Chinese Giant Salamander (Andrias davidianus) Iridovirus Infection Leads to Apoptotic Cell Death through Mitochondrial Damage, Caspases Activation, and Expression of Apoptotic-Related Genes
Abstract
1. Introduction
2. Results
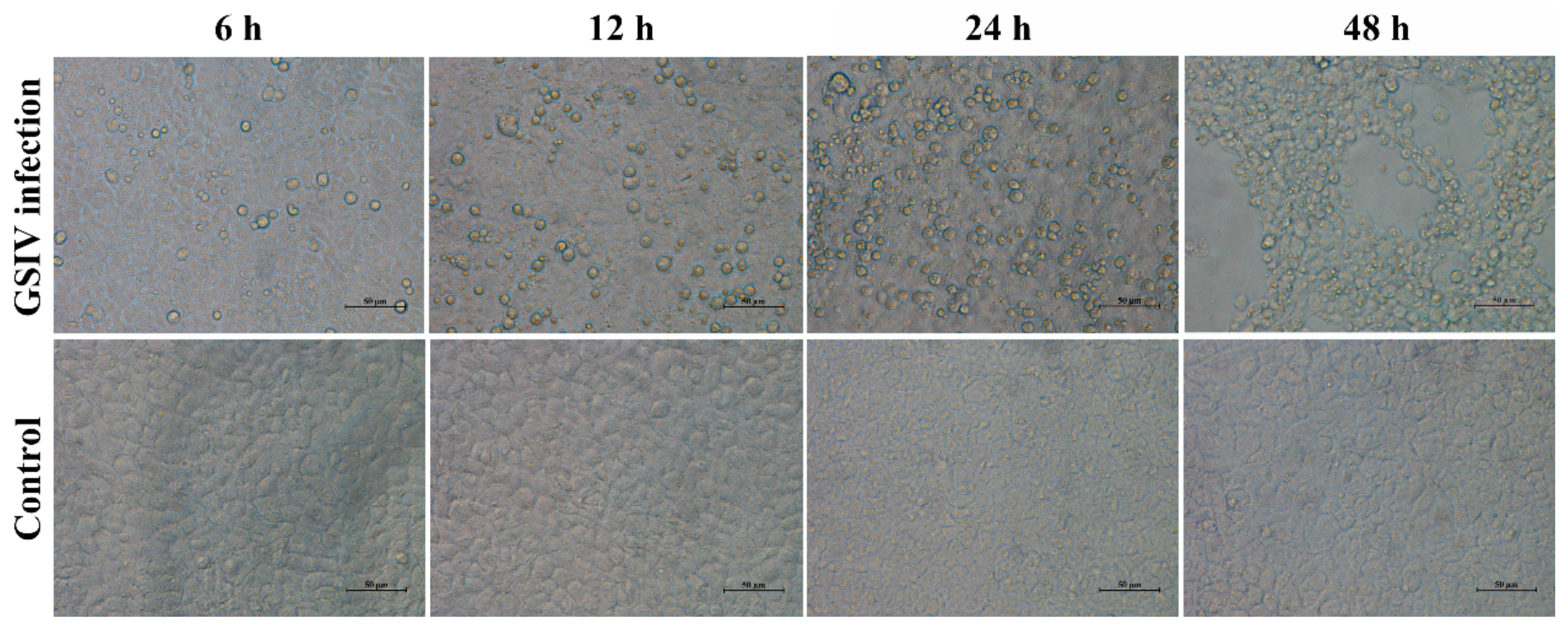
2.1. Cytopathic Effect
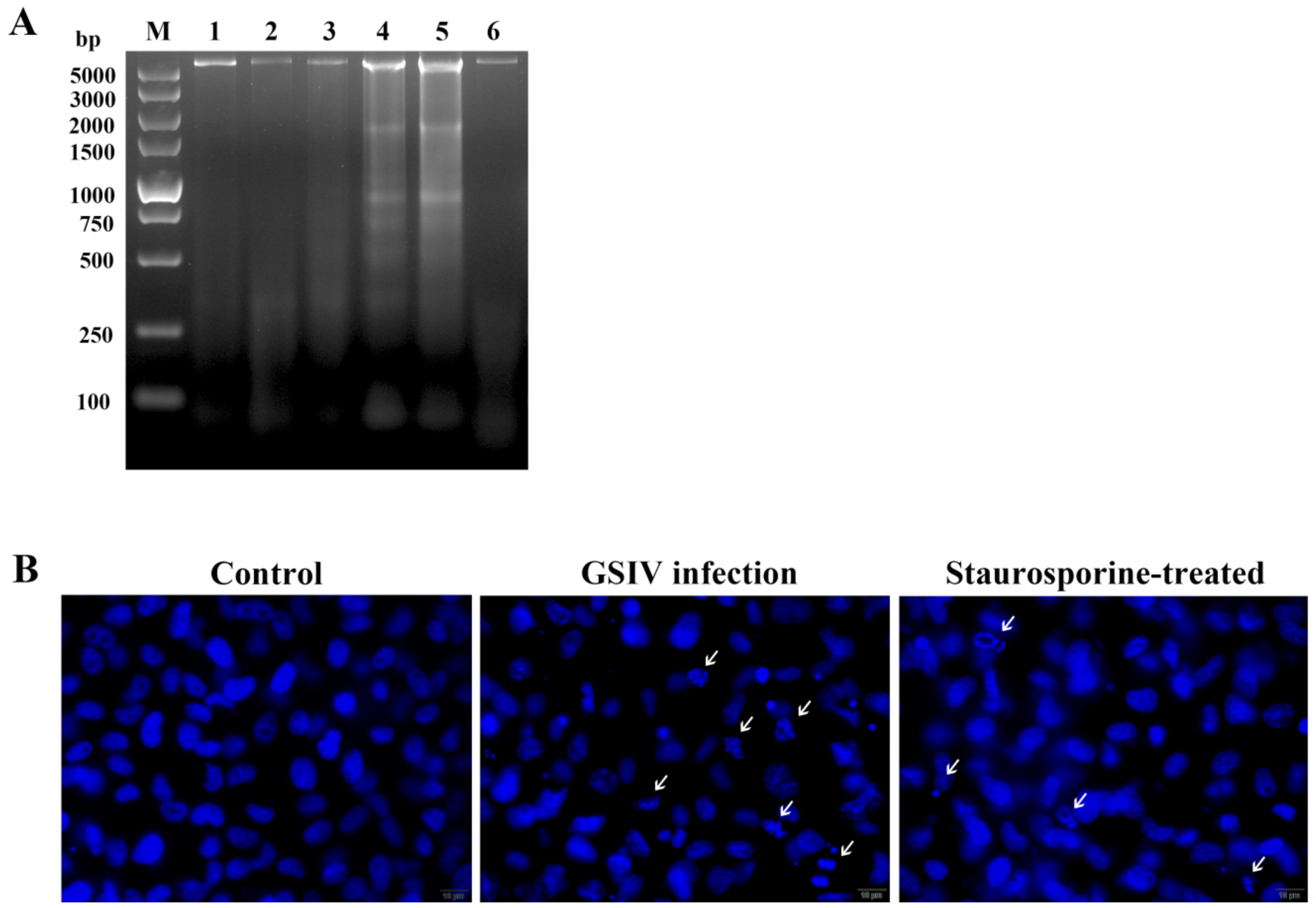
2.2. DNA Fragmentation
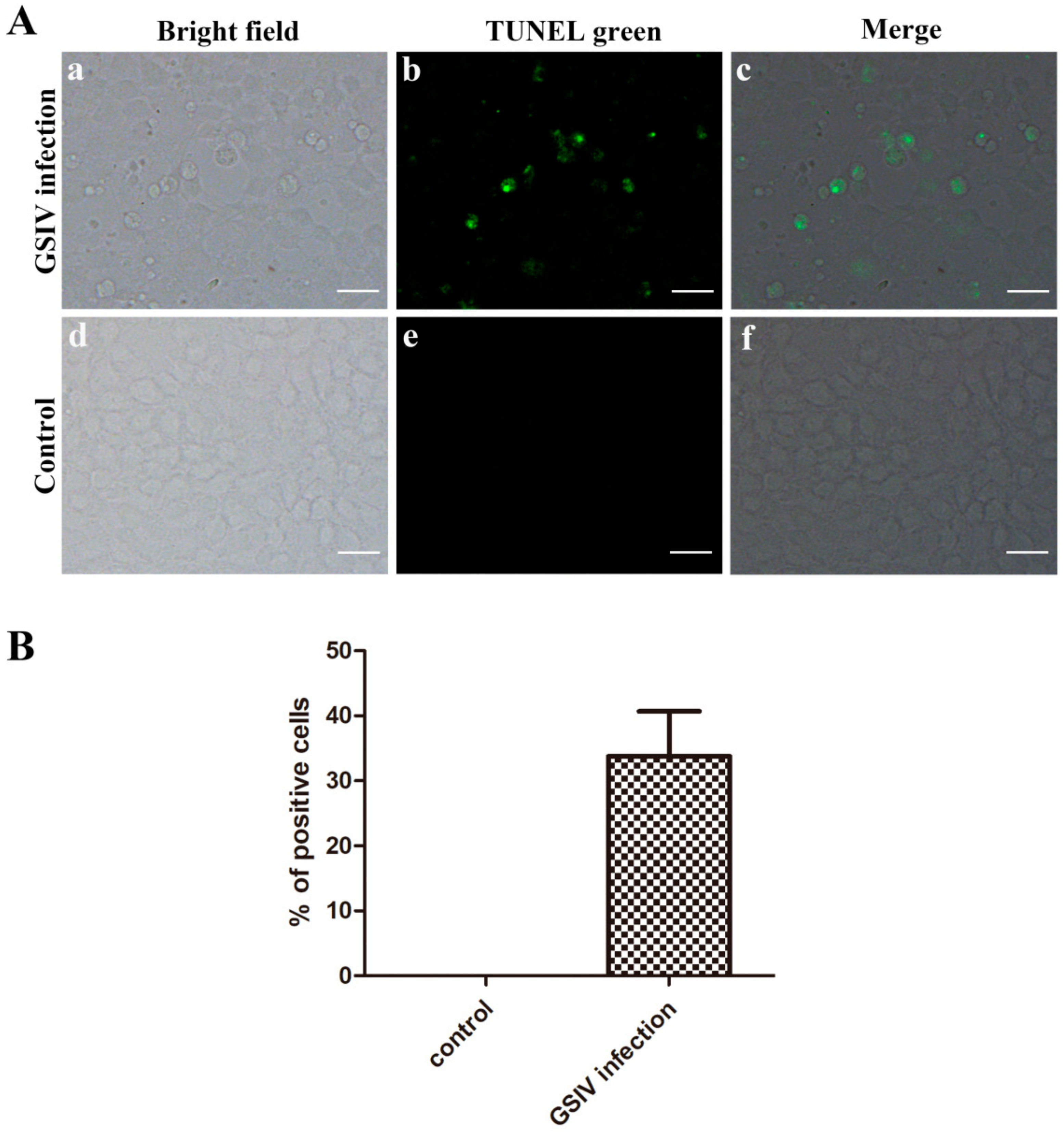
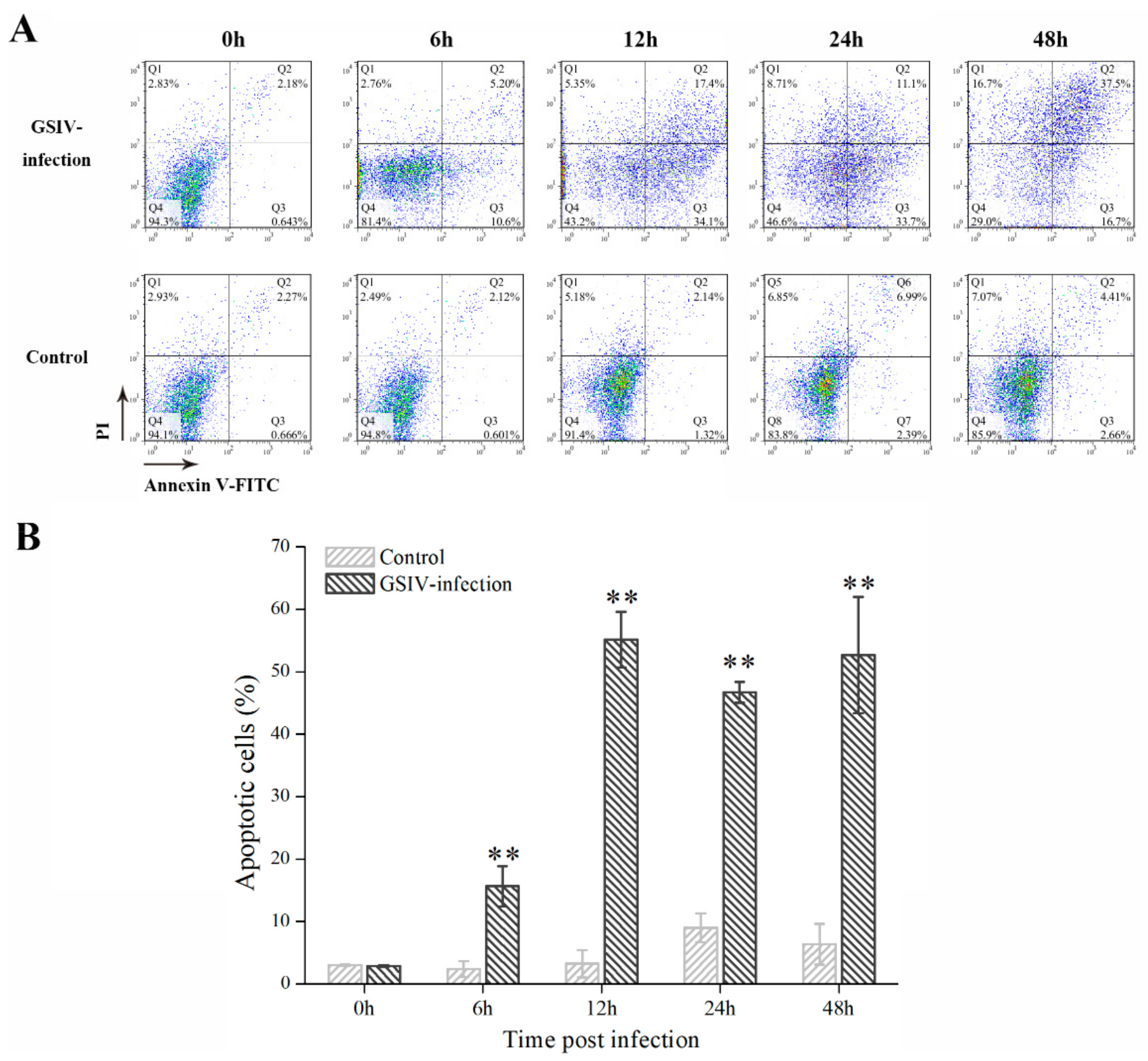
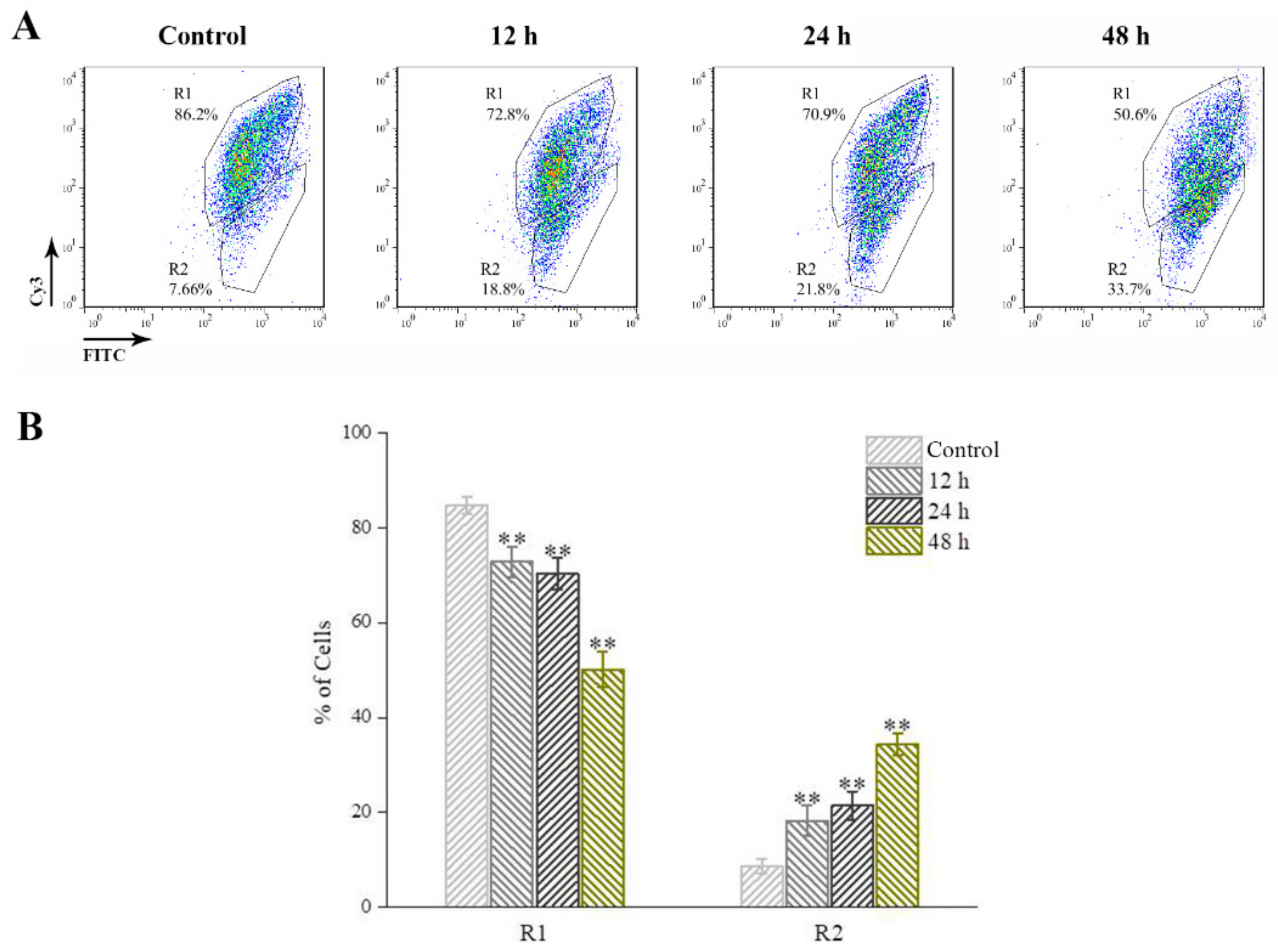
2.3. Typical Apoptotic Cell Death in GSM Cells During GSIV Infection
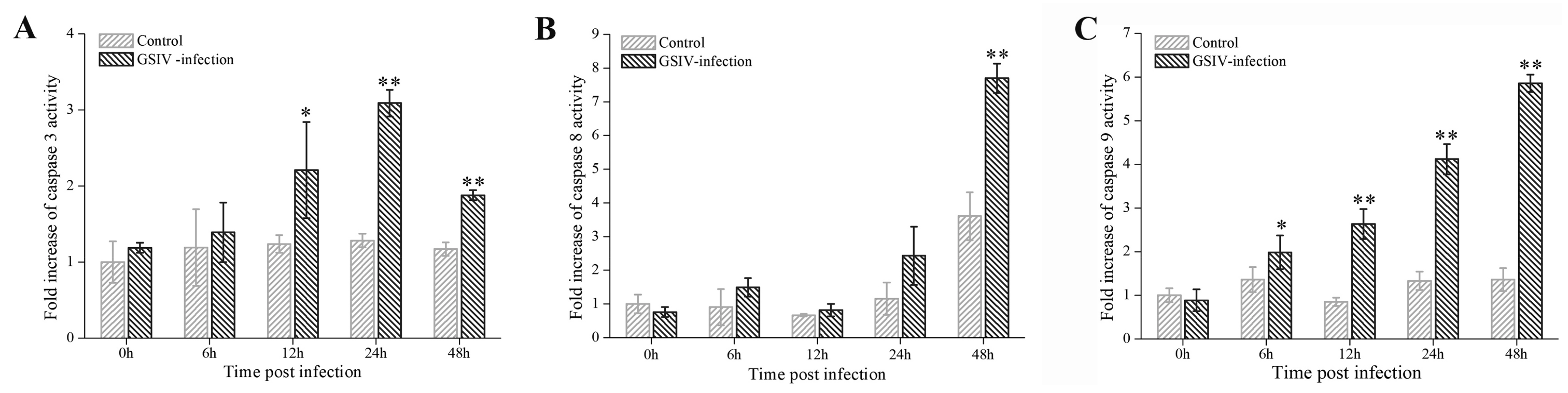
2.4. Caspases Activation
2.5. Mitochondrial Membrane Potential (MMP)
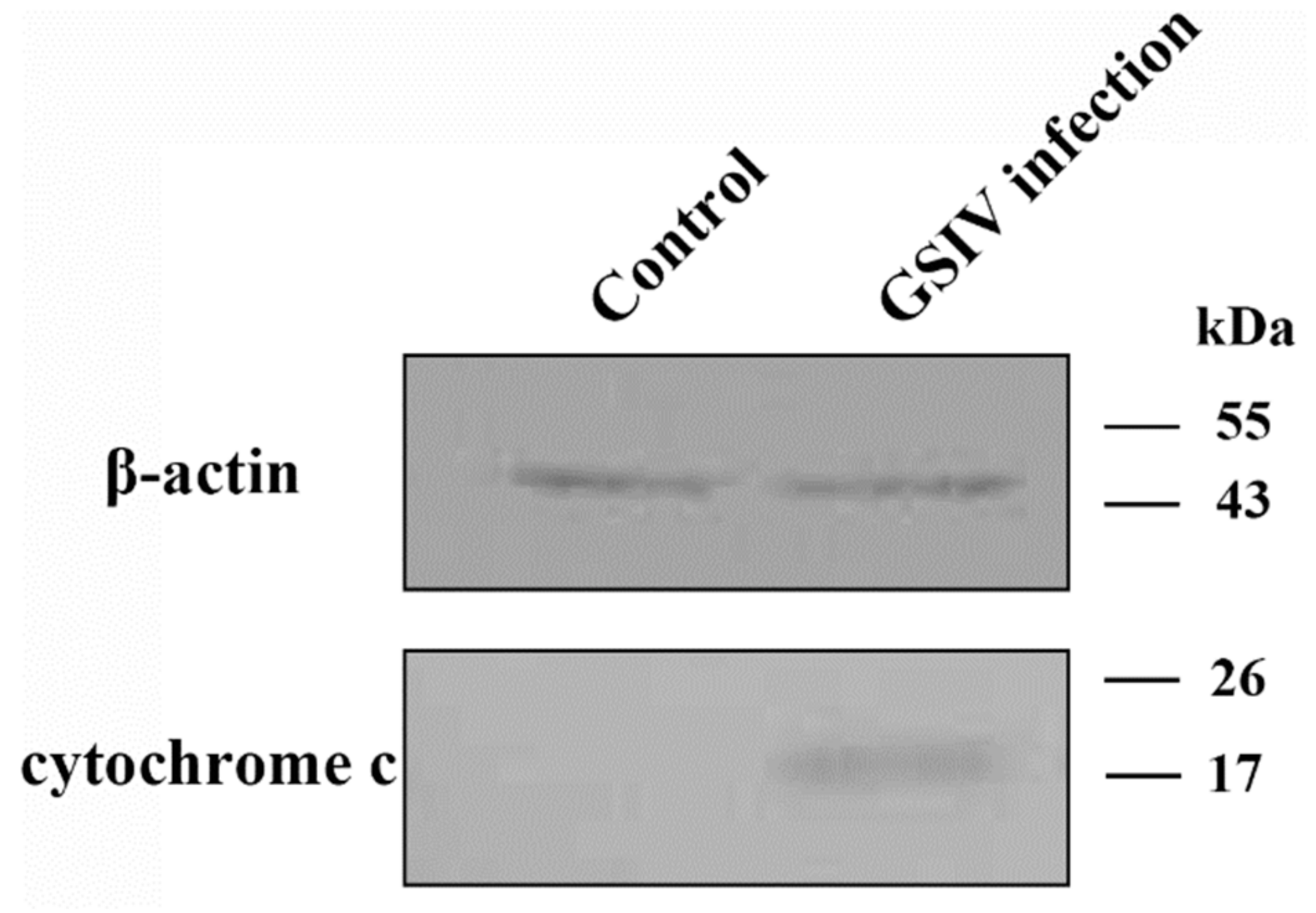
2.6. Cytochrome c Release
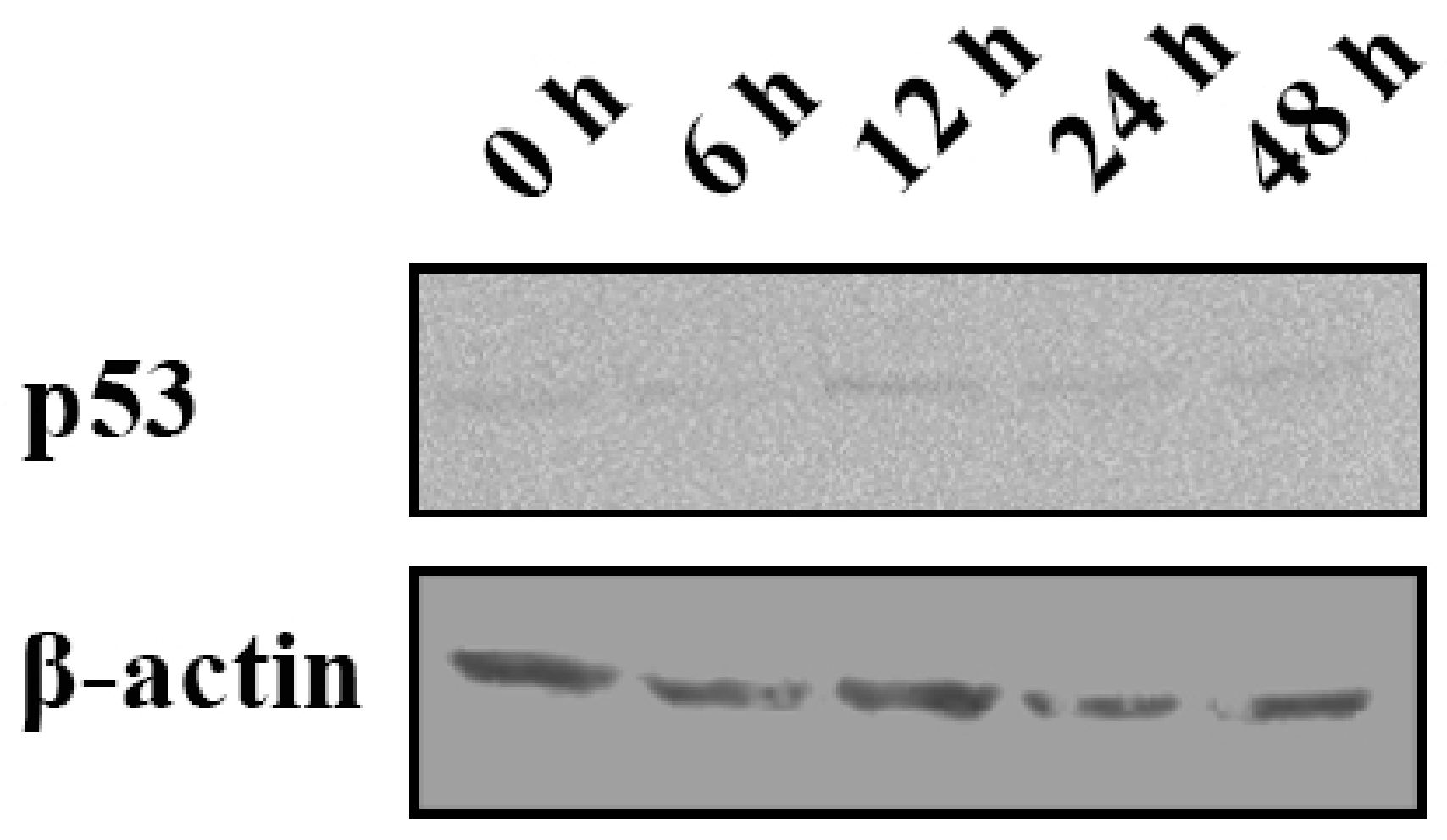
2.7. p53 Expression
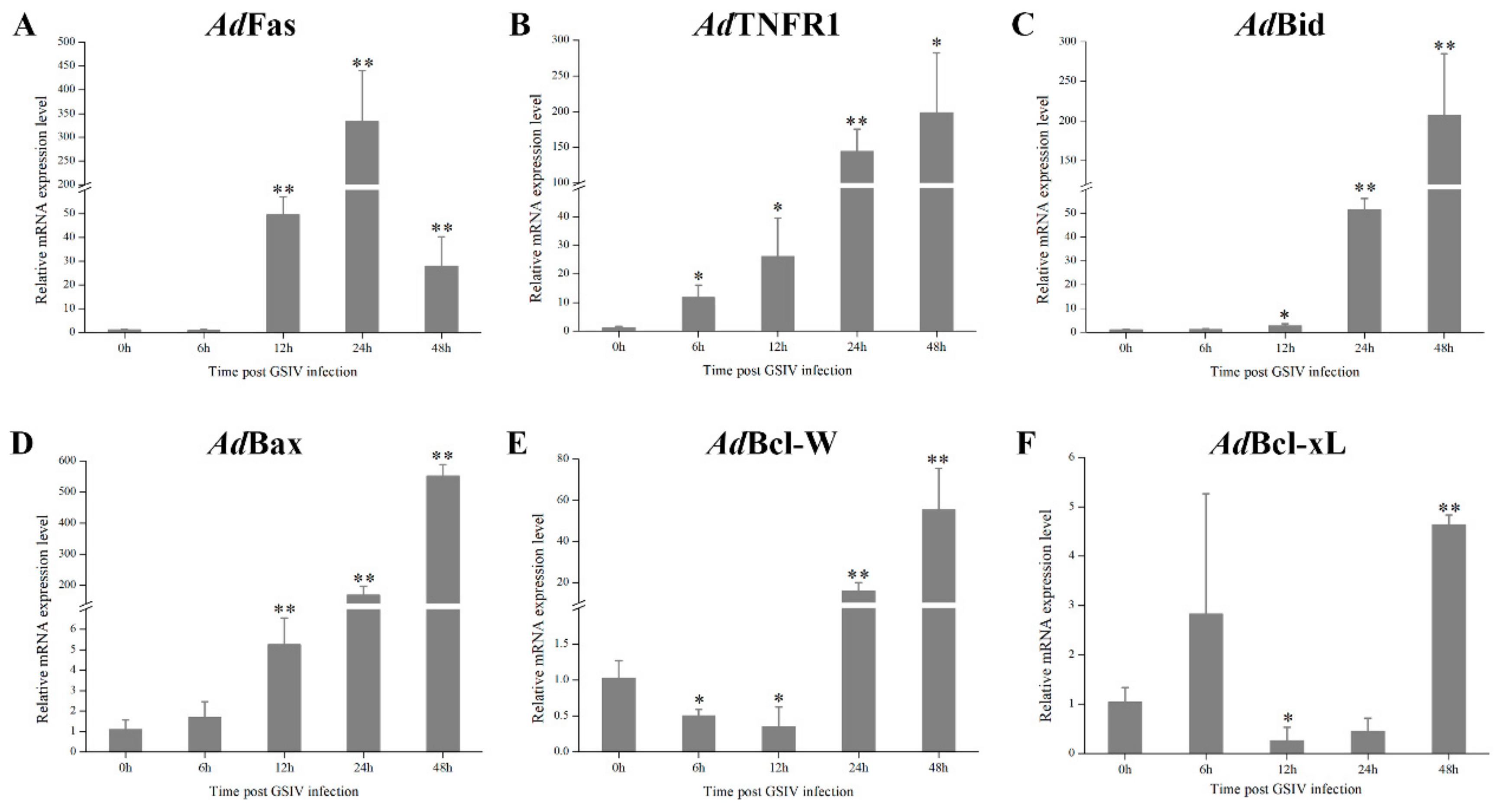
2.8. mRNA Expression of Apoptosis Associated Genes
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Virus
4.2. Droplet Digital PCR (ddPCR) of GSIV MCP Copy
4.3. Nucleus Staining
4.4. DNA Fragmentation Assay
4.5. TUNEL Assay
4.6. Flow Cytometry
4.6.1. Flow Cytometry Assay for Apoptosis Detection
4.6.2. Flow Cytometry Assay for Mitochondrial Membrane Potential (MMP) Detection
4.6.3. Flow Cytometry Assay for Caspases Activity
4.7. Western Blot
4.7.1. Western Blot Analysis for Cytochrome c Release
4.7.2. Western Blot Analysis for p53 Expression
4.8. Quantitative Real-Time PCR (qRT-PCR) of Apoptosis Related Genes Expression
