Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence
Abstract
1. Introduction
2. Results
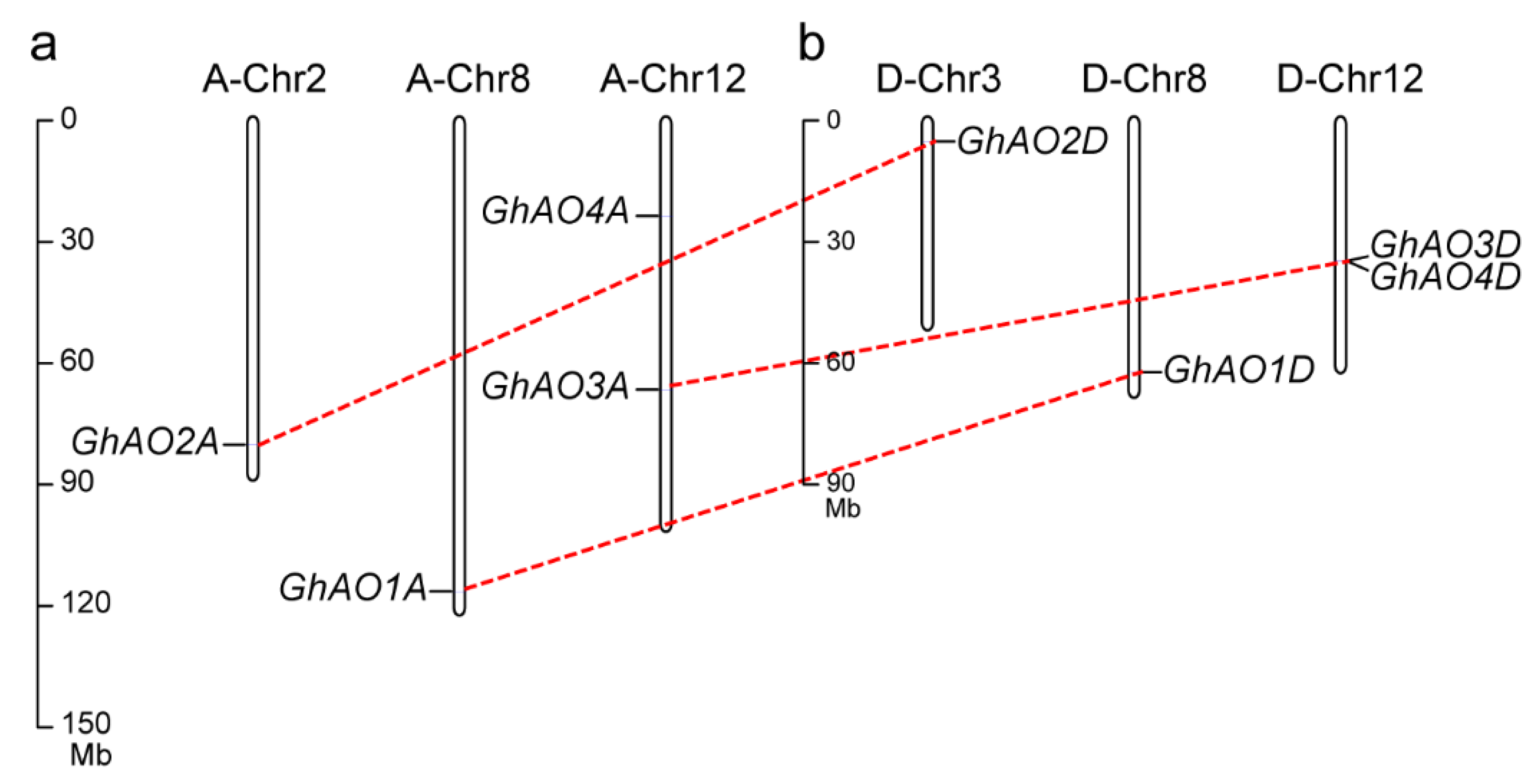
2.1. Identification and Characterization of AO Gene Family Members in G. hirsutum
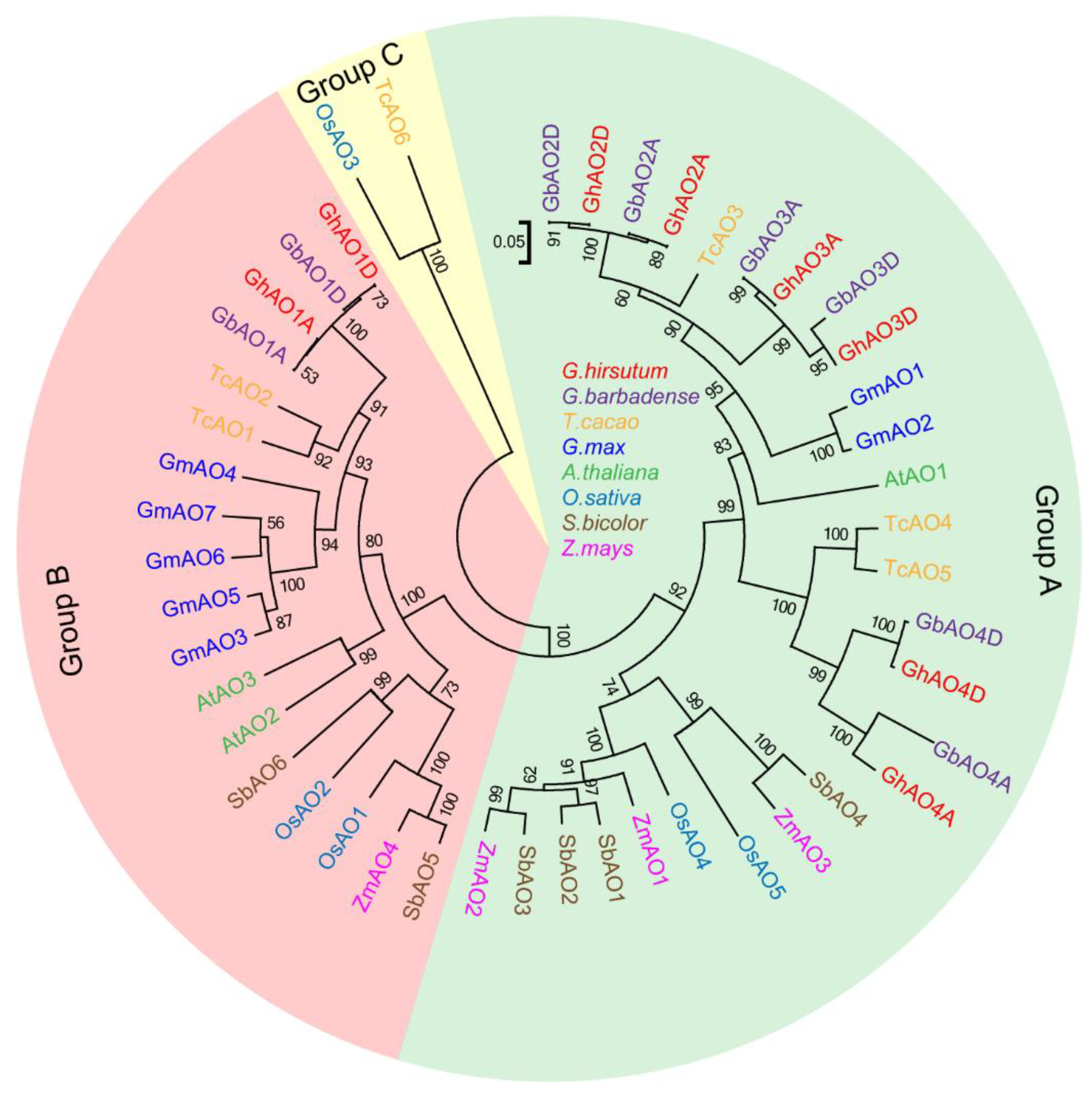
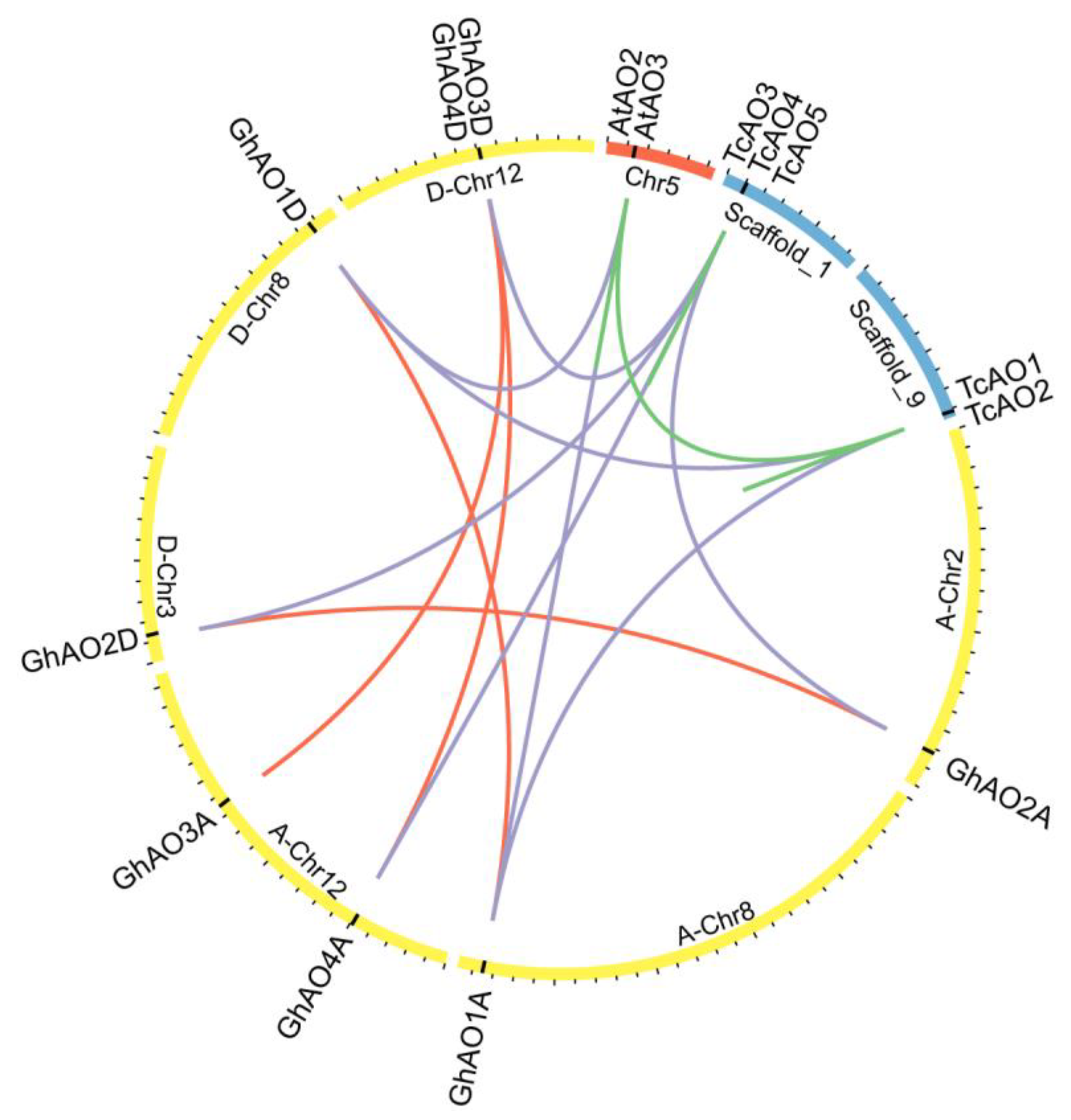
2.2. Phylogenetic and Evolutionary Analyses of GhAO Gene Family
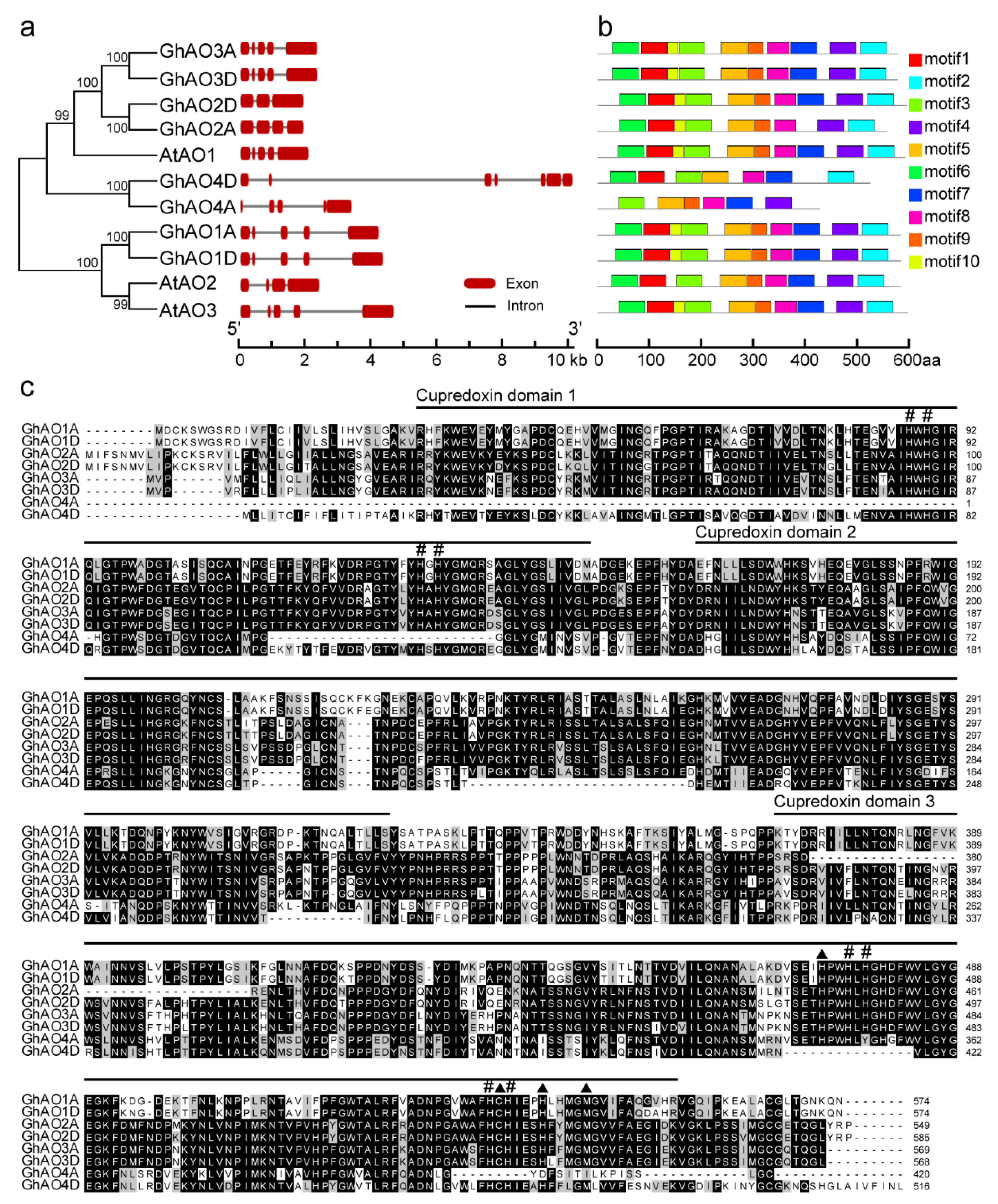
2.3. Gene Structure and Motif Analyses of GhAO Genes
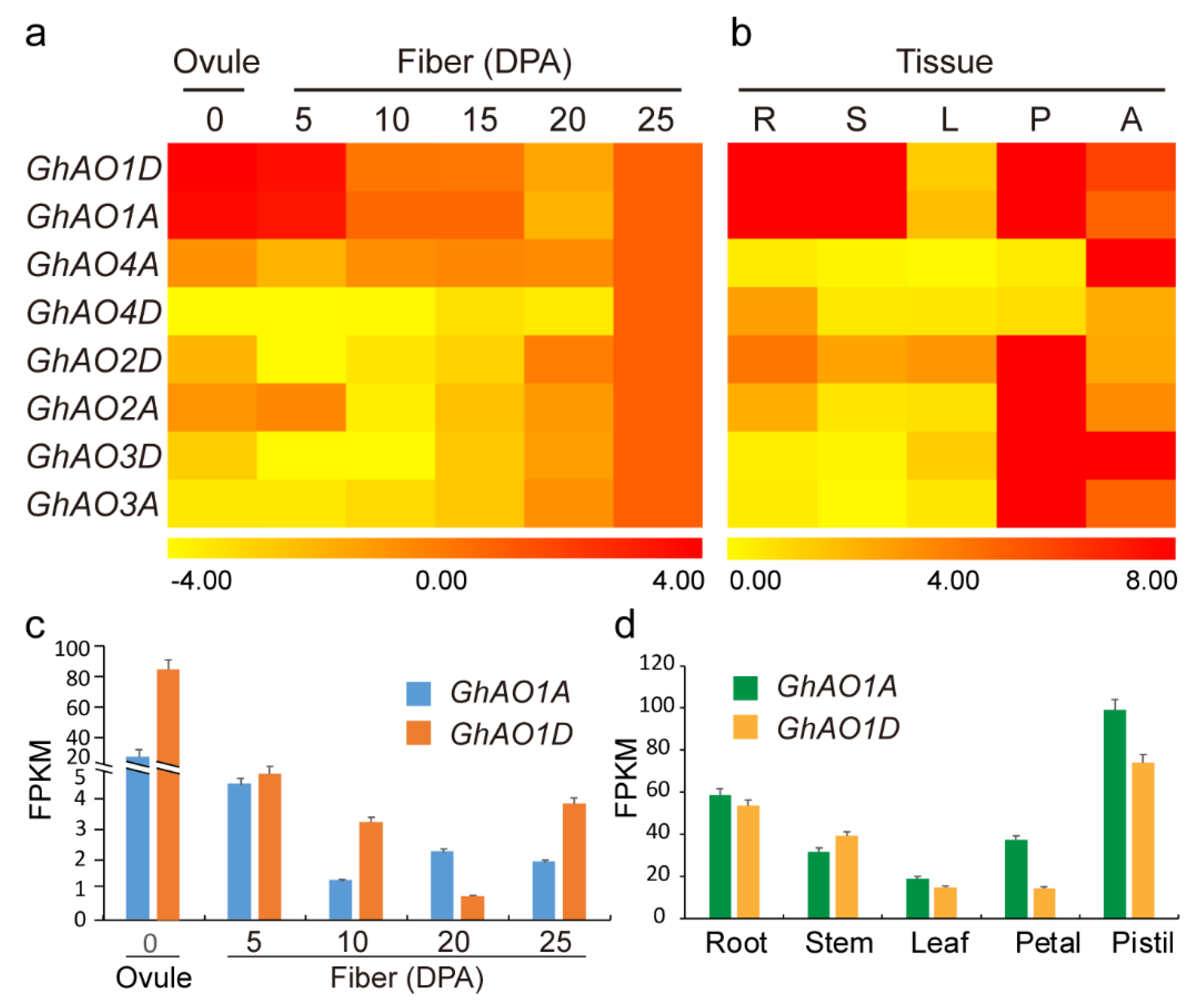
2.4. Expression Analysis of GhAO Genes in Fiber Development and Different Tissues
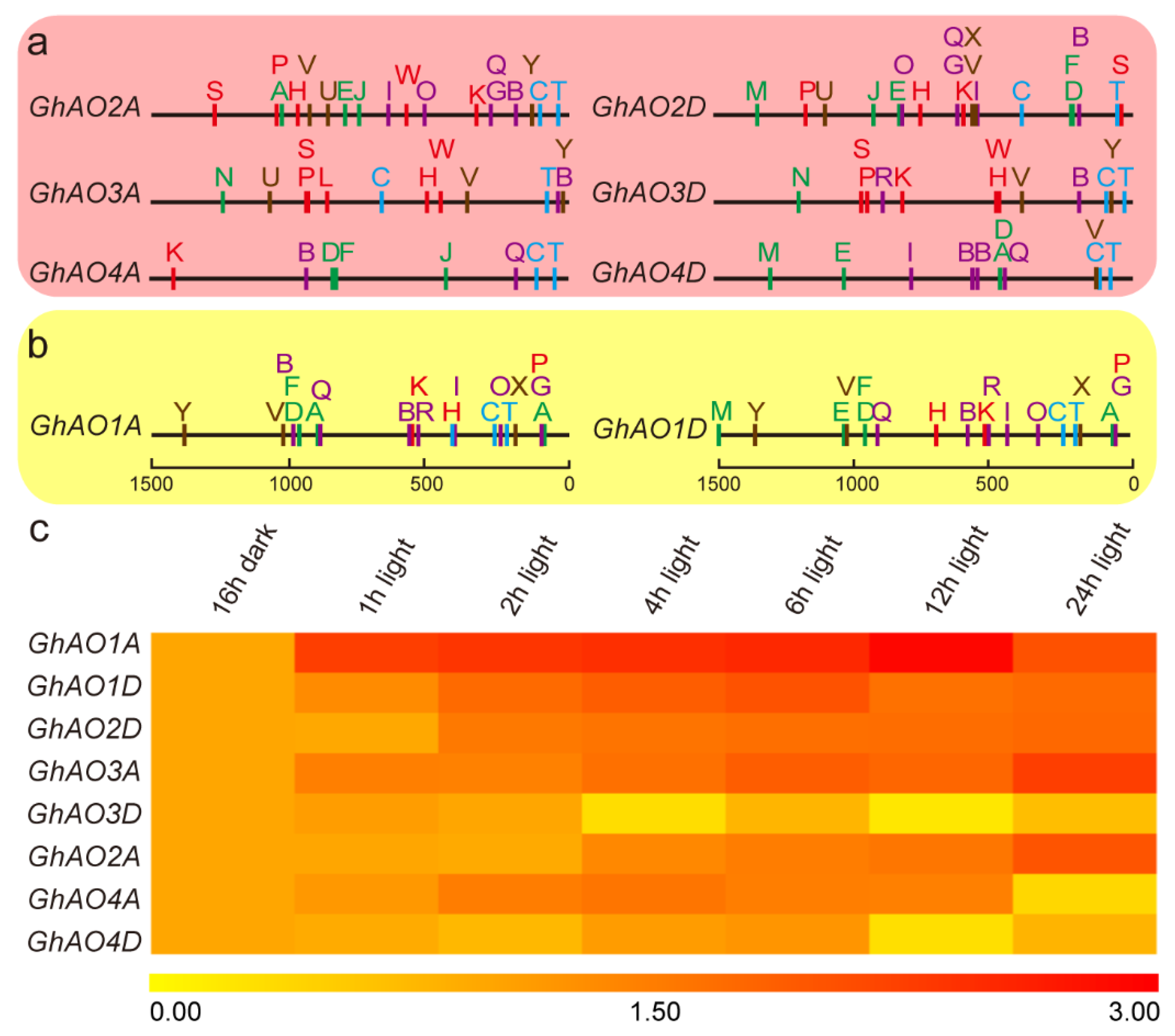
2.5. Cis-Regulatory Element Analysis of the GhAO Gene Promoter Region
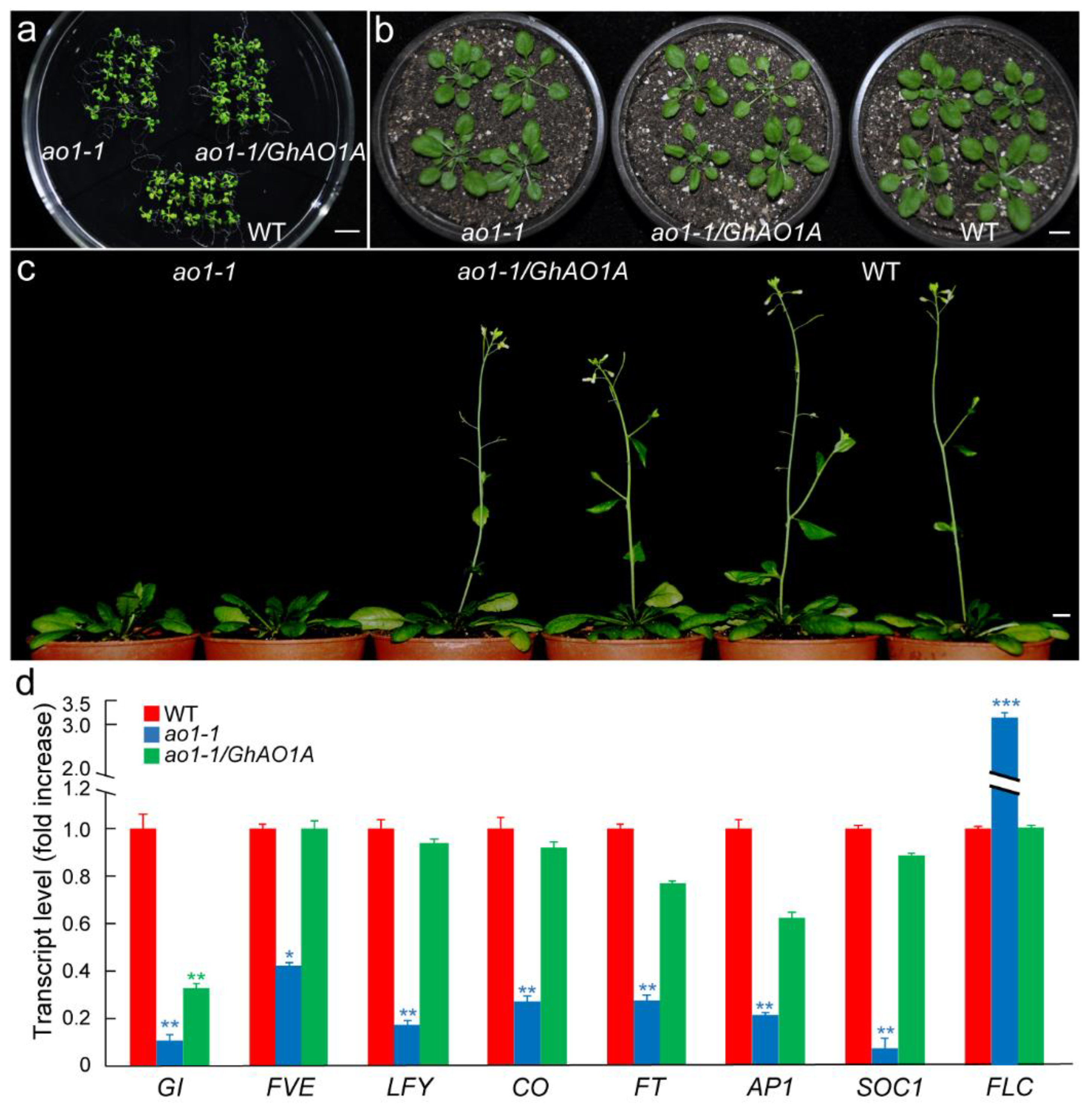
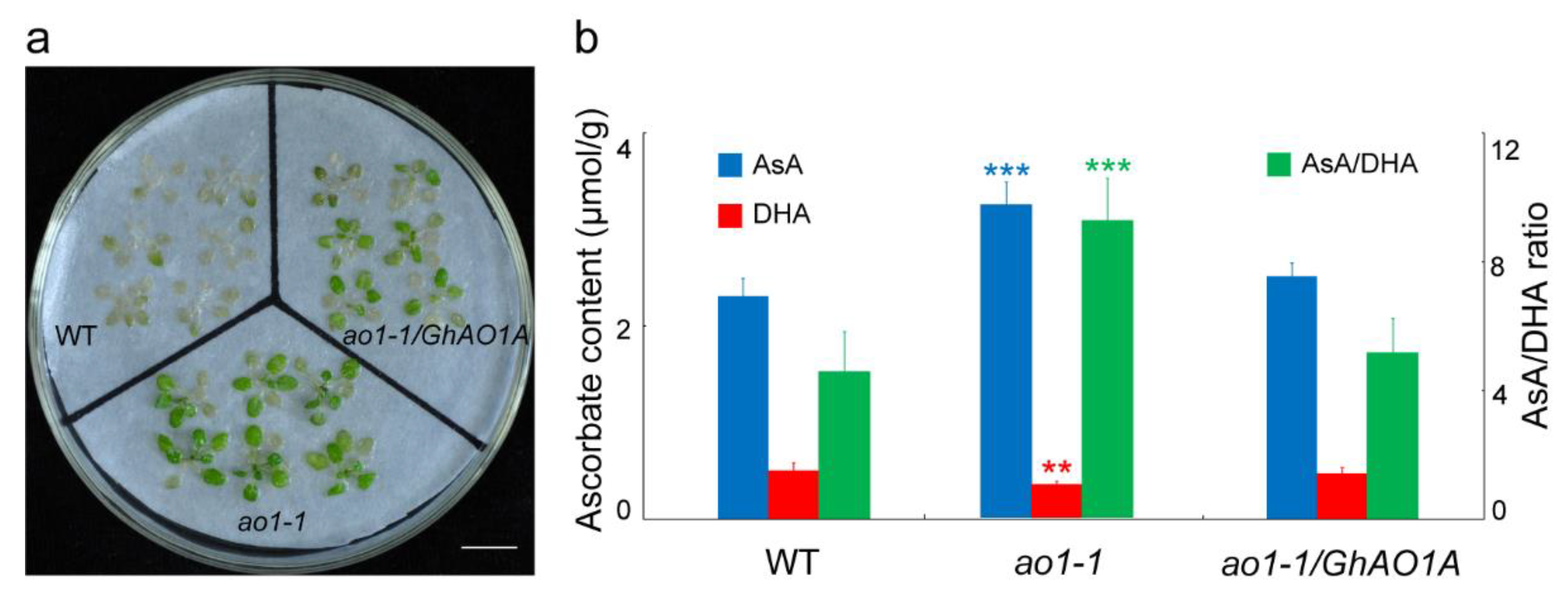
2.6. GhAO1A Rescues the Late-Flowering Phenotype of the Arabidopsis ao1-1 Mutant
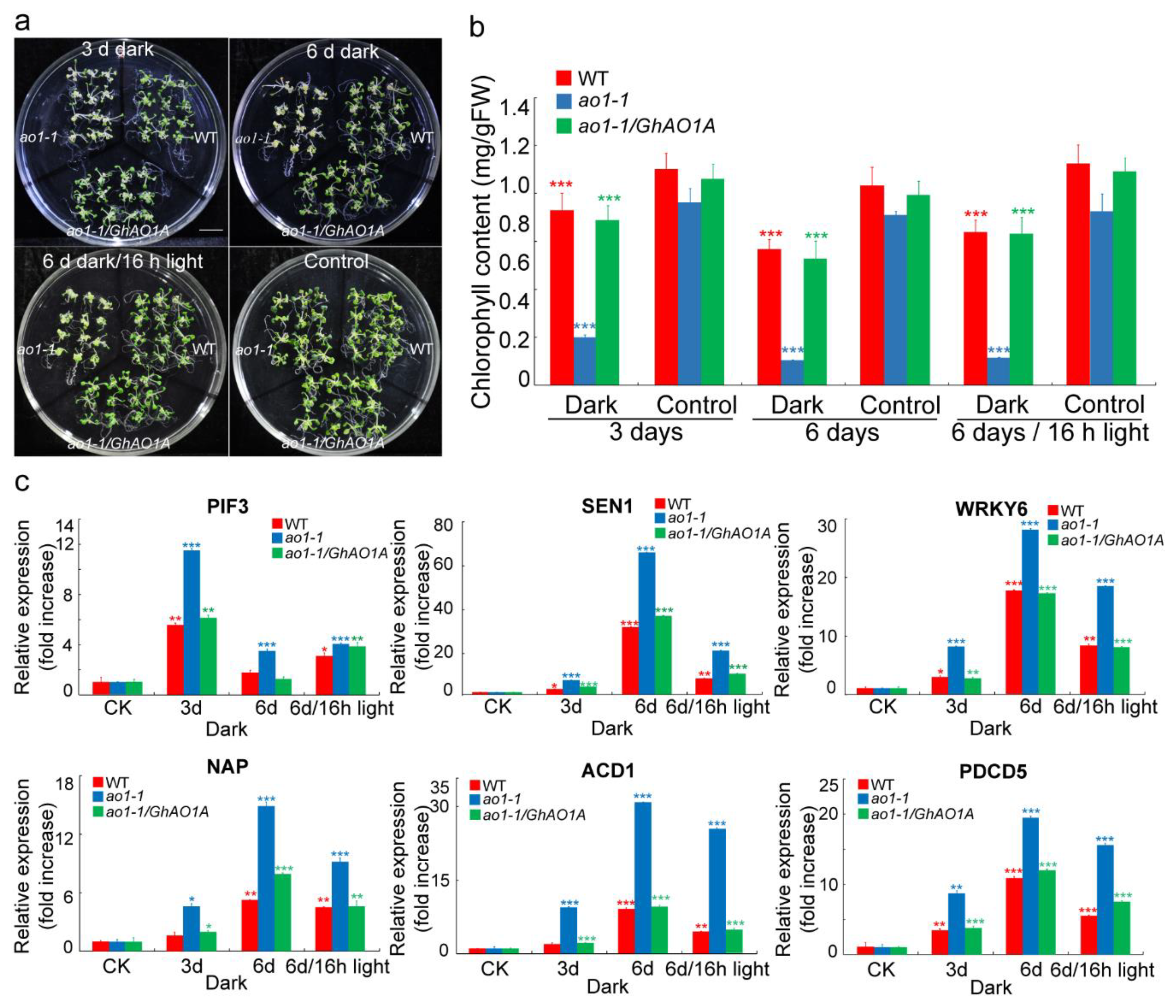
2.7. GhAO1A Delays Dark-Induced Leaf Senescence in Arabidopsis
2.8. GhAO1A Increases the Sensitivity of Arabidopsis to H2O2
3. Discussion
3.1. Identification and Characteristics of AO Gene Family in Gossypium hirsutum
3.2. Expression Patterns of AOs Imply Their Diverse Roles in Plant Growth and Development
3.3. Functional Role of AO Genes in Response to Light Regulation
3.4. Suppressed Expression of AO Increases Arabidopsis Tolerance to H2O2
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of AO Gene Family in G. hirsutum
4.3. Chromosomal Distribution and Duplication Analyses of the GhAO Genes
4.4. Analyses of Phylogenetic Relationship, Gene Structure, and Motif Distribution
4.5. RNA Extraction, Expression Detection, and Promoter Analysis of the GhAO Genes
4.6. Vector Construction and Arabidopsis Transformation
4.7. Functional Analysis of GhAO1A in Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| aa | Amino acid |
| ACD1 | Accelerated cell death 1 |
| AO | Ascorbate oxidase |
| AP1 | APETALA 1 |
| AP2-EREBP | APETALA 2/ethylene-responsive element binding protein |
| APX | Ascorbate peroxidase |
| AsA | Ascorbic acid |
| bp | Base pair |
| CDS | Coding sequence |
