisher, E.M.C.; Parkinson, G.; Isaacs, A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012, 2, 1–6. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Olejniczak, M.; Galka-Marciniak, P.; Jazurek, M.; Krzyzosiak, W.J. RAN translation and frameshifting as translational challenges at simple repeats of human neurodegenerative disorders. Nucleic Acids Res. 2014, 42, 11849–11864. [Google Scholar] [CrossRef]
- Banez-Coronel, M.; Ayhan, F.; Tarabochia, A.D.; Zu, T.; Perez, B.A.; Khoramian Tusi, S.; Pletnikova, O.; Borchelt, D.R.; Ross, C.A.; Margolis, R.L.; et al. RAN Translation in Huntington Disease. Neuron 2015, 88, 667–677. [Google Scholar] [CrossRef]
- Moseley, M.L.; Zu, T.; Ikeda, Y.; Gao, W.; Mosemiller, A.K.; Daughters, R.S.; Chen, G.; Weatherspoon, M.R.; Clark, H.B.; Ebner, T.J.; et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006, 38, 758–769. [Google Scholar] [CrossRef]
- Liquori, C.L.; Ricker, K.; Moseley, M.L.; Jacobsen, J.F.; Kress, W.; Naylor, S.L.; Day, J.W.; Ranum, L.P.W. Myotonic Dystrophy Type 2 Caused by a CCTG Expansion in Intron 1 of ZNF9. Science 2001, 293, 864–867. [Google Scholar] [CrossRef] [PubMed]
- La Spada, A.R.; Wilson, E.M.; Lubahn, D.B.; Harding, A.E.; Fischbeck, K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 1991, 352, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Farg, M.A.; Sundaramoorthy, V.; Sultana, J.M.; Yang, S.; Atkinson, R.A.K.; Levina, V.; Halloran, M.A.; Gleeson, P.A.; Blair, I.P.; Soo, K.Y.; et al. C9ORF72, implicated in amyotrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014, 23, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Lagier-Tourenne, C.; Baughn, M.; Rigo, F.; Sun, S.; Liu, P.; Li, H.-R.; Jiang, J.; Watt, A.T.; Chun, S.; Katz, M.; et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA 2013, 110, E4530–E4539. [Google Scholar] [CrossRef] [PubMed]
- Mizielinska, S.; Gronke, S.; Niccoli, T.; Ridler, C.E.; Clayton, E.L.; Devoy, A.; Moens, T.; Norona, F.E.; Woollacott, I.O.C.; Pietrzyk, J.; et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 2014, 345, 1192–1194. [Google Scholar] [CrossRef]
- Zu, T.; Liu, Y.; Banez-Coronel, M.; Reid, T.; Pletnikova, O.; Lewis, J.; Miller, T.M.; Harms, M.B.; Falchook, A.E.; Subramony, S.H.; et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA 2013, 110, E4968–E4977. [Google Scholar] [CrossRef]
- Gendron, T.F.; Bieniek, K.F.; Zhang, Y.-J.; Jansen-West, K.; Ash, P.E.A.; Caulfield, T.; Daughrity, L.; Dunmore, J.H.; Castanedes-Casey, M.; Chew, J.; et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013, 126, 829–844. [Google Scholar] [CrossRef]
- Tran, H.; Almeida, S.; Moore, J.; Gendron, T.F.; Chalasani, U.; Lu, Y.; Du, X.; Nickerson, J.A.; Petrucelli, L.; Zhiping, W.; et al. Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS. Neuron 2015, 87, 1207–1214. [Google Scholar] [CrossRef]
