The Retinoid and Non-Retinoid Ligands of the Rod Visual G Protein-Coupled Receptor
Abstract
:1. Introduction
GPCRs as Drug Targets
2. Rhodopsin as a Drug Target
2.1. Rhodopsin Structure and Signaling
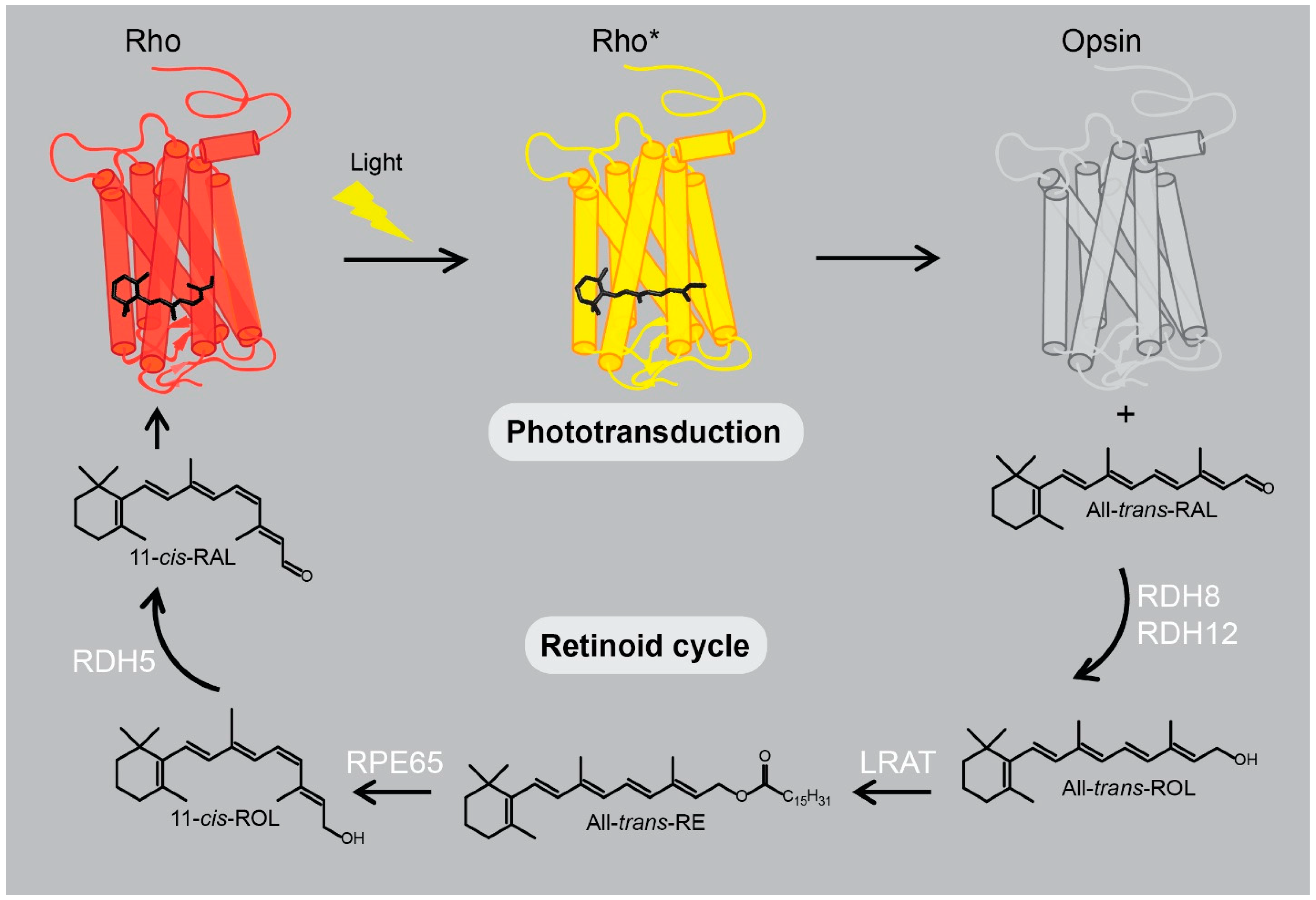
2.2. Rhodopsin’s Orthosteric Binding Pocket - the Retinal Chromophore-Binding Pocket
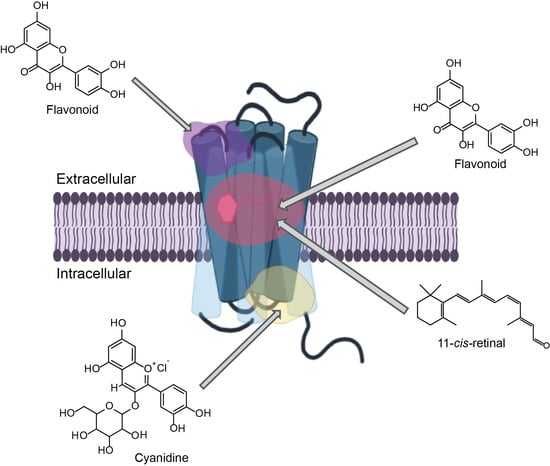
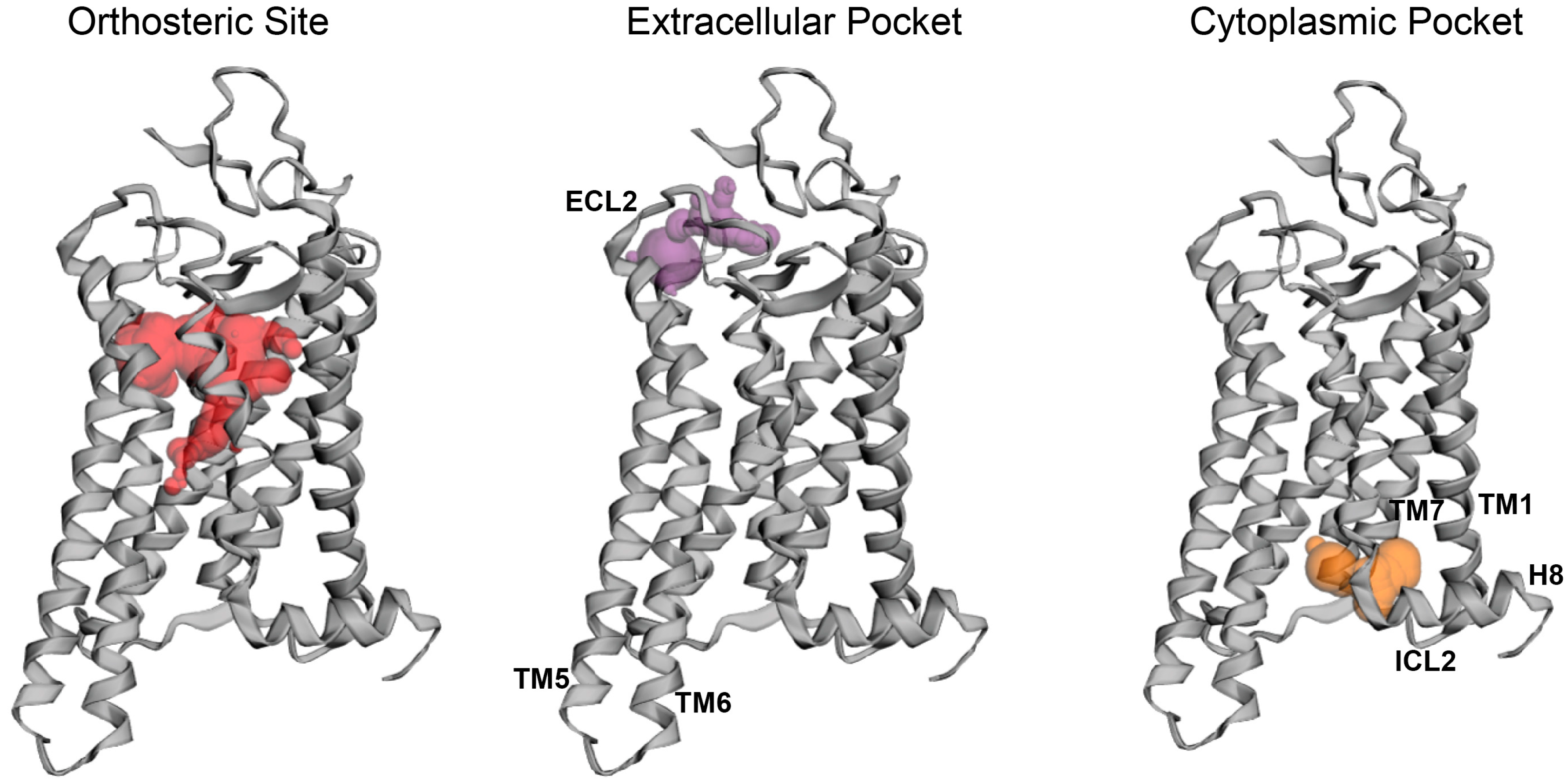
2.3. Potential Rhodopsin Allosteric Binding Pockets
3. Eye-Related Pathologies Associated with Defective Synthesis of Retinal Chromophore
3.1. Leber Congenital Amaurosis
Retinoids as Pharmacological Supplements for Treatment of LCA
3.2. Age-related Macular Degeneration
Retinoids as Pharmacological Supplements for Treatment of AMD
4. Eye-Related Pathologies Associated with Mutations in RHO Gene
4.1. Retinoids as Pharmacological Chaperones to Treat Retinitis Pigmentosa
4.2. Non-retinal Small Molecule Modulators of Retinitis Pigmentosa-Linked Rhodopsin Mutants
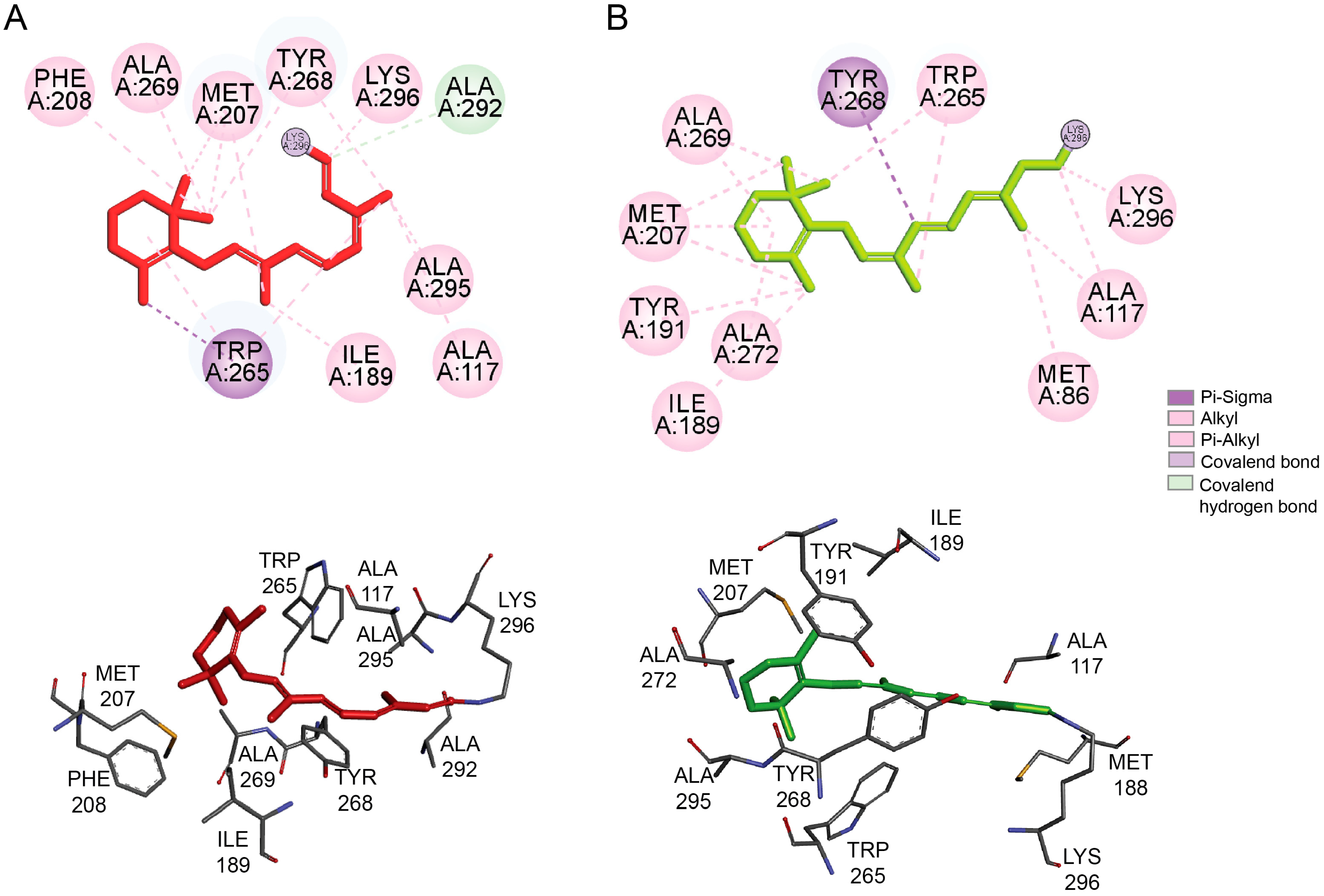
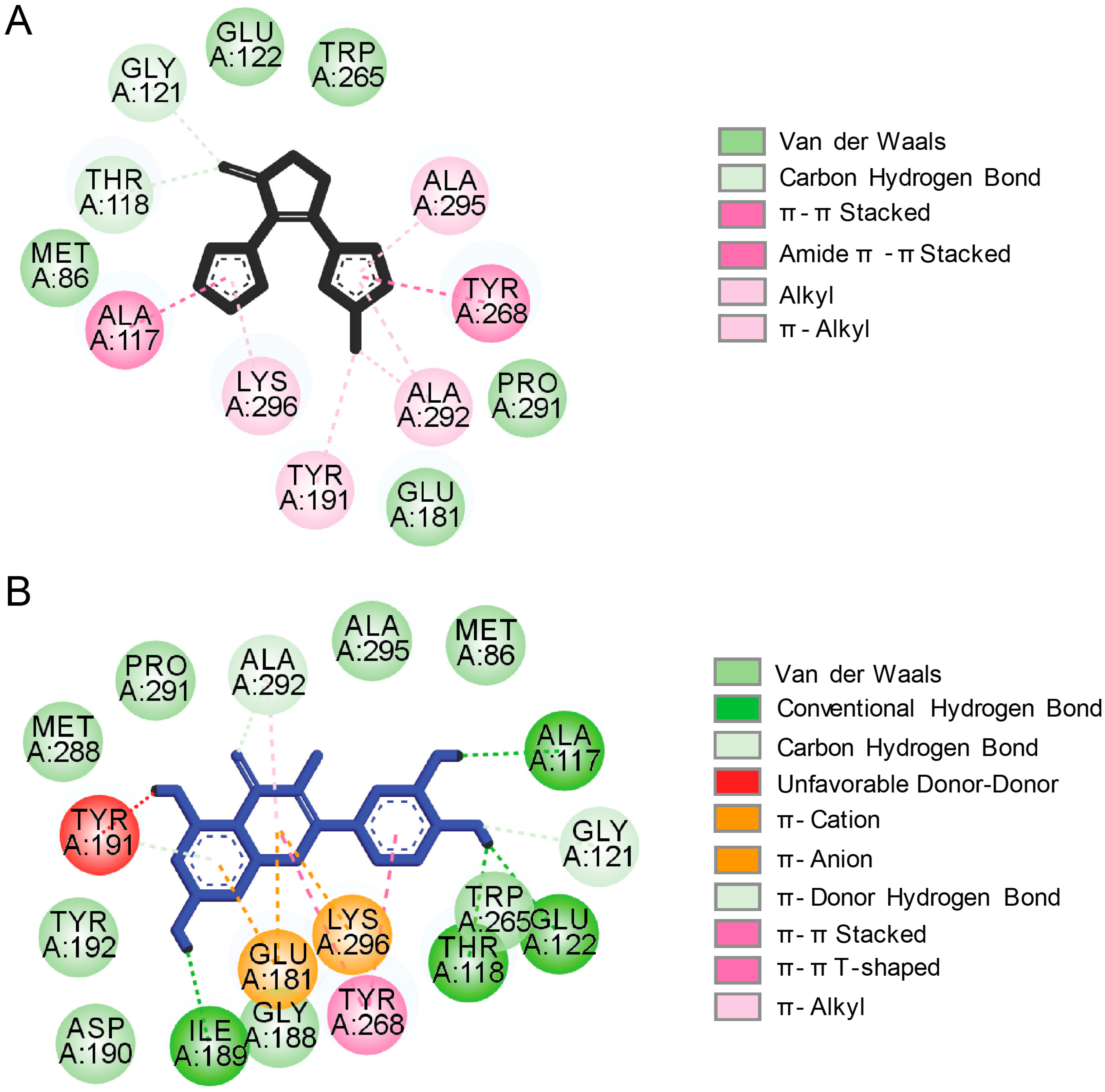
4.3. Natural Products-Derived Compounds as Rhodopsin Modulators and their Potential for Treatment of Retinitis Pigmentosa-Related Retinopathies
5. Protective Effects of Natural Products in In Vitro and In Vivo Models of Visual Diseases
6. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCA4 | ATP-binding cassette subfamily A, member 4 |
| adRP | Autosomal dominant retinitis pigmentosa |
| AMD | Age-related macular degeneration |
| arRP | Autosomal recessive retinitis pigmentosa |
| ATP | Adenosine triphosphate |
| CryoEM | Cryoelectron microscopy |
| CSNB | Congenital stationary night blindness |
| ECL | Extracellular loop |
| ER | Endoplasmic reticulum |
| ERG | Electroretinography |
| FDA | Food and Drugs Administration |
| GPCR | G protein–coupled receptor |
| GRK1 | Rhodopsin kinase |
| Gt | G protein transducin |
| ICL | Intracellular loop |
| i.p | Intraperitoneal |
| IRBP | Interphotoreceptor retinoid-binding protein |
| LCA | Leber Congenital Amaurosis |
| LRAT | Lecithin retinol acyltransferase |
| Meta II | Metarhodopsin II |
| ONL | Outer nuclear layer |
| OS | Outer segments |
| RDH8 | Retinol dehydrogenase 8 |
| ROS | Reactive oxygen species |
| RP | Retinitis pigmentosa |
| RPE | Retinal pigmented epithelium |
| RPE65 | Retinyl pigment epithelium-specific protein 65 |
| TM | Transmembrane |
| WT | Wild-type |
References
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schioth, H.B.; Fredriksson, R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 2005, 142, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Manglik, A.; Kruse, A.C. Structural Basis for G Protein-Coupled Receptor Activation. Biochemistry 2017, 56, 5628–5634. [Google Scholar] [CrossRef]