| CK | Control check |
| CO | CONSTANS |
| DHA | Dehydroascorbate |
| DPA | Days post anthesis |
| FLC | FLOWERING LOCUS C |
| FPKM | Fragments Per Kilobase of exon model per Million mapped reads |
| FT | FLOWERING LOCUS T |
| FVE | FLOWERING LOCUS VE |
| GI | GIGANTEA |
| GSDS | Gene Structure Display Server |
| Ka | Nonsynonymous substitution rate |
| Ks | Synonymous substitution rate |
| LFY | LEAFY |
| MCO | Multicopper oxidase |
| MDHA | Monodehydroascorbate |
| MEME | Multiple Em for Motif Elicitation |
| MeV | MultiExperiment Viewer |
| MW | Molecular weight |
| NAC2 | NAC DOMAIN CONTAINING PROTEIN 2 |
| NAP | NAC DOMAIN CONTAINING PROTEIN |
| NJ | Neighbor-joining |
| ORF | Open reading frame |
| PCD | Programmed cell death |
| PDCD5 | PROGRAMMED CELL DEATH PROTEIN 5 |
| PPH | PHENOPHYTIN PHEOPHORBIDE HYDROLASE |
| pI | Isoelectric point |
| PIF | PHYTOCHROME INTERACTING FACTOR |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| rRNA | Ribosomal RNA |
| redox | Reduction/oxidation |
| RT-PCR | Reverse transcription polymerase chain reaction |
| ROS | Reactive oxygen species |
| SAG | Senescence-associated gene |
| SEN | SENESCENCE |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| TF | Transcription factor |
| WRKY6 | WRKY DNA-BINDING PROTEIN 6 |
| WT | Wild-type |
References
- Hoegger, P.J.; Kilaru, S.; James, T.Y.; Thacker, J.R.; Kües, U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006, 273, 2308–2326. [Google Scholar] [CrossRef] [PubMed]
- De Tullio, M.C.; Guether, M.; Balestrini, R. Ascorbate oxidase is the potential conductor of a symphony of signaling pathways. Plant Signal Behav. 2013, 8, e23213. [Google Scholar] [CrossRef] [PubMed]
- Batth, R.; Singh, K.; Kumari, S.; Mustafiz, A. Transcript profiling reveals the presence of abiotic stress and developmental stage specific ascorbate oxidase genes in plants. Front. Plant Sci. 2017, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Messerschmidt, A.; Rossi, A.; Ladenstein, R.; Huber, R.; Bolognesi, M.; Gatti, G.; Marchesini, A.; Petruzzelli, R.; Finazzi-Agró, A. X-ray crystal structure of the blue oxidase ascorbate oxidase from zucchini: Analysis of the polypeptide fold and a model of the copper sites and ligands. J. Mol. Biol. 1989, 206, 515–527. [Google Scholar] [CrossRef]