- Kwon, I.; Xiang, S.; Kato, M.; Wu, L.; Theodoropoulos, P.; Wang, T.; Kim, J.; Yun, J.; Xie, Y.; McKnight, S. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 2014, 345, 1139–1145. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Jansen-West, K.; Xu, Y.-F.; Gendron, T.F.; Bieniek, K.F.; Lin, W.-L.; Sasaguri, H.; Caulfield, T.; Hubbard, J.; Daughrity, L.; et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014, 128, 505–524. [Google Scholar] [CrossRef]
- Yang, D.; Abdallah, A.; Li, Z.; Lu, Y.; Almeida, S.; Gao, F.-B. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 2015, 130, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Freibaum, B.D.; Lu, Y.; Lopez-Gonzalez, R.; Kim, N.C.; Almeida, S.; Lee, K.-H.; Badders, N.; Valentine, M.; Miller, B.L.; Wong, P.C.; et al. GGGGCC repeat expansion in C9ORF72 compromises nucleocytoplasmic transport. Nature 2015, 525, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Jovičić, A.; Mertens, J.; Boeynaems, S.; Bogaert, E.; Chai, N.; Yamada, S.B.; Paul, J.W., III; Sun, S.; Herdy, J.R.; Bieri, G.; et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015, 18, 1226–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Donnelly, C.J.; Haeusler, A.R.; Grima, J.C.; Machamer, J.B.; Steinwald, P.; Daley, E.L.; Miller, S.J.; Cunningham, K.M.; Vidensky, S.; et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015, 525, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.Y.; Mori, E.; Nizami, Z.F.; Kato, M.; Xiang, S.; Wu, L.C.; Ding, M.; Yu, Y.; Gall, J.G.; McKnight, S.L. Toxic PRn Poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. USA 2017, 114, E111–E117. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Lopez-Gonzalez, R.; Kunz, R.C.; Gangopadhyay, J.; Borufka, C.; Gygi, S.P.; Gao, F.-B.; Reed, R. Evidence that C9ORF72 Dipeptide Repeat Proteins Associate with U2 snRNP to Cause Mis-splicing in ALS/FTD Patients. Cell Rep. 2017, 19, 2244–2256. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.A.; Frick, P.; Grässer, F.A.; Gendron, T.F.; Petrucelli, L.; Cashman, N.R.; Edbauer, D.; Kremmer, E.; Prudlo, J.; Troost, D.; et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015, 130, 845–861. [Google Scholar] [CrossRef]
- Lehmer, C.; Oeckl, P.; Weishaupt, J.H.; Volk, A.E.; Diehl-Schmid, J.; Schroeter, M.L.; Lauer, M.; Kornhuber, J.; Levin, J.; Fassbender, K.; et al. Poly-GP in cerebrospinal fluid links C9orf72-associated dipeptide repeat expression to the asymptomatic phase of ALS/FTD. EMBO Mol. Med. 2017, 9, 859–868. [Google Scholar] [CrossRef]
- Kanekura, K.; Harada, Y.; Fujimoto, M.; Yago, T.; Hayamizu, Y.; Nagaoka, K.; Kuroda, M. Characterization of membrane penetration and cytotoxicity of C9orf72-encoding arginine-rich dipeptides. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- White, M.R.; Miltrea, D.M.; Zhang, P.; Stanley, C.B.; Cassidy, D.E.; Nourse, A.; Phillips, A.H.; Tolbert, M.; Taylor, J.P.; Kriwacki, R.W. C9orf72 Poly(PR) Dipeptide Repeats Disturb Biomolecular Phase Separation and Disrupt Nucleolar Function. Mol. Cell 2019, 74, 1–16. [Google Scholar] [CrossRef]