- Sriram, K.; Insel, P.A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Campbell, A.P.; Smrcka, A.V. Targeting G protein-coupled receptor signalling by blocking G proteins. Nat. Rev. Drug Discov. 2018, 17, 789–803. [Google Scholar] [CrossRef]
- Basith, S.; Cui, M.; Macalino, S.J.Y.; Park, J.; Clavio, N.A.B.; Kang, S.; Choi, S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front. Pharmacol. 2018, 9, 128. [Google Scholar] [CrossRef]
- Coudrat, T.; Christopoulos, A.; Sexton, P.M.; Wootten, D. Structural features embedded in G protein-coupled receptor co-crystal structures are key to their success in virtual screening. PLoS ONE 2017, 12, e0174719. [Google Scholar] [CrossRef] [Green Version]
- Insel, P.A.; Sriram, K.; Gorr, M.W.; Wiley, S.Z.; Michkov, A.; Salmeron, C.; Chinn, A.M. GPCRomics: An Approach to Discover GPCR Drug Targets. Trends Pharmacol. Sci. 2019, 40, 378–387. [Google Scholar] [CrossRef]
- Palczewski, K.; Kumasaka, T.; Hori, T.; Behnke, C.A.; Motoshima, H.; Fox, B.A.; Le Trong, I.; Teller, D.C.; Okada, T.; Stenkamp, R.E.; et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 2000, 289, 739–745. [Google Scholar] [CrossRef] [Green Version]
- Murray, A.R.; Fliesler, S.J.; Al-Ubaidi, M.R. Rhodopsin: The functional significance of asn-linked glycosylation and other post-translational modifications. Ophthalmic Genet. 2009, 30, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimata, N.; Pope, A.; Eilers, M.; Opefi, C.A.; Ziliox, M.; Hirshfeld, A.; Zaitseva, E.; Vogel, R.; Sheves, M.; Reeves, P.J.; et al. Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nat. Commun. 2016, 7, 12683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA—An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 2004, 18, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Parmar, T.; Palczewska, G.; Dong, Z.; Golczak, M.; Palczewski, K.; Jastrzebska, B. Protective Effect of a Locked Retinal Chromophore Analog against Light-Induced Retinal Degeneration. Mol. Pharmacol. 2018, 94, 1132–1144. [Google Scholar] [CrossRef]
- Spalink, J.D.; Reynolds, A.H.; Rentzepis, P.M.; Sperling, W.; Applebury, M.L. Bathorhodopsin intermediates from 11-cis-rhodopsin and 9-cis-rhodopsin. Proc. Natl. Acad. Sci. USA 1983, 80, 1887–1891. [Google Scholar] [CrossRef] [Green Version]
