Simultaneous Enhancement of Thermostability and Catalytic Activity of a Metagenome-Derived β-Glucosidase Using Directed Evolution for the Biosynthesis of Butyl Glucoside
Abstract
:1. Introduction
2. Results
2.1. First and Second Round of Error Prone Polymerase Chain Reaction (epPCR)
2.2. Directed Evolution
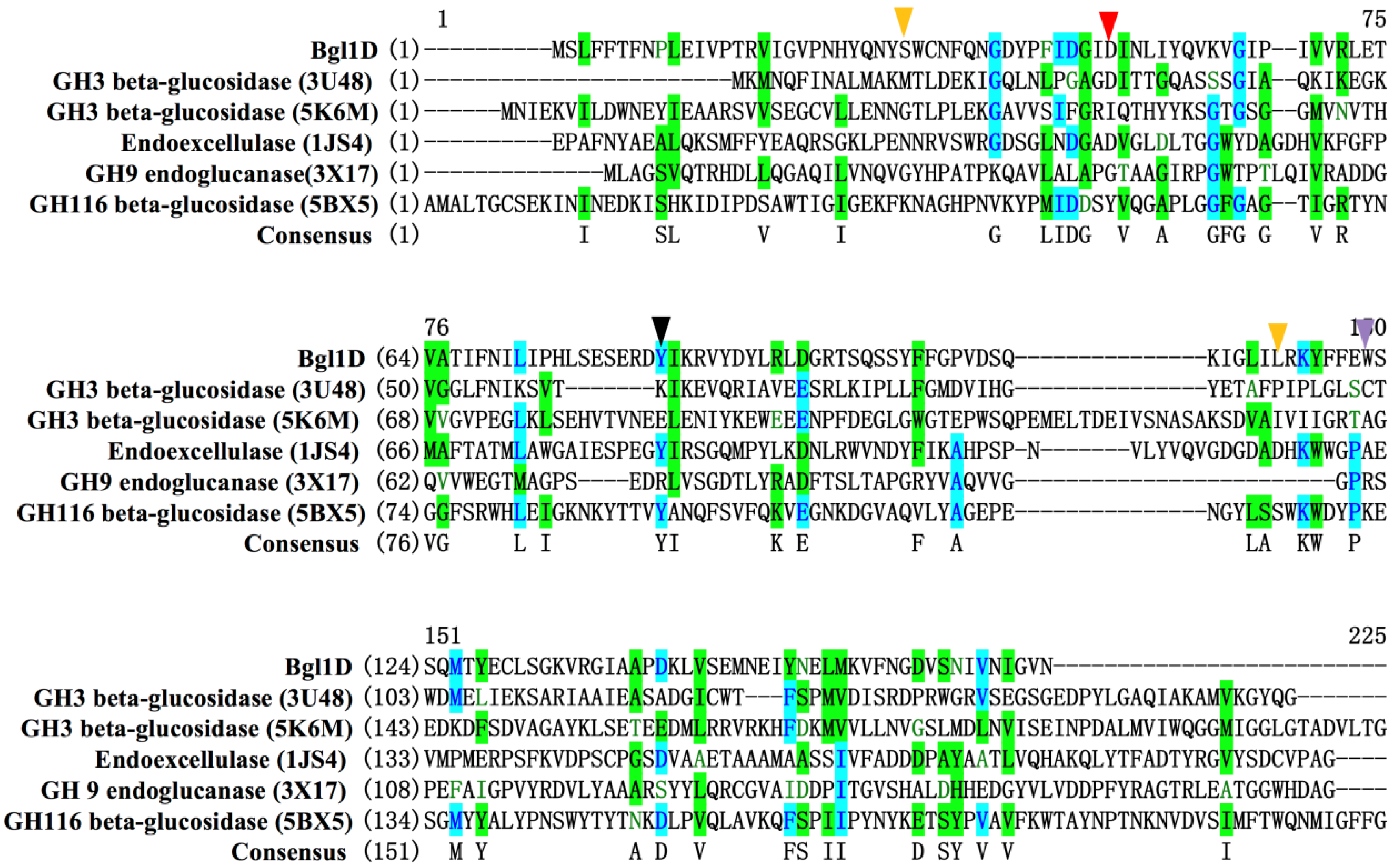
2.3. Site Directed Mutagenesis
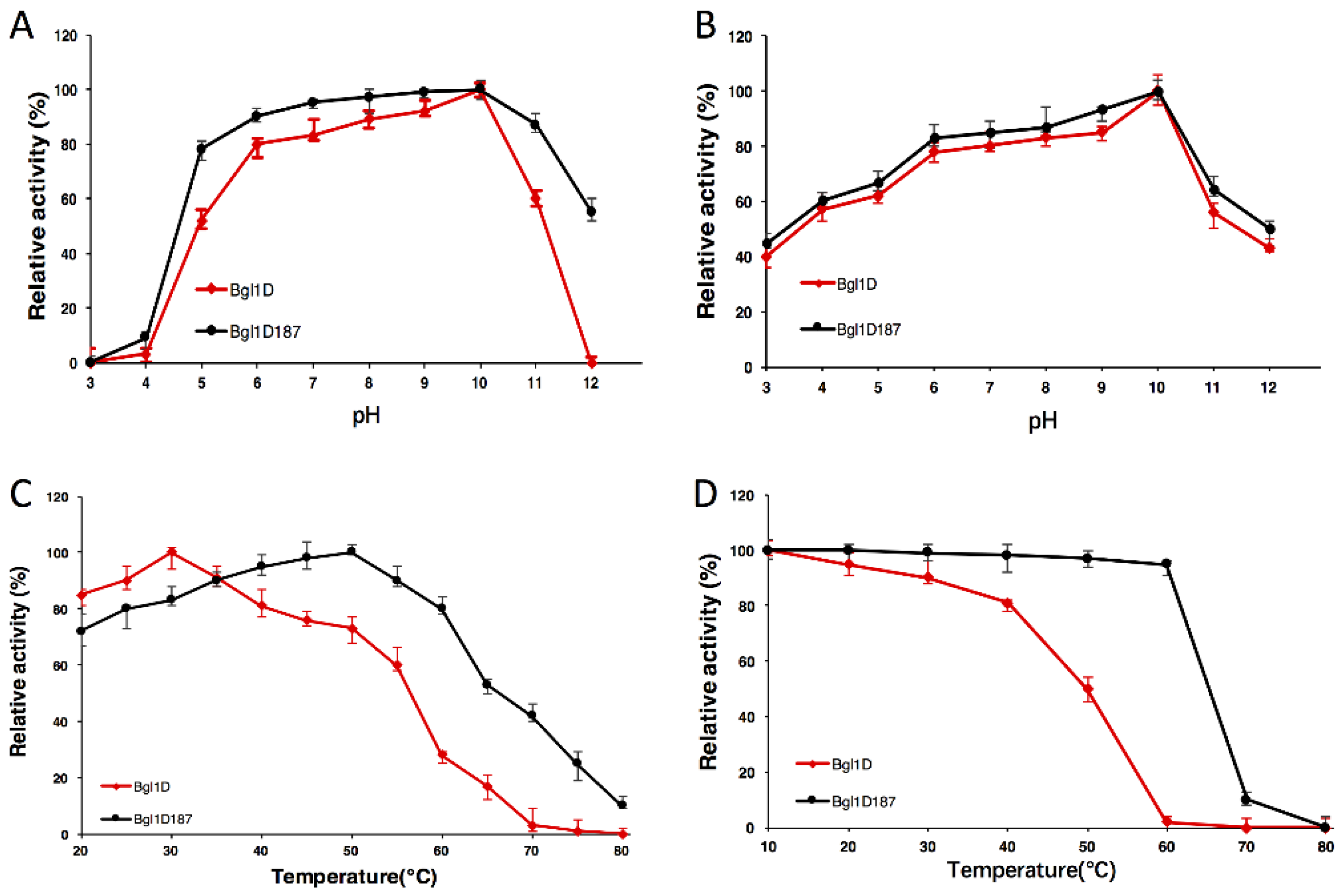
2.4. Comparison of Wild-Type Bgl1D and Bgl1D187 for pH and Temperature
2.5. Comparison of Wild-Type Bgl1D and Bgl1D187 for Kinetic Constants
2.6. Comparison of Wild-Type Bgl1D and Bgl1D187 for Substrate Specificity
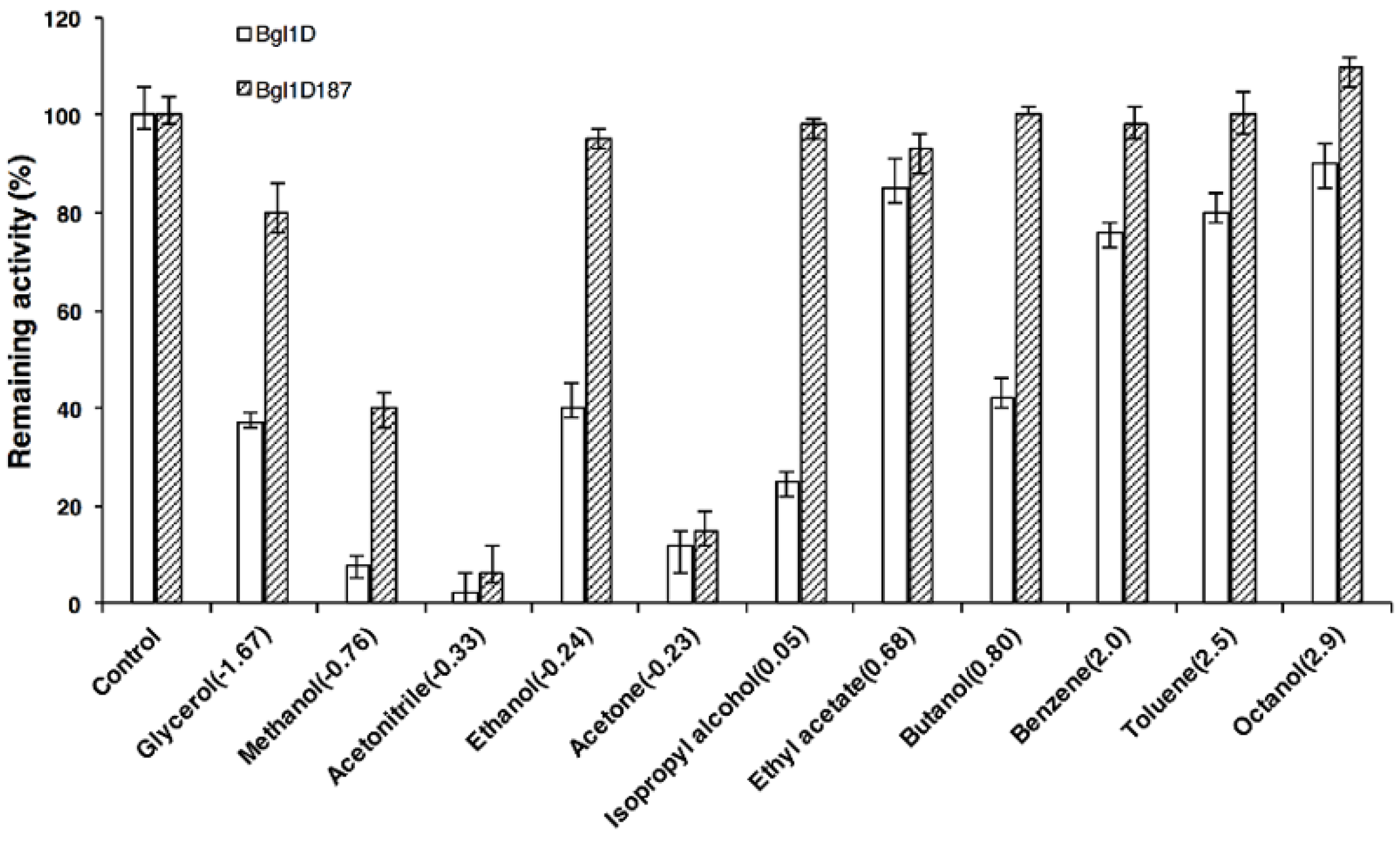
2.7. Comparison of Wild Type and Bgl1D187 for Organic Solvent Tolerance
2.8. Transglycosylation Production of Alcohols
3. Discussion
4. Materials and Methods
4.1. Construction and Screening of Mutant Libraries
4.2. Construction of Site-Directed Mutants
4.3. Overexpression and Purification of the Proteins
4.4. Determination of Enzyme Activity of the Proteins
4.5. Determination of the Effect of pH and Temperature on the Enzyme Activity
4.6. Kinetic Parameters of the Enzyme Proteins
4.7. Substrate Specificity Assays of Enzyme Proteins
4.8. Transglycosylation Activity of the Most Thermostable Enzyme
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| epPCR | Error prone polymerase chain reaction |
| ρNPG | 4-Nitrophenyl-β-d-glucopyranoside |
| IPTG | Isopropyl β-d-thiogalactopyranoside |
| HPLC | High performance liquid chromatography |
| ABE | Acetone-butanol-ethanol |
References
- Volkov, P.V.; Rozhkova, A.M.; Gusakov, A.V.; Sinitsyn, A.P. Homologous cloning, purification and characterization of highly active cellobiohydrolase I (Cel7A) from Penicillium canescens. Protein Expr. Purif. 2014, 103, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Singhania, R.R.; Patel, A.K.; Pandey, A.; Ganansounou, E. Genetic modification: A tool for enhancing beta-glucosidase production for biofuel application. Bioresour. Technol. 2017, 245, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Svensson, D.; Ulvenlund, S.; Adlercreutz, P. Efficient synthesis of a long carbohydrate chain alkyl glycoside catalyzed by cyclodextrin glycosyltransferase (CGTase). Biotechnol. Bioeng. 