- Gupta, R.; Lan, M.; Mojsilovic-Petrovic, J.; Choi, W.H.; Safren, N.; Barmada, S.; Lee, M.J.; Kalb, R. The Proline/Arginine Dipeptide from Hexanucleotide Repeat Expanded C9ORF72 Inhibits the Proteasome. Eneuro 2017, 4, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.J.; Haney, M.S.; Morgens, D.W.; Jovicic, A.; Couthouis, J.; Li, A.; Ousey, J.; Ma, R.; Bieri, G.; Tsui, C.K.; et al. CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide repeat-protein toxicity. Nat. Genet. 2018, 50, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.J.; Jeng, U.S.; Chiang, Y.L.; Hwang, I.S.; Chen, Y.R. The Glycine-Alanine Dipeptide Repeat from C9orf72 Hexanucleotide Expansions Forms Toxic Amyloids Possessing Cell-to-Cell Transmission Properties. J. Biol. Chem. 2016, 291, 4903–4911. [Google Scholar] [CrossRef] [PubMed]
- Hounoum, B.M.; Vourc’h, P.; Romain, F.; Corcia, P.; Patin, F.; Gueguinou, M.; Potier-Cartereau, M.; Vandier, C.; Raoul, C.; Andres, C.R.; et al. NSC-34 Motor Neuron-Like Cells Are Unsuitable as Experimental Model for Glutamate-Mediated Excitotoxicity. Front. Cell. Neurosci. 2016, 10, 1–12. [Google Scholar]
- Chen, P.C.; Ruan, J.S.; Wu, S.N. Evidence of Decreased Activity in Intermediate-Conductance Calcium-Activated Potassium Channels During Retinoic Acid-Induced Differentiation in Motor Neuron-Like NSC-34 Cells. Cell Physiol. Biochem. 2018, 48, 2374–2388. [Google Scholar] [CrossRef] [PubMed]
- Maier, O.; Bohm, J.; Dahm, M.; Bruck, S.; Beyer, C.; Johann, S. Differentiated NSC-34 motoneuron-like cells as experimental model for cholinergic neurodegeneration. Neurochem. Int. 2013, 62, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Johann, S.; Dahm, M.; Kipp, M.; Zahn, U.; Beyer, C. Regulation of Choline Acetyltransferase Expression by 17β-Oestradiol in NSC-34 Cells and in the Spinal Cord. J. Neuroendocrinol. 2011, 23, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Breydo, L.; Rivas, E.G.; Gebru, N.T.; Zheng, D.; Baker, J.D.; Blair, L.J.; Dickey, C.A.; Koren, J., III; Uversky, V.N. Repeated repeat problems: Combinatorial effect of C9orf72-derived dipeptide repeat proteins. Int. J. Biol. Macromol. 2019, 127, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Kapur, M.; Monaghan, C.E.; Ackerman, S.L. Regulation of mRNA Translation in Neurons—A Matter of Life and Death. Neuron 2017, 96, 616–637. [Google Scholar] [CrossRef]
- Shani, T.; Levy, M.; Isrealson, A. Assay to Measure Nucleocytoplasmic Transport in Real Time within Motor Neuron-like NSC-34 Cells. J. Vis. Exp. 2017, 123. [Google Scholar] [CrossRef]
- Eldeiry, M.; Yamanaka, K.; Reece, T.B.; Aftab, M. Spinal Cord Neurons Isolation and Culture from Neonatal Mice. J. Vis. Exp. 2017, 125. [Google Scholar] [CrossRef] [PubMed]
| DRP | Endpoint | 3 µM | 1 µM | 300 nM | 100 nM | 30 nM | 10 nM |
|---|---|---|---|---|---|---|---|
| Day 14 | **** | ns | **** | **** | **** | **** | |
| GR15 | Day 21 | **** | **** | **** | **** | **** | **** |
| Day 28 | **** | **** | **** | **** | **** | **** | |
| Day 14 | ** | ns | **** | **** | *** | **** | |
| PR15 | Day 21 | **** | **** | **** | **** | **** | **** |
| Day 28 | **** | **** | **** | **** | **** | **** | |
| Day 14 | ns | ns | ns | ns | *** | ns | |
| PA15 | Day 21 | ns | ns | ns | ns | ** | ns |