- Vogel, R.; Ludeke, S.; Siebert, F.; Sakmar, T.P.; Hirshfeld, A.; Sheves, M. Agonists and partial agonists of rhodopsin: Retinal polyene methylation affects receptor activation. Biochemistry 2006, 45, 1640–1652. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Jastrzebska, B.; Golczak, M.; Gulati, S.; Tang, H.; Seibel, W.; Li, X.; Jin, H.; Han, Y.; et al. A novel small molecule chaperone of rod opsin and its potential therapy for retinal degeneration. Nat. Commun. 2018, 9, 1976. [Google Scholar] [CrossRef]
- Ortega, J.T.; Parmar, T.; Jastrzebska, B. Flavonoids enhance rod opsin stability, folding, and self-association by directly binding to ligand-free opsin and modulating its conformation. J. Biol. Chem. 2019, 294, 8101–8122. [Google Scholar] [CrossRef]
- Massink, A.; Amelia, T.; Karamychev, A.; IJezerman, A.P. Allosteric modulation of G protein-coupled receptors by amiloride and its derivatives. Perspectives for drug discovery? Med. Res. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, D.; Kaczor, A.A.; Matosiuk, D. Opportunities and Challenges in the Discovery of Allosteric Modulators of GPCRs. Methods Mol. Biol. 2018, 1705, 297–319. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, A.E.; Mason, J.S.; Vajda, S.; Keseru, G.M. Analysis of tractable allosteric sites in G protein-coupled receptors. Sci. Rep. 2019, 9, 6180. [Google Scholar] [CrossRef] [PubMed]
- Wold, E.A.; Chen, J.; Cunningham, K.A.; Zhou, J. Allosteric Modulation of Class A GPCRs: Targets, Agents, and Emerging Concepts. J. Med. Chem. 2019, 62, 88–127. [Google Scholar] [CrossRef]
- Taylor, C.M.; Barda, Y.; Kisselev, O.G.; Marshall, G.R. Modulating G-protein coupled receptor/G-protein signal transduction by small molecules suggested by virtual screening. J. Med. Chem. 2008, 51, 5297–5303. [Google Scholar] [CrossRef] [Green Version]
- Tirupula, K.C.; Balem, F.; Yanamala, N.; Klein-Seetharaman, J. pH-dependent interaction of rhodopsin with cyanidin-3-glucoside. 2. Functional aspects. Photochem. Photobiol. 2009, 85, 463–470. [Google Scholar] [CrossRef]
- Herrera-Hernandez, M.G.; Ramon, E.; Lupala, C.S.; Tena-Campos, M.; Perez, J.J.; Garriga, P. Flavonoid allosteric modulation of mutated visual rhodopsin associated with retinitis pigmentosa. Sci. Rep. 2017, 7, 11167. [Google Scholar] [CrossRef] [Green Version]
- Yanamala, N.; Gardner, E.; Riciutti, A.; Klein-Seetharaman, J. The cytoplasmic rhodopsin-protein interface: Potential for drug discovery. Curr. Drug Targets 2012, 13, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Yanamala, N.; Tirupula, K.C.; Balem, F.; Klein-Seetharaman, J. pH-dependent interaction of rhodopsin with cyanidin-3-glucoside. 1. Structural aspects. Photochem. Photobiol. 2009, 85, 454–462. [Google Scholar] [CrossRef]