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GSIV | Chinese giant salamander iridovirus |
| GSM | Chinese giant salamander muscle cell line |
| MMP | mitochondria membrane potential |
| MOI | multiplicity of infection |
References
- Zhou, X.; Jiang, W.; Liu, Z.; Liu, S.; Liang, X. Virus Infection and Death Receptor-Mediated Apoptosis. Viruses 2017, 9, 316. [Google Scholar] [CrossRef]
- Danthi, P. Viruses and the Diversity of Cell Death. Annu. Rev. Virol. 2016, 3, 533–553. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, J.G.; Branton, P.E. Regulation of apoptosis by viral gene products. J. Virol. 1997, 71, 1739–1746. [Google Scholar] [PubMed]
- Everett, H.; McFadden, G. Apoptosis: An innate immune response to virus infection. Trends Microbiol. 1999, 7, 160–165. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Brenner, C.; Morselli, E.; Touat, Z.; Kroemer, G. Viral Control of Mitochondrial Apoptosis. PLoS Pathog. 2008, 4, e1000018. [Google Scholar] [CrossRef]
- Shibahara, T.; Sato, K.; Ishikawa, Y.; Kadota, K. Porcine circovirus induces B lymphocyte depletion in pigs with wasting disease syndrome. J. Vet. Med. Sci. 2000, 62, 1125–1131. [Google Scholar] [CrossRef]
- Gupta, S.K.; Sahoo, A.P.; Rosh, N.; Gandham, R.K.; Saxena, L.; Singh, A.K.; Harish, D.R.; Tiwari, A.K. Canine parvovirus NS1 induced apoptosis involves mitochondria, accumulation of reactive oxygen species and activation of caspases. Virus Res. 2016, 213, 46–61. [Google Scholar] [CrossRef]