2009, 104, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Soultani, S.; Ghoul, M. Optimization of the enzymatic synthesis of butyl glucoside using response surface methodology. Biotechnol. Prog. 1998, 14, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.Y.; Nordberg Karlsson, E.; Adlercreutz, P. Complexation of alkyl glycosides with alpha-cyclodextrin can have drastically different effects on their conversion by glycoside hydrolases. J. Biotechnol. 2015, 200, 52–58. [Google Scholar] [CrossRef]
- Boissou, F.; Sayoud, N.; De Oliveira Vigier, K.; Barakat, A.; Marinkovic, S.; Estrine, B.; Jerome, F. Acid-assisted ball milling of cellulose as an efficient pretreatment process for the production of butyl glycosides. ChemSusChem 2015, 8, 3263–3269. [Google Scholar] [CrossRef]
- Xu, X.Q.; Shi, Y.; Wu, X.B.; Zhan, X.L.; Zhou, H.T.; Chen, Q.X. Heat inactivation kinetics of Hypocrea orientalis beta-glucosidase with enhanced thermal stability by glucose. Int. J. Biol. Macromol. 2015, 81, 1012–1018. [Google Scholar] [CrossRef] [Green Version]
- Terefe, N.S.; Sheean, P.; Fernando, S.; Versteeg, C. The stability of almond beta-glucosidase during combined high pressure-thermal processing: A kinetic study. Appl. Microbiol. Biotechnol 2013, 97, 2917–2928. [Google Scholar] [CrossRef]
- Song, J.K.; Rhee, J.S. Simultaneous enhancement of thermostability and catalytic activity of phospholipase A(1) by evolutionary molecular engineering. Appl. Environ. Microbiol. 2000, 66, 890–894. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Fang, X.; Su, F.; Chen, Y.; Xu, L.; Yan, Y. Enhancing the thermostability of Rhizomucor miehei lipase with a limited screening library by rational-design point mutations and disulfide bonds. Appl. Environ. Microbiol. 2018, 84, e02129-17. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, H.; Oda-Ueda, N.; Ueda, T.; Ohkuri, T. A novel engineered interchain disulfide bond in the constant region enhances the thermostability of adalimumab Fab. Biochem. Biophys Res. Commun. 2018, 495, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Bian, F.; Yue, S.; Peng, Z.; Zhang, X.; Chen, G.; Yu, J.; Xuan, N.; Bi, Y. A comprehensive alanine-scanning mutagenesis study reveals roles for salt bridges in the structure and activity of Pseudomonas aeruginosa elastase. PLoS ONE 2015, 10, e0121108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, K.; Kinoshita, M. Physicochemical origin of high correlation between thermal stability of a protein and its packing efficiency: A theoretical study for Staphylococcal nuclease mutants. Biophys. Physicobiol. 