- Canter, P.H.; Ernst, E. Anthocyanosides of Vaccinium myrtillus (bilberry) for night vision—A systematic review of placebo-controlled trials. Surv. Ophthalmol. 2004, 49, 38–50. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Kahremany, S.; Zhang, J.; Jastrzebska, B.; Querubin, J.; Petersen-Jones, S.M.; Palczewski, K. Retinal-chitosan Conjugates Effectively Deliver Active Chromophores to Retinal Photoreceptor Cells in Blind Mice and Dogs. Mol. Pharmacol. 2018, 93, 438–452. [Google Scholar] [CrossRef] [Green Version]
- Klein, R.; Klein, B.E.; Linton, K.L. Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology 1992, 99, 933–943. [Google Scholar] [CrossRef]
- Steinmetz, R.L.; Haimovici, R.; Jubb, C.; Fitzke, F.W.; Bird, A.C. Symptomatic abnormalities of dark adaptation in patients with age-related Bruch’s membrane change. Br. J. Ophthalmol. 1993, 77, 549–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvatore, S.; Fishman, G.A.; McAnany, J.J.; Genead, M.A. Association of dark-adapted visual function with retinal structural changes in patients with Stargardt disease. Retina 2014, 34, 989–995. [Google Scholar] [CrossRef] [Green Version]
- Fishman, G.A.; Farbman, J.S.; Alexander, K.R. Delayed rod dark adaptation in patients with Stargardt’s disease. Ophthalmology 1991, 98, 957–962. [Google Scholar] [CrossRef]
- Kemp, C.M.; Jacobson, S.G.; Roman, A.J.; Sung, C.H.; Nathans, J. Abnormal rod dark adaptation in autosomal dominant retinitis pigmentosa with proline-23-histidine rhodopsin mutation. Am. J. Ophthalmol. 1992, 113, 165–174. [Google Scholar] [CrossRef]
- Kemp, C.M.; Jacobson, S.G.; Faulkner, D.J.; Walt, R.W. Visual function and rhodopsin levels in humans with vitamin A deficiency. Exp. Eye Res. 1988, 46, 185–197. [Google Scholar] [CrossRef]
- Steinmetz, R.L.; Polkinghorne, P.C.; Fitzke, F.W.; Kemp, C.M.; Bird, A.C. Abnormal dark adaptation and rhodopsin kinetics in Sorsby’s fundus dystrophy. Invest. Ophthalmol. Vis. Sci. 1992, 33, 1633–1636. [Google Scholar]
- Maeda, T.; Maeda, A.; Leahy, P.; Saperstein, D.A.; Palczewski, K. Effects of long-term administration of 9-cis-retinyl acetate on visual function in mice. Invest. Ophthalmol. Vis. Sci. 2009, 50, 322–333. [Google Scholar] [CrossRef]
- Hofmann, L.; Gulati, S.; Sears, A.; Stewart, P.L.; Palczewski, K. An effective thiol-reactive probe for differential scanning fluorimetry with a standard real-time polymerase chain reaction device. Anal. Biochem. 2016, 499, 63–65. [Google Scholar] [CrossRef] [Green Version]