- Calabrese, F.; Pontisso, P.; Pettenazzo, E.; Benvegnu, L.; Vario, A.; Chemello, L.; Alberti, A.; Valente, M. Liver cell apoptosis in chronic hepatitis C correlates with histological but not biochemical activity or serum HCV-RNA levels. Hepatology 2000, 31, 1153–1159. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, R.; Zhang, Z.; Jin, Y.; Cardona, C.J.; Xing, Z. Intrinsic apoptosis and proinflammatory cytokines regulated in human astrocytes infected with enterovirus 71. J. Gen Virol. 2015, 96, 3010–3022. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, A.; Hayakawa, S.; Yanai, H.; Stoiber, D.; Negishi, H.; Kikuchi, H.; Sasaki, S.; Imai, K.; Shibue, T.; Honda, K.; et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003, 424, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Fontela, C.; Garcia, M.A.; Garcia-Cao, I.; Collado, M.; Arroyo, J.; Esteban, M.; Serrano, M.; Rivas, C. Resistance to viral infection of super p53 mice. Oncogene 2005, 24, 3059–3062. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.L.; Kraft, R.M.; Aubert, M.; Goodwin, E.; DiMaio, D.; Blaho, J.A. p53 and hTERT determine sensitivity to viral apoptosis. J Virol. 2007, 81, 12985–12995. [Google Scholar] [CrossRef] [PubMed]