2015, 12, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uria, A.R.; Zilda, D.S. Metagenomics-guided mining of commercially useful biocatalysts from marine microorganisms. Adv. Food Nutr. Res. 2016, 78, 1–26. [Google Scholar] [PubMed]
- Meneses, C.; Silva, B.; Medeiros, B.; Serrato, R.; Johnston-Monje, D. A metagenomic advance for the cloning and characterization of a cellulase from red rice crop residues. Molecules 2016, 21, 831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [Green Version]
- Madhavan, A.; Sindhu, R.; Parameswaran, B.; Sukumaran, R.K.; Pandey, A. Metagenome analysis: A powerful tool for enzyme bioprospecting. Appl. Biochem. Biotechnol. 2017, 183, 636–651. [Google Scholar] [CrossRef]
- Bao, L.; Huang, Q.; Chang, L.; Sun, Q.; Zhou, J.; Lu, H. Cloning and characterization of two beta-glucosidase/xylosidase enzymes from yak rumen metagenome. Appl. Biochem. Biotechnol. 2012, 166, 72–86. [Google Scholar] [CrossRef]
- Kumari, M.; Pandey, S.; Mishra, S.K.; Nautiyal, C.S.; Mishra, A. Effect of biosynthesized silver nanoparticles on native soil microflora via plant transport during plant-pathogen-nanoparticles interaction. 3 Biotech 2017, 7, 345. [Google Scholar] [CrossRef]
- Nakamura, K.; Iizuka, R.; Nishi, S.; Yoshida, T.; Hatada, Y.; Takaki, Y.; Iguchi, A.; Yoon, D.H.; Sekiguchi, T.; Shoji, S.; et al. Culture-independent method for identification of microbial enzyme-encoding genes by activity-based single-cell sequencing using a water-in-oil microdroplet platform. Sci. Rep. 2016, 6, 22259. [Google Scholar] [CrossRef]
- Uchiyama, T.; Miyazaki, K.; Yaoi, K. Characterization of a novel beta-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J. Biol. Chem. 2013, 288, 18325–18334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, J.; Liu, X.; Zhang, C.; Zhu, R.; Mao, H. Gene cloning and expression of fungal lignocellulolytic enzymes from the rumen of gayal (Bos frontalis). J. Gen. Appl. Microbiol. 2018, 64, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Duan, C.J.; Pang, H.; Mo, X.C.; Wu, C.F.; Yu, Y.; Hu, Y.L.; Wei, J.; Tang, J.L.; Feng, J.X. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl. Microbiol. Biotechnol. 