Structure and Location Studies on Key Enzymes in Saponins Biosynthesis of Panax notoginseng
Abstract
1. Introduction
2. Results
2.1. Isolation of Four Full-Length cDNA Sequences Involved in Saponin Biosynthesis
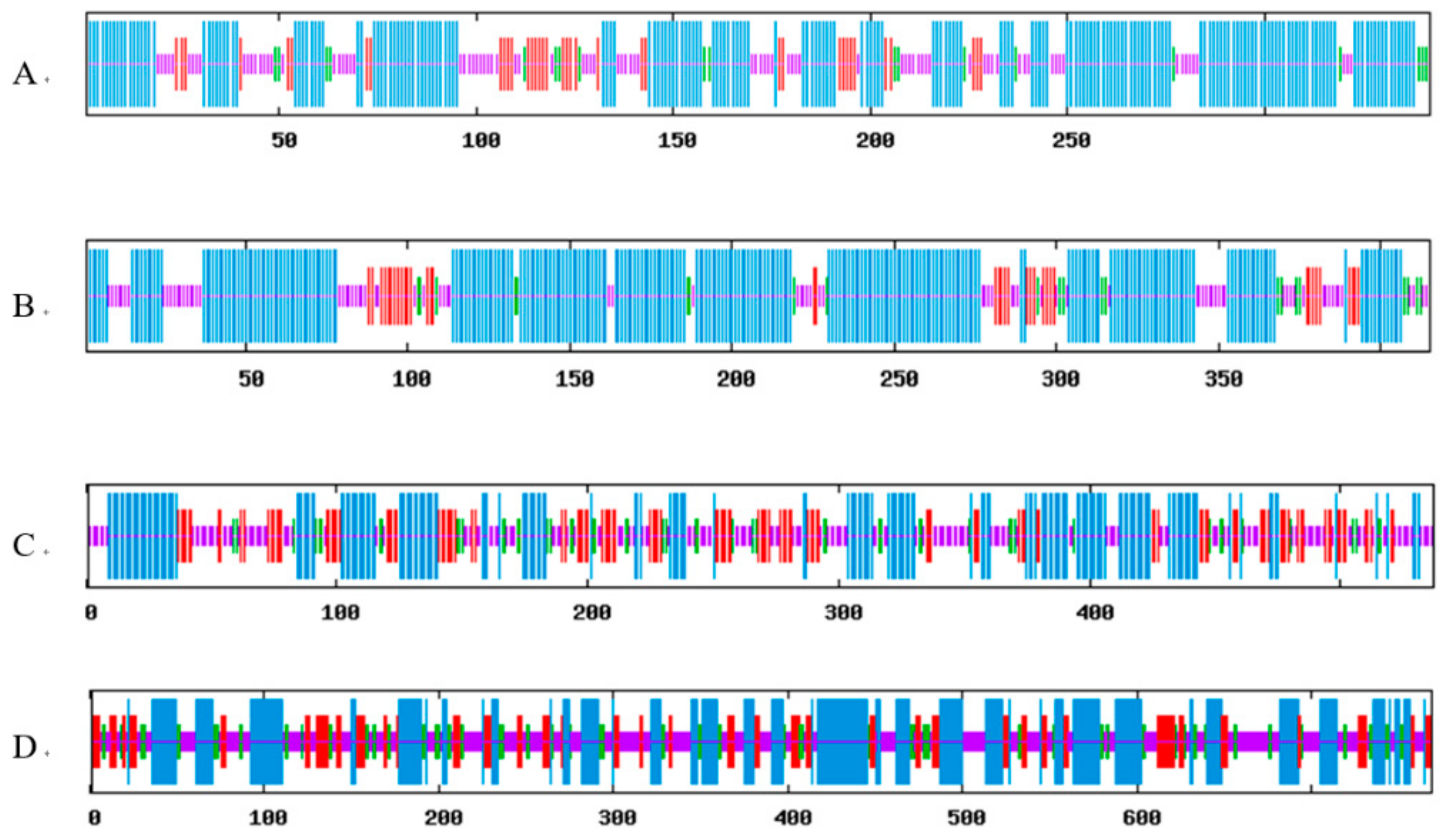
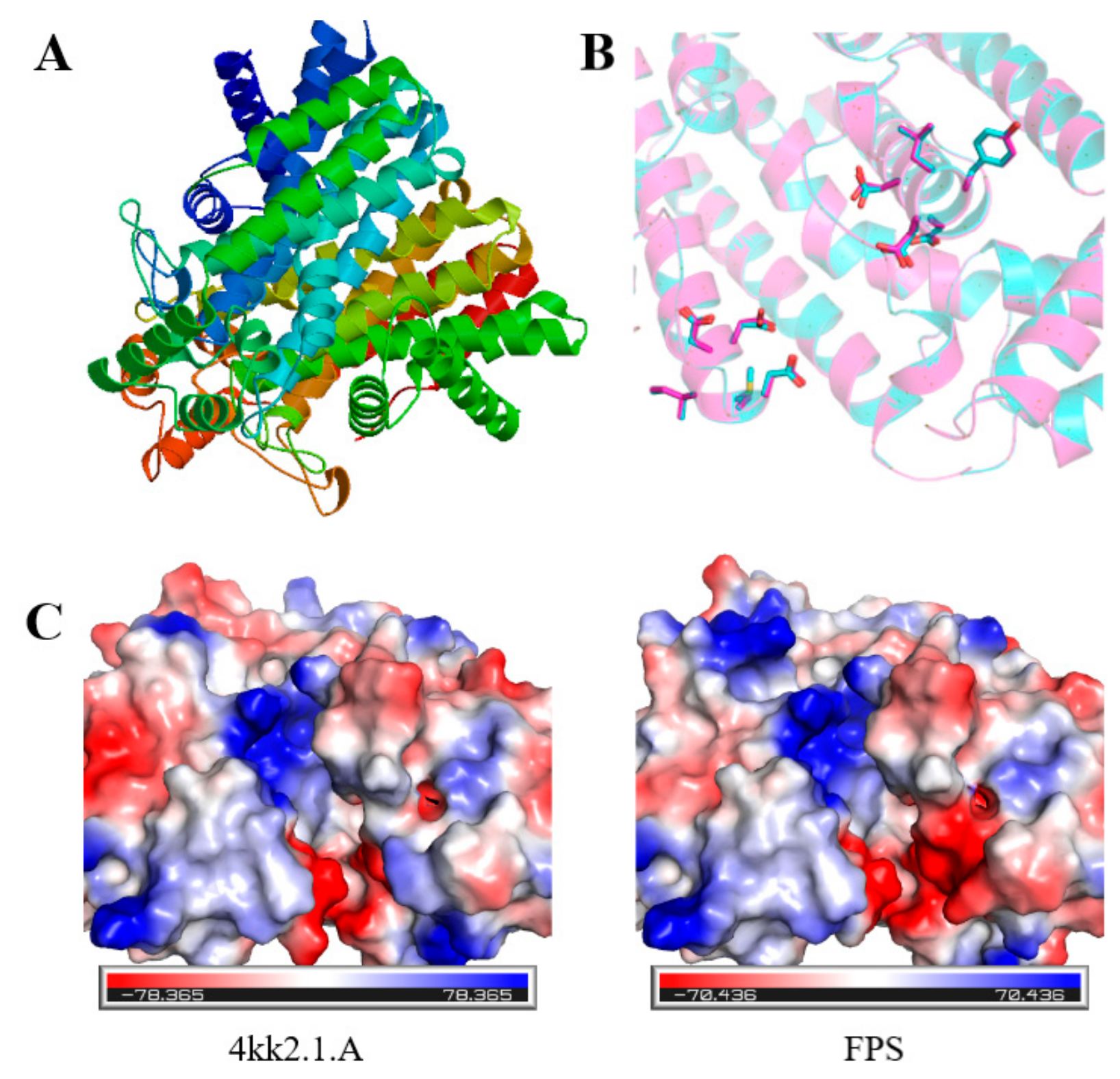