- Reshi, L.; Wu, H.C.; Wu, J.L.; Wang, H.V.; Hong, J.R. GSIV serine/threonine kinase can induce apoptotic cell death via p53 and pro-apoptotic gene Bax upregulation in fish cells. Apoptosis Int. J. Programmed Cell Death 2016, 21, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Robert, J. Antiviral immunity in amphibians. Viruses 2011, 3, 2065–2086. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Wen, C.M.; Hui, C.F.; Chen, M.C.; Wu, J.L.; Hsueh, T.C.; Lei, W.H.; Hong, J.R. Giant seaperch iridovirus infection upregulates Bas and Bak expression, leading to apoptotic death of fish cells. Fish Shellfish Immunol. 2015, 45, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.; Barbosa-Solomieu, V.; Chinchar, V.G. A decade of advances in iridovirus research. Adv. Virus Res. 2005, 65, 173–248. [Google Scholar]
- Castedo, M.; Perfettini, J.L.; Andreau, K.; Roumier, T.; Piacentini, M.; Kroemer, G. Mitochondrial apoptosis induced by the HIV-1 envelope. Ann. N. Y. Acad. Sci. 2003, 1010, 19–28. [Google Scholar] [CrossRef]
- Essbauer, S.; Ahne, W. The epizootic haematopoietic necrosis virus (Iridoviridae) induces apoptosis in vitro. J. Vet. Med. B Infect Dis. Vet. Public Health 2002, 49, 25–30. [Google Scholar] [CrossRef]
- Hu, G.B.; Cong, R.S.; Fan, T.J.; Mei, X.G. Induction of apoptosis in a flounder gill cell line by lymphocystis disease virus infection. J. Fish Dis. 2004, 27, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Imajoh, M.; Sugiura, H.; Oshima, S. Morphological changes