- Takahama, U.; Oniki, T. The association of ascorbate and ascorbate oxidase in the appoplast with IAA-enhanced elongation of epicotyls from Vigna angularis. Plant Cell Physiol. 1994, 35, 257–266. [Google Scholar]
- Smirnoff, N. Ascorbic acid: Metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 2000, 3, 229–235. [Google Scholar] [CrossRef]
- Li, R.; Xin, S.; Tao, C.C.; Jin, X.; Li, H.B. Cotton ascorbate oxidase promotes cell growth in cultured tobacco bright yellow-2 cells through generation of apoplast oxidation. Int. J. Mol. Sci. 2017, 18, 1346. [Google Scholar] [CrossRef]
- Farver, O.; Wherland, S.; Pecht, I. Intramolecular electron transfer in ascorbate oxidase is enhanced in the presence of oxygen. J. Biol. Chem. 1994, 269, 22933–22936. [Google Scholar]
- Esaka, M.; Fujisawa, K.; Goto, M.; Kisu, Y. Regulation of ascorbate oxidase expression in pumpkin by auxin and copper. Plant Physiol. 1992, 100, 231–237. [Google Scholar] [CrossRef]
- Pignocchi, C.; Fletcher, J.M.; Wilkinson, J.E.; Barnes, J.D.; Foyer, C.H. The function of ascorbate oxidase in Tobacco. Plant Physiol. 2003, 132, 1631–1641. [Google Scholar] [CrossRef]
- Sanmartin, M.; Drogoudi, P.A.; Lyons, T.; Pateraki, I.; Barnes, J.; Kanellis, A.K. Over-expression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta 2003, 216, 918–928. [Google Scholar] [PubMed]
- Yamamoto, A.; Bhuiyan, M.N.; Waditee, R.; Tanaka, Y.; Esaka, M.; Oba, K.; Jagendorf, A.T.; Takabe, T. Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J. Exp. Bot. 2005, 56, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Sanmartin, M.; Pateraki, I.; Chatzopoulou, F.; Kanellis, A.K. Differential expression of the ascorbate oxidase multigene family during fruit development and in response to stress. Planta 2007, 225, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Esaka, M. cDNA cloning and gene expression of ascorbate oxidase in tobacco. Plant Mol. Biol. 1996, 30, 833–837. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Li, H.X.; Shu, W.B.; Zhang, C.J.; Zhang, W.; Ye, Z.B. Suppressed expression of ascorbate oxidase gene promotes ascorbic acid accumulation in tomato fruit. Plant Mol. Biol. Rep. 2011, 29, 638–645. [Google Scholar] [CrossRef]
- Garchery, C.; Gest, N.; Do, P.T.; Alhagdow, M.; Baldet, P.; Menard, G.; Rothan, C.; Massot, C.; Gautier, H.; Aarrouf, J.; et al. A diminution in ascorbate oxidase activity affects carbon allocation and improves yield in tomato under water deficit. Plant Cell Environ. 2013, 36, 159–175. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Francis, D.; De Gara, L. The redox state of the ascorbate-dehydroascorbate pair as a specifc sensor of cell division in tobacco BY-2 cells. Protoplasma 1999, 209, 90–97. [Google Scholar] [CrossRef]
- Potters, G.; Horemans, N.; Caubergs, R.J.; Asard, H. Ascorbate and dehydroascorbate influence cell cycle progression in a tobacco cell suspension. Plant Physiol. 2000, 124, 17–20. [Google Scholar] [CrossRef]
- Potters, G.; Horemans, N.; Bellone, S.; Caubergs, R.J.; Trost, P.; Guisez, Y.; Asard, H. Dehydroascorbate influences the plant cell cycle through a glutathione independent reduction mechanism. Plant Physiol. 2004, 134, 1479–1487. [Google Scholar] [CrossRef]
- Balestrini, R.; Ott, T.; Güther, M.; Bonfante, P.; Udvardi, M.K.; De Tullio, M.C. Ascorbate oxidase: The unexpected involvement of a ‘wasteful enzyme’ in the symbioses with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi. Plant Physiol. Biochem. 2012, 59, 71–79. [Google Scholar] [CrossRef]
- Shalata, A.; Neumann, P.M. Exogenous ascorbic acid (vitamin C) increases resistance to salt stress and reduces lipid peroxidation. J. Exp. Bot. 2001, 52, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Fotopoulos, V.; Kanellis, A.K. Altered apoplastic ascorbate redox state in tobacco plants via ascorbate oxidase overexpression results in delayed dark-induced senescence in detached leaves. Plant Physiol. Biochem. 2013, 73, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Li, H.B.; Qin, Y.M.; Pang, Y.; Song, W.Q.; Mei, W.Q.; Zhu, Y.X. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 2007, 175, 462–471. [Google Scholar] [CrossRef]