2007, 75, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Schroder, C.; Elleuche, S.; Blank, S.; Antranikian, G. Characterization of a heat-active archaeal beta-glucosidase from a hydrothermal spring metagenome. Enzym. Microb. Technol. 2014, 57, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Niu, Y.; Li, C.; Zhu, D.; Wang, W.; Liu, X.; Cheng, B.; Ma, C.; Xu, P. Characterization of a novel metagenome-derived 6-phospho-beta-glucosidase from black liquor sediment. Appl. Environ. Microbiol. 2013, 79, 2121–2127. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Qian, C.; Zhang, X.Z.; Liu, N.; Yan, X.; Zhou, Z. Characterization of a novel thermostable beta-glucosidase from a metagenomic library of termite gut. Enzym. Microb. Technol. 2012, 51, 319–324. [Google Scholar] [CrossRef]
- Del Pozo, M.V.; Fernandez-Arrojo, L.; Gil-Martinez, J.; Montesinos, A.; Chernikova, T.N.; Nechitaylo, T.Y.; Waliszek, A.; Tortajada, M.; Rojas, A.; Huws, S.A.; et al. Microbial beta-glucosidases from cow rumen metagenome enhance the saccharification of lignocellulose in combination with commercial cellulase cocktail. Biotechnol. Biofuels 2012, 5, 73. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Feng, Y.; Mo, X.; Duan, C.; Tang, J.; Feng, J. Cloning and expression of a beta-glucosidase gene umcel3G from metagenome of buffalo rumen and characterization of the translated product. Chin. J. Biotechnol. 2008, 24, 232–238. [Google Scholar]
- Kim, S.J.; Lee, C.M.; Kim, M.Y.; Yeo, Y.S.; Yoon, S.H.; Kang, H.C.; Koo, B.S. Screening and characterization of an enzyme with beta-glucosidase activity from environmental DNA. J. Microbiol. Biotechnol. 2007, 17, 905–912. [Google Scholar]
- Jiang, C.; Li, S.X.; Luo, F.F.; Jin, K.; Wang, Q.; Hao, Z.Y.; Wu, L.L.; Zhao, G.C.; Ma, G.F.; Shen, P.H.; et al. Biochemical characterization of two novel beta-glucosidase genes by metagenome expression cloning. Bioresour. Technol. 2011, 102, 3272–3278. [Google Scholar] [CrossRef]
- Jiang, C.J.; Chen, G.; Huang, J.; Huang, Q.; Jin, K.; Shen, P.H.; Li, J.F.; Wu, B. A novel beta-glucosidase with lipolytic activity from a soil metagenome. Folia Microbiol. (Praha) 2011, 56, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Hansson, T.; Kaper, T.; van Der Oost, J.; de Vos, W.M.; Adlercreutz, P. Improved oligosaccharide synthesis by protein engineering of beta-glucosidase CelB from hyperthermophilic Pyrococcus furiosus. Biotechnol. Bioeng. 2001, 73, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Qu, G.; Li, A.; Sun, Z.; Acevedo-Rocha, C.G.; Reetz, M.T. The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chem. Int. Ed. 2019. [Google Scholar] [CrossRef] [PubMed]