contribute to apoptotic cell death and are affected by caspase-3 and caspase-6 inhibitors during red sea bream iridovirus permissive replication. Virology 2004, 322, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Paul, E.R.; Chitnis, N.S.; Henderson, C.W.; Kaul, R.J.; D‘Costa, S.M.; Bilimoria, S.L. Induction of apoptosis by iridovirus virion protein extract. Arch. Virol. 2007, 152, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Huang, X.H.; Gui, J.F.; Zhang, Q.Y. Mitochondrion-mediated apoptosis induced by Rana grylio virus infection in fish cells. Apoptosis Int. J. Programmed Cell Death 2007, 12, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, W.; Huang, Y.; Xu, L.; Qin, Q. Involvement of the PI3K and ERK signaling pathways in largemouth bass virus-induced apoptosis and viral replication. Fish Shellfish Immunol. 2014, 41, 371–379. [Google Scholar] [CrossRef]
- Ma, J.; Zeng, L.; Zhou, Y.; Jiang, N.; Zhang, H.; Fan, Y.; Meng, Y.; Xu, J. Ultrastructural morphogenesis of an amphibian iridovirus isolated from Chinese giant salamander (Andrias davidianus). J. Comp. Pathol. 2014, 150, 325–331. [Google Scholar] [CrossRef]
- Geng, Y.; Wang, K.Y.; Zhou, Z.Y.; Li, C.W.; Wang, J.; He, M.; Yin, Z.Q.; Lai, W.M. First report of a ranavirus associated with morbidity and mortality in farmed Chinese giant salamanders (Andrias davidianus). J. Comp. Pathol. 2011, 145, 95–102. [Google Scholar] [CrossRef]