- Raven, E.L. Peroxidase-catalyzed oxidation of ascorbate. Structural, spectroscopic and mechanistic correlations in ascorbate peroxidase. Subcell. Biochem. 2000, 35, 317–349. [Google Scholar]
- Mosery, O.; Kanellis, A.K. Ascorbate oxidase of Cucumis melo L. var. reticulatus: Purification, characterization and antibody production. J. Exp. Bot. 1994, 45, 717–724. [Google Scholar] [CrossRef]
- Ohkawa, J.; Ohya, T.; Ito, T.; NoJzawa, H.; Nishi, Y.; Okada, N.; Yoshida, K.; Takano, M.; Shinmyo, A. Structure of the genomic DNA encoding cucumber ascorbate oxidase and its expression in transgenic plants. Plant Cell Rep. 1994, 13, 481–488. [Google Scholar] [CrossRef]
- Kato, N.; Esaka, M. Changes in ascorbate oxidase gene expression and ascorbate levels in cell division and cell elongation in tobacco cells. Plant Physiol. 2010, 105, 321–329. [Google Scholar] [CrossRef]
- Kato, N.; Esaka, M. Expansion of transgenic tobacco protoplasts expressing pumpkin ascorbate oxidase is more rapid than that of wild-type protoplasts. Planta 2000, 210, 1018–1022. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. FVE, an Arabidopsis homologue of the retinoblastoma-associated protein that regulates flowering time and cold response, binds to chromatin as a large multiprotein complex. Mol. Cells 2011, 32, 227–234. [Google Scholar] [CrossRef]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef]
- Song, Y.; Yang, C.W.; Gao, S.; Zhang, W.; Li, L.; Kuai, B.K. Age-triggered and dark-induced leaf Senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef]
- Niu, Y.H.; Guo, F.Q. Nitric oxide regulates dark-induced leaf senescence through EIN2 in Arabidopsis. J. Integr. Plant Biol. 2012, 54, 516–525. [Google Scholar] [CrossRef]
- Guo, Y. Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 2013, 82, 519–528. [Google Scholar] [CrossRef]