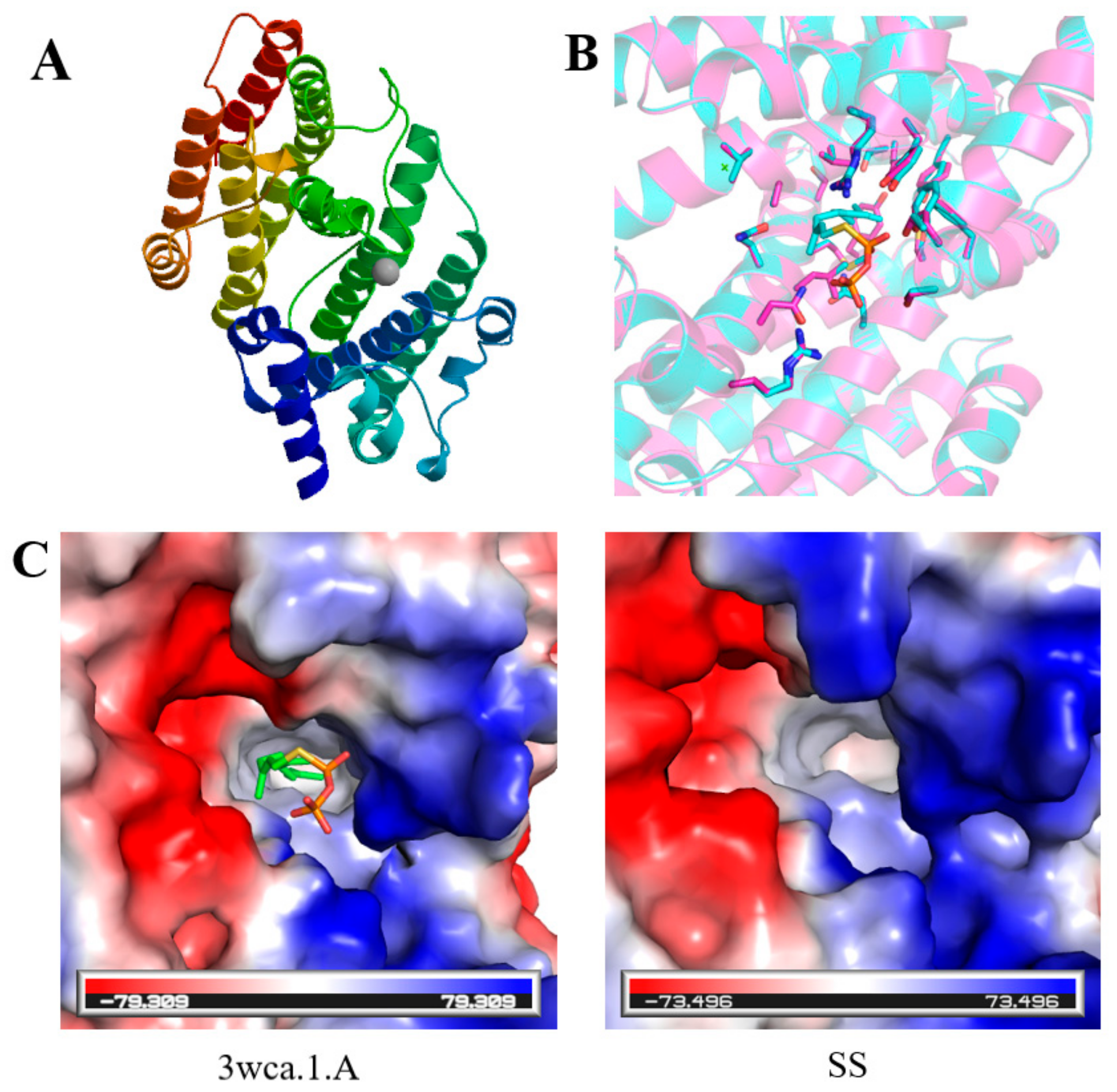
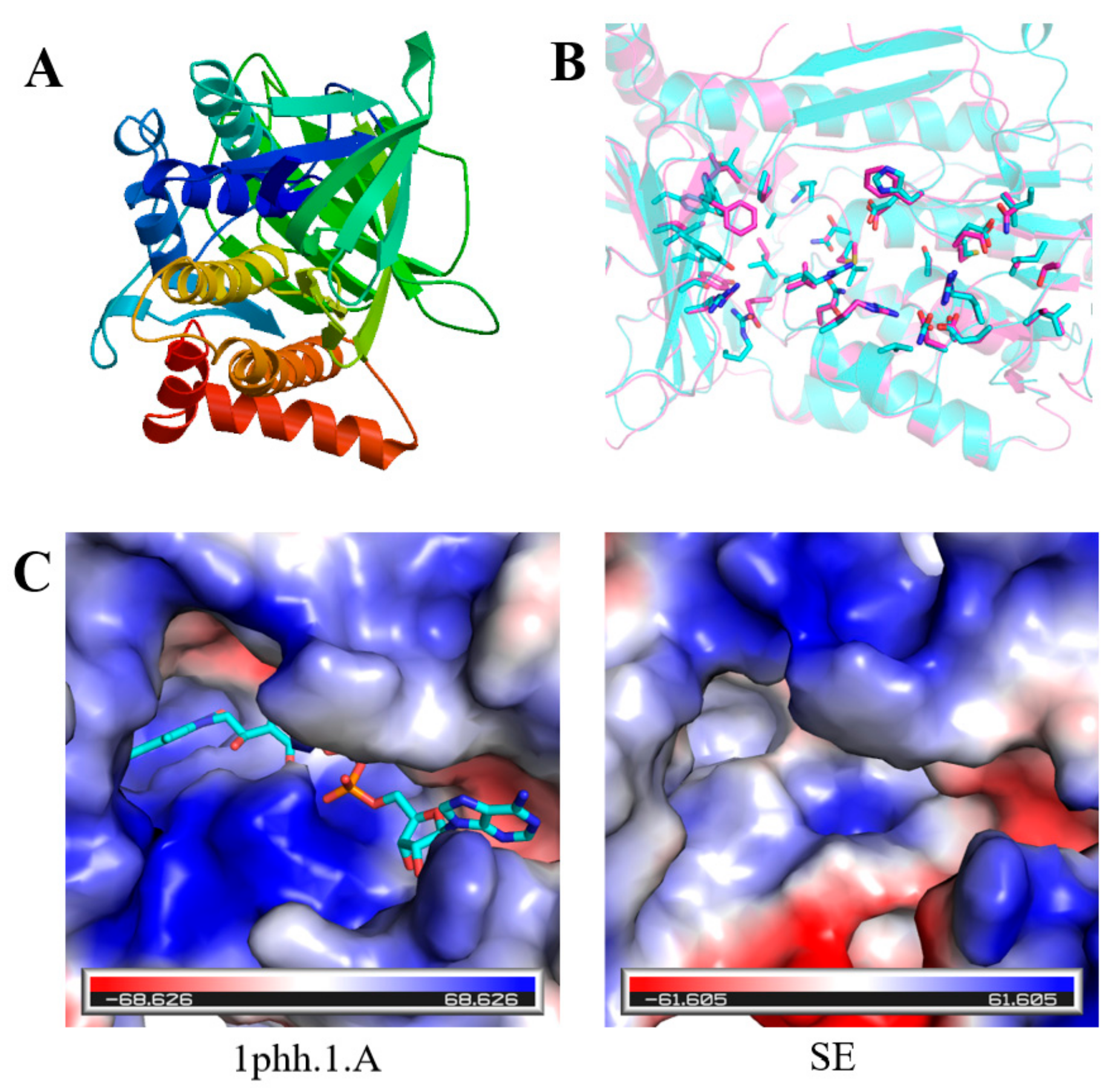