- Noorwez, S.M.; Malhotra, R.; McDowell, J.H.; Smith, K.A.; Krebs, M.P.; Kaushal, S. Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H. J. Biol. Chem. 2004, 279, 16278–16284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, H.F.; Cheetham, M.E. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum. Mol. Genet. 2008, 17, 3043–3054. [Google Scholar] [CrossRef] [Green Version]
- Mendes, H.F.; Zaccarini, R.; Cheetham, M.E. Pharmacological manipulation of rhodopsin retinitis pigmentosa. Adv. Exp. Med. Biol. 2010, 664, 317–323. [Google Scholar] [CrossRef]
- Noorwez, S.M.; Kuksa, V.; Imanishi, Y.; Zhu, L.; Filipek, S.; Palczewski, K.; Kaushal, S. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J. Biol. Chem. 2003, 278, 14442–14450. [Google Scholar] [CrossRef] [Green Version]
- Gragg, M.; Park, P.S. Misfolded rhodopsin mutants display variable aggregation properties. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2938–2948. [Google Scholar] [CrossRef]
- Behnen, P.; Felline, A.; Comitato, A.; Di Salvo, M.T.; Raimondi, F.; Gulati, S.; Kahremany, S.; Palczewski, K.; Marigo, V.; Fanelli, F. A Small Chaperone Improves Folding and Routing of Rhodopsin Mutants Linked to Inherited Blindness. iScience 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Samardzija, M.; Wenzel, A.; Naash, M.; Reme, C.E.; Grimm, C. Rpe65 as a modifier gene for inherited retinal degeneration. Eur. J. Neurosci. 2006, 23, 1028–1034. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Edwards, P.C.; Burghammer, M.; Villa, C.; Schertler, G.F. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004, 343, 1409–1438. [Google Scholar] [CrossRef]
- Deupi, X.; Edwards, P.; Singhal, A.; Nickle, B.; Oprian, D.; Schertler, G.; Standfuss, J. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl. Acad. Sci. USA 2012, 109, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Mattle, D.; Kuhn, B.; Aebi, J.; Bedoucha, M.; Kekilli, D.; Grozinger, N.; Alker, A.; Rudolph, M.G.; Schmid, G.; Schertler, G.F.X.; et al. Ligand channel in pharmacologically stabilized rhodopsin. Proc. Natl. Acad. Sci. USA 2018, 115, 3640–3645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinheiro, G.; Araujo Filho, I.; Araujo Neto, I.; Rego, A.C.M.; Azevedo, E.P.; Pinheiro, F.I.; Lima Filho, A.A.S. Nature as a source of drugs for ophthalmology. Arq. Bras. Oftalmol. 2018, 81, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; de Bruyn Kops, C.; Kirchmair, J. Data Resources for the Computer-Guided Discovery of Bioactive Natural Products. J. Chem. Inf. Model. 2017, 57, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Milea, D.; Aung, T. Flavonoids and glaucoma: Revisiting therapies from the past. Graefes Arch. Clin. Exp. Ophthalmol. 2015, 253, 1839–1840. [Google Scholar] [CrossRef] [Green Version]