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999, 11, 949–956. [Google Scholar] [CrossRef]
- De Tullio, M.C.; Ciraci, S.; Liso, S.; Arrigoni, O. Ascorbic acid oxidase is dynamically regulated by light and oxygen. A tool for oxygen management in plants. J. Plant Physiol. 2007, 164, 39–46. [Google Scholar] [CrossRef]
- Teng, K.; Chang, Z.; Li, X.; Sun, X.; Liang, X.; Xu, L.; Chao, Y.; Han, L. Functional and RNA-Sequencing Analysis Revealed Expression of a Novel Stay-Green Gene from Zoysia japonica (ZjSGR) Caused Chlorophyll Degradation and Accelerated Senescence in Arabidopsis. Front. Plant Sci. 2016, 7, 1894. [Google Scholar] [CrossRef]
- Xin, S.; Tao, C.C.; Li, H.B. Cloning and Functional Analysis of the Promoter of an Ascorbate Oxidase Gene from Gossypium hirsutum. PLoS ONE 2016, 11, e0161695. [Google Scholar] [CrossRef]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef]
- Kim, J.I.; Murphy, A.S.; Baek, D.; Lee, S.W.; Yun, D.J.; Bressan, R.A.; Narasimhan, M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3981–3992. [Google Scholar] [CrossRef]
- Mueller-Roeber, B.; Balazadeh, S. Auxin and its role in plant senescence. J. Plant Growth Regul. 2014, 33, 21–33. [Google Scholar] [CrossRef]
- Schippers, J.H.; Schmidt, R.; Wagstaff, C.; Jing, H. Living to Die and Dying to Live: The Survival Strategy behind Leaf Senescence. Plant Physiol. 2015, 169, 914–930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ju, H.W.; Chung, M.S.; Huang, P.; Ahn, S.J.; Kim, C.S. The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 2011, 52, 138–148. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Chen, Y.; He, J.X.; Bi, Y. Phytochrome-interacting factor 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 2015, 237, 57–68. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Casati, P. AtPDCD5 plays a role during dark-senescence in Arabidopsis. Plant Signal. Behav. 2016, 11, e1176820. [Google Scholar] [CrossRef]