- Shukor, H.; Abdeshahian, P.; Al-Shorgani, N.K.; Hamid, A.A.; Rahman, N.A.; Kalil, M.S. Saccharification of polysaccharide content of palm kernel cake using enzymatic catalysis for production of biobutanol in acetone-butanol-ethanol fermentation. Bioresour. Technol. 2016, 202, 206–213. [Google Scholar] [CrossRef]
- Mattos, C. Protein-water interactions in a dynamic world. Trends Biochem. Sci. 2002, 27, 203–208. [Google Scholar] [CrossRef]
- Vieille, C.; Burdette, D.S.; Zeikus, J.G. Thermozymes. Biotechnol. Annu. Rev. 1996, 2, 1–83. [Google Scholar]
- Pei, X.Q.; Yi, Z.L.; Tang, C.G.; Wu, Z.L. Three amino acid changes contribute markedly to the thermostability of beta-glucosidase BglC from Thermobifida fusca. Bioresour. Technol. 2011, 102, 3337–3342. [Google Scholar] [CrossRef]
- Vogt, G.; Woell, S.; Argos, P. Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 1997, 269, 631–643. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Escudero, M.; Del Pozo, M.V.; Marin-Navarro, J.; Gonzalez, B.; Golyshin, P.N.; Polaina, J.; Ferrer, M.; Sanz-Aparicio, J. Structural and functional characterization of a ruminal beta-glycosidase defines a novel subfamily of glycoside hydrolase family 3 with permuted domain topology. J. Biol. Chem. 2016, 291, 24200–24214. [Google Scholar] [CrossRef] [Green Version]
- Sakon, J.; Irwin, D.; Wilson, D.B.; Karplus, P.A. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 1997, 4, 810–818. [Google Scholar] [CrossRef]
- Okano, H.; Kanaya, E.; Ozaki, M.; Angkawidjaja, C.; Kanaya, S. Structure, activity, and stability of metagenome-derived glycoside hydrolase family 9 endoglucanase with an N-terminal Ig-like domain. Protein Sci. 2015, 24, 408–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charoenwattanasatien, R.; Pengthaisong, S.; Breen, I.; Mutoh, R.; Sansenya, S.; Hua, Y.; Tankrathok, A.; Wu, L.; Songsiriritthigul, C.; Tanaka, H.; et al. Bacterial beta-Glucosidase reveals the structural and functional basis of genetic defects in human glucocerebrosidase 2 (GBA2). ACS Chem. Biol. 2016, 11, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Liu, S.; Wang, S.; Wang, L. Ligand-binding specificity and promiscuity of the main lignocellulolytic enzyme families as revealed by active-site architecture analysis. Sci. Rep. 2016, 6, 23605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiesmann, C.; Hengstenberg, W.; Schulz, G.E. Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis. J. Mol. Biol. 1997, 269, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Berry, A. A thermostable variant of fructose bisphosphate aldolase constructed by directed evolution also shows increased stability in organic solvents. Protein Eng. Des. Sel. 2004, 17, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Madan, B.; Mishra, P. Directed evolution of Bacillus licheniformis lipase for improvement of thermostability. Biochem. Eng. J. 2014, 91, 276–282. [Google Scholar] [CrossRef]
- Chamoli, S.; Kumar, P.; Navani, N.K.; Verma, A.K. Secretory expression, characterization and docking study of glucose-tolerant beta-glucosidase from B. subtilis. Int. J. Biol. Macromol. 2016, 85, 425–433. [Google Scholar] [CrossRef]
- Hong, J.; Tamaki, H.; Kumagai, H. Unusual hydrophobic linker region of beta-glucosidase (BGLII) from Thermoascus aurantiacus is required for hyper-activation by organic solvents. Appl. Microbiol. Biotechnol. 2006, 73, 80–88. [Google Scholar] [CrossRef]
- Watt, D.K.; Ono, H.; Hayashi, K. Agrobacterium tumefaciens beta-glucosidase is also an effective beta-xylosidase, and has a high transglycosylation activity in the presence of alcohols. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1998, 1385, 78–88. [Google Scholar] [CrossRef]
- Florindo, R.N.; Souza, V.P.; Mutti, H.S.; Camilo, C.; Manzine, L.R.; Marana, S.R.; Polikarpov, I.; Nascimento, A.S. Structural insights into beta-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum. New Biotechnol. 2018, 40, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Sujittra, S.; Vichitphan, K.; Han, J.; Vichitphan, S.; Swangkaew, J. Hanseniaspora thailandica BC9 beta-glucosidase for the production of beta-D-hexyl glucoside. J. Microbiol. Biotechnol. 2018, 28, 579–587. [Google Scholar] [PubMed]