- Rhone, M.; Basu, A. Phytochemicals and age-related eye diseases. Nutr. Rev. 2008, 66, 465–472. [Google Scholar] [CrossRef]
- Majumdar, S.; Srirangam, R. Potential of the bioflavonoids in the prevention/treatment of ocular disorders. J. Pharm. Pharmacol. 2010, 62, 951–965. [Google Scholar] [CrossRef] [Green Version]
- Huvaere, K.; Skibsted, L.H. Flavonoids protecting food and beverages against light. J. Sci. Food Agric. 2015, 95, 20–35. [Google Scholar] [CrossRef]
- Kalt, W.; Hanneken, A.; Milbury, P.; Tremblay, F. Recent research on polyphenolics in vision and eye health. J. Agric. Food Chem. 2010, 58, 4001–4007. [Google Scholar] [CrossRef]
- Park, J.H.; Scheerer, P.; Hofmann, K.P.; Choe, H.W.; Ernst, O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 2008, 454, 183–187. [Google Scholar] [CrossRef] [Green Version]
- Jastrzebska, B.; Fotiadis, D.; Jang, G.F.; Stenkamp, R.E.; Engel, A.; Palczewski, K. Functional and structural characterization of rhodopsin oligomers. J. Biol. Chem. 2006, 281, 11917–11922. [Google Scholar] [CrossRef] [Green Version]
- Jastrzebska, B.; Chen, Y.; Orban, T.; Jin, H.; Hofmann, L.; Palczewski, K. Disruption of Rhodopsin Dimerization with Synthetic Peptides Targeting an Interaction Interface. J. Biol. Chem. 2015, 290, 25728–25744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, H.; Nakamura, Y.; Tachibanaki, S.; Kawamura, S.; Hirayama, M. Stimulatory effect of cyanidin 3-glycosides on the regeneration of rhodopsin. J. Agric. Food Chem. 2003, 51, 3560–3563. [Google Scholar] [CrossRef] [PubMed]
- Nomi, Y.; Iwasaki-Kurashige, K.; Matsumoto, H. Therapeutic Effects of Anthocyanins for Vision and Eye Health. Molecules 2019, 24, 3311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, D.E.; Hurst, R.D. Polyphenolic phytochemicals—Just antioxidants or much more? Cell. Mol. Life Sci. 2007, 64, 2900–2916. [Google Scholar] [CrossRef]
- Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar] [CrossRef] [Green Version]
- Muangnoi, C.; Sharif, U.; Ratnatilaka Na Bhuket, P.; Rojsitthisak, P.; Paraoan, L. Protective Effects of Curcumin Ester Prodrug, Curcumin Diethyl Disuccinate against H2O2-Induced Oxidative Stress in Human Retinal Pigment Epithelial Cells: Potential Therapeutic Avenues for Age-Related Macular Degeneration. Int. J. Mol. Sci. 2019, 20, 3367. [Google Scholar] [CrossRef] [Green Version]