- Li, L.H.; Xing, Y.F.; Chang, D.; Fang, S.S.; Cui, B.Y.; Li, Q.; Wang, X.J.; Guo, S.; Yang, X.; Men, S.Z.; et al. CaM/BAG5/Hsc70 signaling complex dynamically regulates leaf senescence. Sci. Rep. 2016, 6, 31889. [Google Scholar] [CrossRef]
- Balazadeh, S.; Riaño-Pachón, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 1, 63–75. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 2014, 5, 650. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Khanna-Chopra, R. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 2012, 249, 469–481. [Google Scholar] [CrossRef]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.; Dijkwel, P.P. Circadian clock-associated 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134. [Google Scholar] [CrossRef]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Jin, X.; Zhu, L.P.; Yao, Q.; Meng, X.R.; Ding, G.H.; Wang, D.; Xie, Q.L.; Tong, Z.; Tao, C.C.; Yu, L.; et al. Expression profiling of mitogen-activated protein kinase genes reveals their evolutionary and functional diversity in different rubber tree (Hevea brasiliensis) cultivars. Genes 2017, 8, 261. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Ma, W.; Zhao, T.; Li, J.; Liu, B.L.; Fang, L.; Hu, Y.; Zhang, T.Z. Identification and characterization of the GhHsp20 gene family in Gossypium hirsutum. Sci. Rep. 2016, 6, 32517. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutional genetics analysis using maximum likelihood, evolutional distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef]
- Jin, X.; Wang, L.M.; He, L.P.; Feng, W.Q.; Wang, X.C. Two-dimensional gel electrophoresis-based analysis provides global insights into the cotton ovule and fiber proteomes. Sci. China Life Sci. 2016, 59, 154–163. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Springer: Boston, MA, USA, 2010; pp. 267–277. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Li, L.; Kubiszewski-Jakubiak, S.; Radomiljac, J.; Wang, Y.; Law, S.R.; Keech, O.; Narsai, R.; Berkowitz, O.; Duncan, O.; Murcha, M.W.; et al. Characterization of a novel β-barrel protein (AtOM47) from the mitochondrial outer membrane of Arabidopsis thaliana. J. Exp. Bot. 2016, 21, 6061–6075. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID | Chromosomal Location | ORF (bp) | Protein a | |||