- Boudabbous, M.; Ben Hmad, I.; Saibi, W.; Mssawra, M.; Belghith, H.; Gargouri, A. Trans-glycosylation capacity of a highly glycosylated multi-specific beta-glucosidase from Fusarium solani. Bioprocess Biosyst. Eng. 2017, 40, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. The importance of additive and non-additive mutational effects in protein engineering. Angew. Chem. Int. Ed. Engl. 2013, 52, 2658–2666. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, R.C.; Joyce, G.F. Mutagenic PCR. PCR Methods Appl. 1994, 3, 136–140. [Google Scholar] [CrossRef] [PubMed]
| Enzyme | Mutations | t1/2 (min) | Relative t1/2 |
|---|---|---|---|
| Bgl1D | None | 45 | 1 |
| Bgl1D58 | Q25L/K117N/M148K | 46.32 | 1 |
| Bgl1D 94 | S28P/I57K/E154G | 45.71 | 1 |
| Bgl1D 47 | F68L/I70M | 45.98 | 1 |
| Bgl1D2 | Q25L/S28T/L115Q/K117N/M148K | 298.02 | 7 |
| Bgl1D6 | S28P/I57K/Y82S/W122G/E154G | 45.48 | 1 |
| Bgl1D20 | Y37H/D44E/F68L/I70M/R91G | 46.34 | 1 |
| Bgl1D28 | S28T | 105.9 | 2 |
| Bgl1D115q | L115Q | 136.74 | 3 |
| Bgl1D115 | L115N | 183.75 | 4 |
| Bgl1D28115 | S28T/L115N | 447.85 | 10 |
| Bgl1D187 | S28T/Y37H/D44E/R91G/L115N | 468.21 | 10 |
| Enzyme | Mutations | kcat (s−1) | Km(mM) | Vmax (U/mg) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| Bgl1D | None | 13.40 ± 0.19 | 0.54 ± 0.03 | 20.10 ± 0.06 | 24.81 ± 0.22 |
| Bgl1D58 | Q25L/K117N/M148K | 23.84 ± 1.92 | 1.25 ± 0.12 | 21.73 ± 2.46 | 14.45 ± 0.95 |
| Bgl1D94 | S28P/I57K/E154G | 48.14 ± 2.34 | 1.17 ± 0.05 | 52.94 ± 4.23 | 41.13 ±2.67 |
| Bgl1D47 | F68L/I70M | 29.38 ± 3.53 | 0.53 ± 0.08 | 25.82 ± 6.36 | 55.43 ± 3.22 |
| Bgl1D2 | Q25L/S28T/L115Q/K117N/M148K | 31.22 ± 1.04 | 1.74 ± 0.14 | 37.46 ± 0.65 | 17.94 ± 0.03 |
| Bgl1D6 | S28P/I57K/Y82S/W122G/E154G | 55.80 ± 6.05 | 1.67 ± 0.12 | 42.50 ± 3.80 | 33.41 ± 2.60 |
| Bgl1D20 | Y37H/D44E/F68L/I70M/R91G | 139.09 ± 8.05 | 0.47 ± 0.02 | 153.70 ± 2.50 | 295.74 ± 1.31 |
| Bgl1D44 | D44G | 0 | 0 | 0 | - |
| Bgl1D82 | Y82S | 5.34 ± 0.08 | 2.13 ± 0.0.73 | 9.53 ± 0.11 | 2.51 ± 0.02 |
| Bgl1D28115 | S28T/L115N | 16.53 ± 2.86 | 1.01 ± 0.45 | 26.33 ± 3.21 | 16.37 ± 3.14 |
| Bgl1D187 | S28T/Y37H/D44E/R91G/L115N | 241.54 ± 10.07 | 0.43 ± 0.04 | 314.30 ± 2.25 | 561.72 ± 4.50 |
| Substrate | Linkage of Glycosyl Group | Specific Activity (U/mg) | |
|---|---|---|---|
| Bgl1D | Bgl1D187 | ||
| Aryl-glycosides | |||
| ρNP-β-d-glucopyranoside | βGlc | 10.8 | 335.88 ± 0.21 |
| ρNP-β-d-galactopyranoside | βGal | 7.34 ± 0.07 | 16.72 ± 0.82 |
| oNP-β-d-galactopyranoside | βGal | 4.86 ± 0.54 | 7.93 ± 1.72 |
| ρNP-α-d-glucopyranoside | αGlc | ND a | ND a |
| ρNP-N-acety-β-d-glucosaminide | βGlc | 0.85 ± 0.83 | 2.87 ± 0.57 |