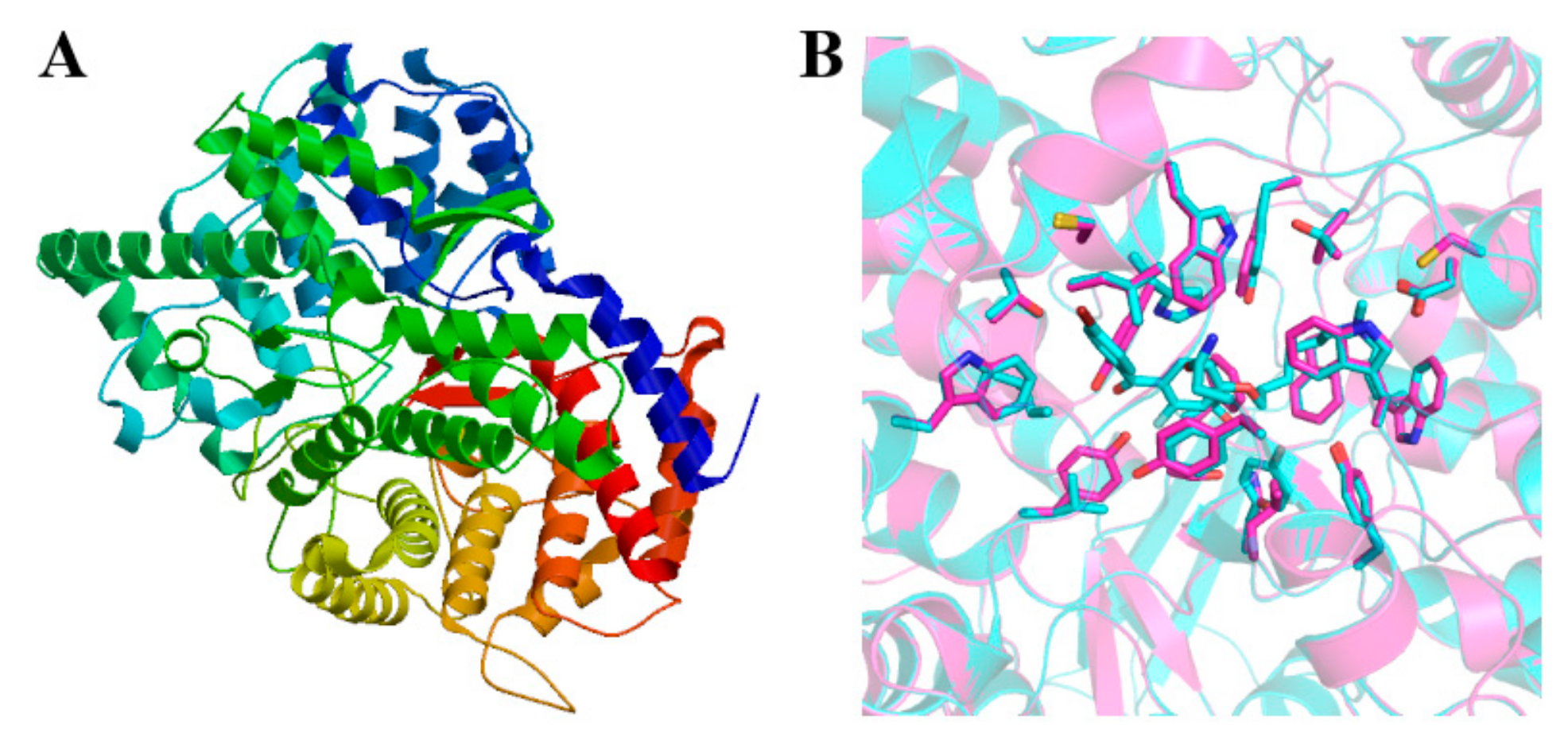
2.2. Prediction of Secondary and Tertiary Structures