|---|---|---|---|---|---|---|---|
| Size (aa) | MW (kD) | pI | Subcellular Localization b | ||||
| GhAO1A | Gh_A08G1967 | A-Chr8 | 1725 | 574 | 64.06 | 8.63 | E |
| GhAO1D | Gh_D08G2139 | D-Chr8 | 1725 | 574 | 64.12 | 7.99 | E |
| GhAO2A | Gh_A02G1327 | A-Chr2 | 1650 | 549 | 61.87 | 7.05 | E |
| GhAO2D | Gh_D03G0441 | D-Chr3 | 1758 | 585 | 65.64 | 7.33 | E |
| GhAO3A | Gh_A12G0933 | A-Chr12 | 1710 | 568 | 64.18 | 7.08 | E |
| GhAO3D | Gh_D12G0952 | D-Chr12 | 1707 | 569 | 64.12 | 8.07 | E |
| GhAO4A | Gh_A12G0719 | A-Chr12 | 1263 | 420 | 46.61 | 5.65 | E |
| GhAO4D | Gh_D12G0953 | D-Chr12 | 1551 | 516 | 58.07 | 5.95 | E |
| Testing Group | Mt a | M1 b | M2 c | X2 | Pd |
|---|---|---|---|---|---|
| GhAO1A/GhAO1D with TcAO1 | 438 | 1 | 2 | 0.33 | 0.5637 |
| GhAO2A/GhAO2D with TcAO3 | 467 | 4 | 7 | 0.82 | 0.3657 |
| GhAO3A/GhAO3D with TcAO3 | 473 | 3 | 7 | 1.6 | 0.2059 |
| GhAO4A/GhAO4D with TcAO5 | 268 | 29 | 13 | 6.1 | 0.0136 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Z.; Chen, L.; Wang, F.; Song, W.; Cao, A.; Xie, S.; Chen, X.; Jin, X.; Li, H. Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence. Int. J. Mol. Sci. 2019, 20, 6167. https://doi.org/10.3390/ijms20246167
Pan Z, Chen L, Wang F, Song W, Cao A, Xie S, Chen X, Jin X, Li H. Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence. International Journal of Molecular Sciences. 2019; 20(24):6167. https://doi.org/10.3390/ijms20246167
Chicago/Turabian StylePan, Ze, Lihua Chen, Fei Wang, Wangyang Song, Aiping Cao, Shuangquan Xie, Xifeng Chen, Xiang Jin, and Hongbin Li. 2019. "Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence" International Journal of Molecular Sciences 20, no. 24: 6167. https://doi.org/10.3390/ijms20246167
APA StylePan, Z., Chen, L., Wang, F., Song, W., Cao, A., Xie, S., Chen, X., Jin, X., & Li, H. (2019). Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence. International Journal of Molecular Sciences, 20(24), 6167. https://doi.org/10.3390/ijms20246167