- Ooe, E.; Kuse, Y.; Yako, T.; Sogon, T.; Nakamura, S.; Hara, H.; Shimazawa, M. Bilberry extract and anthocyanins suppress unfolded protein response induced by exposure to blue LED light of cells in photoreceptor cell line. Mol. Vis. 2018, 24, 621–632. [Google Scholar]
- Liu, L.; Wu, X.W. Nobiletin protects human retinal pigment epithelial cells from hydrogen peroxide-induced oxidative damage. J. Biochem. Mol. Toxicol. 2018, 32, e22052. [Google Scholar] [CrossRef]
- Du, W.; An, Y.; He, X.; Zhang, D.; He, W. Protection of Kaempferol on Oxidative Stress-Induced Retinal Pigment Epithelial Cell Damage. Oxid. Med. Cell. Longev. 2018, 2018, 1610751. [Google Scholar] [CrossRef]
- Liu, S.Y.; Song, J.Y.; Fan, B.; Wang, Y.; Pan, Y.R.; Che, L.; Sun, Y.J.; Li, G.Y. Resveratrol protects photoreceptors by blocking caspase- and PARP-dependent cell death pathways. Free Radic. Biol. Med. 2018, 129, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Wang, C.; Song, D.; Song, Y.; Dunaief, J.L. Intraperitoneal injection of (-)-Epigallocatechin-3-gallate protects against light-induced photoreceptor degeneration in the mouse retina. Mol. Vis. 2017, 23, 171–178. [Google Scholar] [PubMed]
- Henneman, N.F.; Foster, S.L.; Chrenek, M.A.; Sellers, J.T.; Wright, C.B.; Schmidt, R.H.; Nickerson, J.M.; Boatright, J.H. Xanthohumol Protects Morphology and Function in a Mouse Model of Retinal Degeneration. Investig. Ophthalmol. Vis. Sci. 2018, 59, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, S.; Kurihara, T.; Ebinuma, M.; Kubota, M.; Yuki, K.; Sasaki, M.; Noda, K.; Ozawa, Y.; Oike, Y.; Ishida, S.; et al. Resveratrol prevents light-induced retinal degeneration via suppressing activator protein-1 activation. Am. J. Pathol. 2010, 177, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Bian, M.; Zhang, Y.; Du, X.; Xu, J.; Cui, J.; Gu, J.; Zhu, W.; Zhang, T.; Chen, Y. Apigenin-7-diglucuronide protects retinas against bright light-induced photoreceptor degeneration through the inhibition of retinal oxidative stress and inflammation. Brain Res. 2017, 1663, 141–150. [Google Scholar] [CrossRef]
- Kim, J.; Jin, H.L.; Jang, D.S.; Jeong, K.W.; Choung, S.Y. Quercetin-3-O-alpha-l-arabinopyranoside protects against retinal cell death via blue light-induced damage in human RPE cells and Balb-c mice. Food Funct. 2018, 9, 2171–2183. [Google Scholar] [CrossRef]
- Maccarone, R.; Di Marco, S.; Bisti, S. Saffron supplement maintains morphology and function after exposure to damaging light in mammalian retina. Invest. Ophthalmol. Vis. Sci. 2008, 49, 1254–1261. [Google Scholar] [CrossRef] [Green Version]