- Meng, Y.; Ma, J.; Jiang, N.; Zeng, L.B.; Xiao, H.B. Pathological and microbiological findings from mortality of the Chinese giant salamander (Andrias davidianus). Arch. Virol. 2014, 159, 1403–1412. [Google Scholar] [CrossRef]
- Fan, Y.; Chang, M.X.; Ma, J.; LaPatra, S.E.; Hu, Y.W.; Huang, L.; Nie, P.; Zeng, L. Transcriptomic analysis of the host response to an iridovirus infection in Chinese giant salamander, Andrias davidianus. Vet. Res. 2015, 46, 136. [Google Scholar] [CrossRef]
- Chen, Q.; Ma, J.; Fan, Y.; Meng, Y.; Xu, J.; Zhou, Y.; Liu, W.; Zeng, X.; Zeng, L. Identification of type I IFN in Chinese giant salamander (Andrias davidianus) and the response to an iridovirus infection. Mol. Immunol. 2015, 65, 350–359. [Google Scholar] [CrossRef]
- Du, J.; Wang, L.; Wang, Y.; Shen, J.; Pan, C.; Meng, Y.; Yang, C.; Ji, H.; Dong, W. Autophagy and apoptosis induced by Chinese giant salamander (Andrias davidianus) iridovirus (CGSIV). Vet. Microbiol. 2016, 195, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Chinchar, V.G.; Bryan, L.; Wang, J.; Long, S.; Chinchar, G.D. Induction of apoptosis in frog virus 3-infected cells. Virology 2003, 306, 303–312. [Google Scholar] [CrossRef]
- Kuwana, T.; Mackey, M.R.; Perkins, G.; Ellisman, M.H.; Latterich, M.; Schneiter, R.; Green, D.R.; Newmeyer, D.D. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002, 111, 331–342. [Google Scholar] [CrossRef]