| ρNP-β-d-xylopyranoside | βXyl | 5.70 ± 0.22 | ND a |
| Salicin | βGlc | 2.84 ± 0.76 | 38.99 ± 1.55 |
| Saccharides | |||
| Sophorose | Glcβ(1,2)Glc | ND a | ND a |
| Cellobiose | Glcβ(1,4)Glc | 1.40 ± 0.04 | 181.57 ± 0.10 |
| Lactose | Galβ(1,4)Glc | 2.38 ± 0.48 | 31.5 ± 0.10 |
| Trehalose | Glcα(1,1)Glc | ND a | ND a |
| Maltose | Glcα(1,4)Glc | ND a | ND a |
| Isomaltose | Glcα(1,6)Glc | ND a | ND a |
| Mannose | Glcα(1,4)Glc | ND a | ND a |
| Sucrose | Glcα(1,2)Fru | ND a | ND a |
| Xylan | βXyl | 8.64 ± 0.64 | 10.66 ± 0.55 |
| CMC | βGlc | ND a | 19.19 ± 0.36 |
| Soluble starch | αGlc | ND a | ND a |
| Starch from wheat | αGlc | ND a | ND a |
| 4-Methylumbelliferyl-β-d-glucopyranoside | βGlc | −b | −b |
| Acceptors | Donors | |
|---|---|---|
| Glucose | Cellobiose | |
| Methanol | ND a | ND a |
| Ethanol | ND a | ND a |
| n-Propanol | ND a | ND a |
| Butanol | −b | ND a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, B.; Hui, Q.; Kashif, M.; Yu, R.; Chen, S.; Ou, Q.; Wu, B.; Jiang, C. Simultaneous Enhancement of Thermostability and Catalytic Activity of a Metagenome-Derived β-Glucosidase Using Directed Evolution for the Biosynthesis of Butyl Glucoside. Int. J. Mol. Sci. 2019, 20, 6224. https://doi.org/10.3390/ijms20246224
Yin B, Hui Q, Kashif M, Yu R, Chen S, Ou Q, Wu B, Jiang C. Simultaneous Enhancement of Thermostability and Catalytic Activity of a Metagenome-Derived β-Glucosidase Using Directed Evolution for the Biosynthesis of Butyl Glucoside. International Journal of Molecular Sciences. 2019; 20(24):6224. https://doi.org/10.3390/ijms20246224
Chicago/Turabian StyleYin, Bangqiao, Qinyan Hui, Muhammad Kashif, Ran Yu, Si Chen, Qian Ou, Bo Wu, and Chengjian Jiang. 2019. "Simultaneous Enhancement of Thermostability and Catalytic Activity of a Metagenome-Derived β-Glucosidase Using Directed Evolution for the Biosynthesis of Butyl Glucoside" International Journal of Molecular Sciences 20, no. 24: 6224. https://doi.org/10.3390/ijms20246224
APA StyleYin, B., Hui, Q., Kashif, M., Yu, R., Chen, S., Ou, Q., Wu, B., & Jiang, C. (2019). Simultaneous Enhancement of Thermostability and Catalytic Activity of a Metagenome-Derived β-Glucosidase Using Directed Evolution for the Biosynthesis of Butyl Glucoside. International Journal of Molecular Sciences, 20(24), 6224. https://doi.org/10.3390/ijms20246224