- Falsini, B.; Piccardi, M.; Minnella, A.; Savastano, C.; Capoluongo, E.; Fadda, A.; Balestrazzi, E.; Maccarone, R.; Bisti, S. Influence of saffron supplementation on retinal flicker sensitivity in early age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6118–6124. [Google Scholar] [CrossRef]
- Vasireddy, V.; Chavali, V.R.; Joseph, V.T.; Kadam, R.; Lin, J.H.; Jamison, J.A.; Kompella, U.B.; Reddy, G.B.; Ayyagari, R. Rescue of photoreceptor degeneration by curcumin in transgenic rats with P23H rhodopsin mutation. PLoS ONE 2011, 6, e21193. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Sanchez, L.; Lax, P.; Esquiva, G.; Martin-Nieto, J.; Pinilla, I.; Cuenca, N. Safranal, a saffron constituent, attenuates retinal degeneration in P23H rats. PLoS ONE 2012, 7, e43074. [Google Scholar] [CrossRef] [Green Version]
| Name | Structure | Binding Pocket | References |
|---|---|---|---|
| 11-cis-retinal | 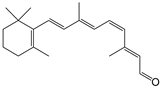 | Orthosteric Site | 10 |
| all-trans-retinal | 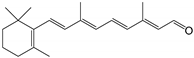 | Orthosteric Site | 10 |
| 9-cis-retinal | 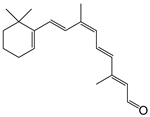 | Orthosteric Site | 14 |
| 11-cis-6-membered-ring retinal | 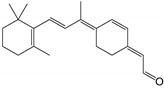 | Orthosteric Site | 13 |
| 5,8-epoxy-13-cis-retinoic acid |  | Orthosteric Site | 43 |
| YC-001 | 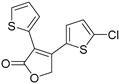 | Orthosteric Site | 16 |
| RS1 | 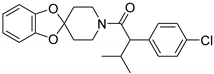 | Orthosteric Site | 47 |
| Quercetin | 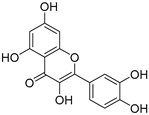 | Orthosteric Site Allosteric Site | 17, 24 |
| Myricetin | 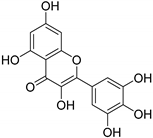 | Orthosteric Site Allosteric Site | 17 |
| Cyanidin | 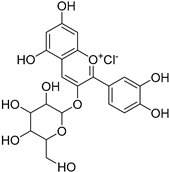 | Allosteric Site | 26, 27 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega, J.T.; Jastrzebska, B. The Retinoid and Non-Retinoid Ligands of the Rod Visual G Protein-Coupled Receptor. Int. J. Mol. Sci. 2019, 20, 6218. https://doi.org/10.3390/ijms20246218
Ortega JT, Jastrzebska B. The Retinoid and Non-Retinoid Ligands of the Rod Visual G Protein-Coupled Receptor. International Journal of Molecular Sciences. 2019; 20(24):6218. https://doi.org/10.3390/ijms20246218
Chicago/Turabian StyleOrtega, Joseph T., and Beata Jastrzebska. 2019. "The Retinoid and Non-Retinoid Ligands of the Rod Visual G Protein-Coupled Receptor" International Journal of Molecular Sciences 20, no. 24: 6218. https://doi.org/10.3390/ijms20246218
APA StyleOrtega, J. T., & Jastrzebska, B. (2019). The Retinoid and Non-Retinoid Ligands of the Rod Visual G Protein-Coupled Receptor. International Journal of Molecular Sciences, 20(24), 6218. https://doi.org/10.3390/ijms20246218