- Wei, M.C.; Zong, W.X.; Cheng, E.H.; Lindsten, T.; Panoutsakopoulou, V.; Ross, A.J.; Roth, K.A.; MacGregor, G.R.; Thompson, C.B.; Korsmeyer, S.J. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science 2001, 292, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Letai, A.; Bassik, M.C.; Walensky, L.D.; Sorcinelli, M.D.; Weiler, S.; Korsmeyer, S.J. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002, 2, 183–192. [Google Scholar] [CrossRef]
- Cory, S.; Huang, D.C.; Adams, J.M. The Bcl-2 family: Roles in cell survival and oncogenesis. Oncogene 2003, 22, 8590–8607. [Google Scholar] [CrossRef] [PubMed]
- Son, K.N.; Pugazhenthi, S.; Lipton, H.L. Activation of tumor suppressor protein p53 is required for Theiler’s murine encephalomyelitis virus-induced apoptosis in M1-D macrophages. J. Virol. 2009, 83, 10770–10777. [Google Scholar] [CrossRef]
- Li, S.; Lu, L.F.; Liu, S.B.; Zhang, C.; Li, Z.C.; Zhou, X.Y.; Zhang, Y.A. Spring viraemia of carp virus modulates p53 expression using two distinct mechanisms. PLoS Pathog. 2019, 15, e1007695. [Google Scholar] [CrossRef]
- Jiang, N.; Fan, Y.; Zhou, Y.; Liu, W.; Robert, J.; Zeng, L. Rag1 and rag2 gene expressions identify lymphopoietic tissues in juvenile and adult Chinese giant salamander (Andrias davidianus). Dev. Comp. Immunol. 2018, 87, 24–35. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Jiang, N.; Fan, Y.; Zhou, Y.; Liu, W.; Xue, M.; Meng, Y.; Zeng, L. Chinese Giant Salamander (Andrias davidianus) Iridovirus Infection Leads to Apoptotic Cell Death through Mitochondrial Damage, Caspases Activation, and Expression of Apoptotic-Related Genes. Int. J. Mol. Sci. 2019, 20, 6149. https://doi.org/10.3390/ijms20246149
Li Y, Jiang N, Fan Y, Zhou Y, Liu W, Xue M, Meng Y, Zeng L. Chinese Giant Salamander (Andrias davidianus) Iridovirus Infection Leads to Apoptotic Cell Death through Mitochondrial Damage, Caspases Activation, and Expression of Apoptotic-Related Genes. International Journal of Molecular Sciences. 2019; 20(24):6149. https://doi.org/10.3390/ijms20246149
Chicago/Turabian StyleLi, Yiqun, Nan Jiang, Yuding Fan, Yong Zhou, Wenzhi Liu, Mingyang Xue, Yan Meng, and Lingbing Zeng. 2019. "Chinese Giant Salamander (Andrias davidianus) Iridovirus Infection Leads to Apoptotic Cell Death through Mitochondrial Damage, Caspases Activation, and Expression of Apoptotic-Related Genes" International Journal of Molecular Sciences 20, no. 24: 6149. https://doi.org/10.3390/ijms20246149
APA StyleLi, Y., Jiang, N., Fan, Y., Zhou, Y., Liu, W., Xue, M., Meng, Y., & Zeng, L. (2019). Chinese Giant Salamander (Andrias davidianus) Iridovirus Infection Leads to Apoptotic Cell Death through Mitochondrial Damage, Caspases Activation, and Expression of Apoptotic-Related Genes. International Journal of Molecular Sciences, 20(24), 6149. https://doi.org/10.3390/ijms20246149