| Day 28 | ns | ns | ns | ns | **** | ns | |
| Day 14 | ns | ns | ns | * | ns | ns | |
| GP15 | Day 21 | ns | ns | ns | ns | ns | ns |
| Day 28 | ns | * | ns | * | ns | ns |
| DRP | Comparison | 3 µM | 1 µM | 300 nM | 100 nM | 30 nM | 10 nM |
|---|---|---|---|---|---|---|---|
| D7 vs. D14 | ** | ns | **** | **** | **** | **** | |
| GR15 | D14 vs. D21 | **** | **** | **** | **** | **** | ns |
| D21 vs. D28 | **** | **** | **** | **** | **** | *** | |
| D7 vs. D14 | ** | ns | **** | **** | *** | **** | |
| PR15 | D14 vs. D21 | **** | **** | **** | **** | **** | ns |
| D21 vs. D28 | **** | ** | ns | ns | *** | **** | |
| D7 vs. D14 | ns | ns | ns | ns | *** | ns | |
| PA15 | D14 vs. D21 | ns | ns | ns | ns | ns | ns |
| D21 vs. D28 | ns | ns | ns | ns | ns | ns | |
| D7 vs. D14 | ns | ns | ns | * | ns | ns | |
| GP15 | D14 vs. D21 | ns | ns | ns | ns | ns | ns |
| D21 vs. D28 | ns | ns | ns | ns | ns | ns |
| Supplier | Sequence | Purity (%) | Molecular Weight (g) | Solubilized in | [Stock] | Stored at (°C) |
|---|---|---|---|---|---|---|
| GenicBio | (GR)15 | 94.53 | 3216.57 | Sterile DMSO | 10 mM | 4 |
| GenicBio | (PR)15 | 93.17 | 3817.53 | Sterile DMSO | 10 mM | 4 |
| GenicBio | (GP)15 | 94.49 | 2330.56 | Sterile DMSO | 10 mM | 4 |
| GenicBio | (PA)15 | 90.75 | 2540.91 | Sterile DMSO | 10 mM | 4 |
| GenicBio | FITC-(GR)15 | 92.15 | 3719.11 | Sterile DMSO | 10 mM | 4 |
| GenicBio | FITC-(PR)15 | 94.52 | 4320.07 | Sterile DMSO | 10 mM | 4 |
| GenicBio | FITC-(GP)15 | 95.50 | 2833.05 | Sterile DMSO | 10 mM | 4 |
| GenicBio | FITC-(PA)15 | 94.69 | 3043.44 | Sterile DMSO | 10 mM | 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gill, A.L.; Wang, M.Z.; Levine, B.; Premasiri, A.; Vieira, F.G. Primary Neurons and Differentiated NSC-34 Cells Are More Susceptible to Arginine-Rich ALS Dipeptide Repeat Protein-Associated Toxicity than Non-Differentiated NSC-34 and CHO Cells. Int. J. Mol. Sci. 2019, 20, 6238. https://doi.org/10.3390/ijms20246238
Gill AL, Wang MZ, Levine B, Premasiri A, Vieira FG. Primary Neurons and Differentiated NSC-34 Cells Are More Susceptible to Arginine-Rich ALS Dipeptide Repeat Protein-Associated Toxicity than Non-Differentiated NSC-34 and CHO Cells. International Journal of Molecular Sciences. 2019; 20(24):6238. https://doi.org/10.3390/ijms20246238
Chicago/Turabian StyleGill, Anna L., Monica Z. Wang, Beth Levine, Alan Premasiri, and Fernando G. Vieira. 2019. "Primary Neurons and Differentiated NSC-34 Cells Are More Susceptible to Arginine-Rich ALS Dipeptide Repeat Protein-Associated Toxicity than Non-Differentiated NSC-34 and CHO Cells" International Journal of Molecular Sciences 20, no. 24: 6238. https://doi.org/10.3390/ijms20246238
APA StyleGill, A. L., Wang, M. Z., Levine, B., Premasiri, A., & Vieira, F. G. (2019). Primary Neurons and Differentiated NSC-34 Cells Are More Susceptible to Arginine-Rich ALS Dipeptide Repeat Protein-Associated Toxicity than Non-Differentiated NSC-34 and CHO Cells. International Journal of Molecular Sciences, 20(24), 6238. https://doi.org/10.3390/ijms20246238