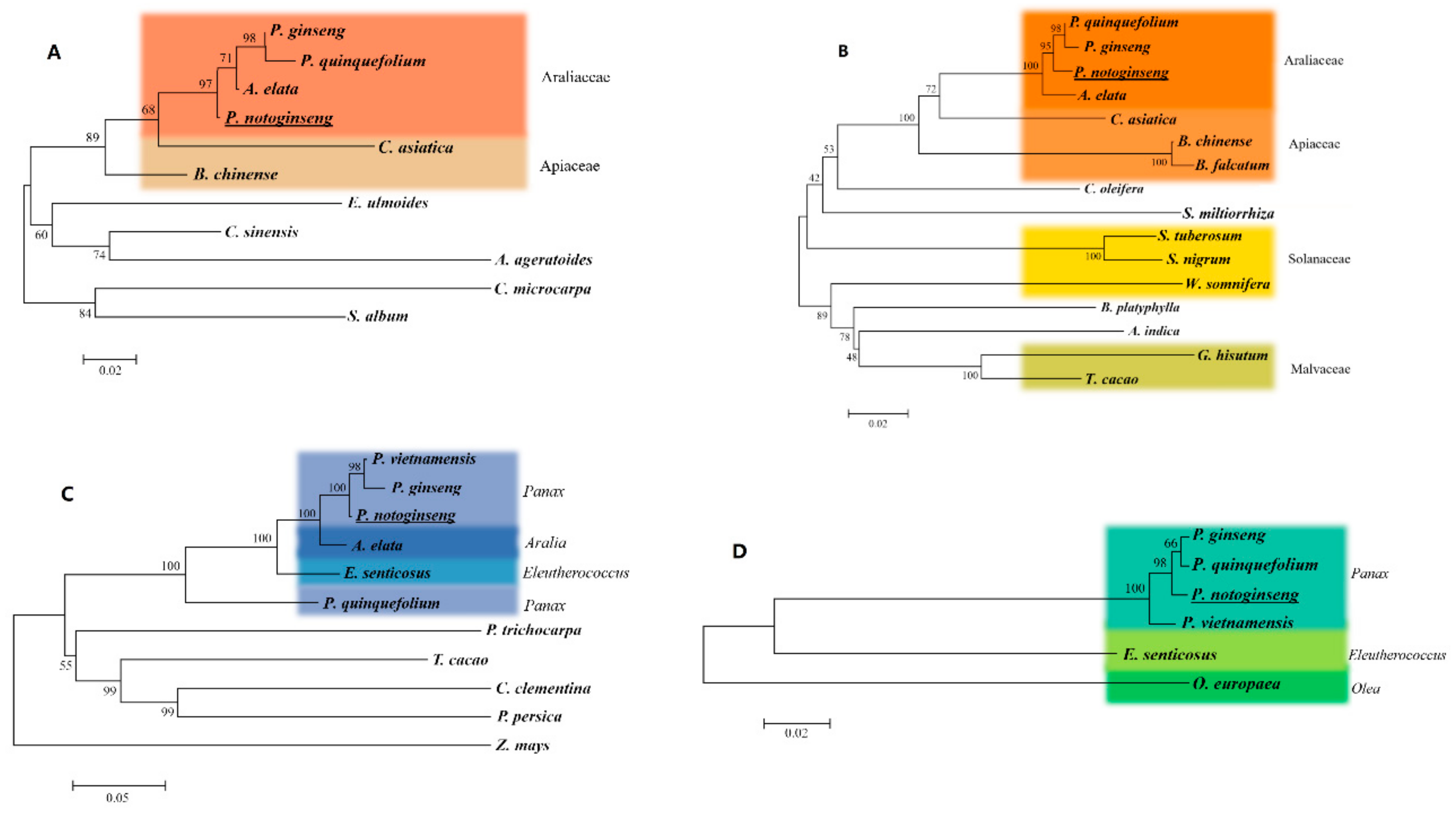
2.3. Sequence Homology and Phylogenetic Analysis
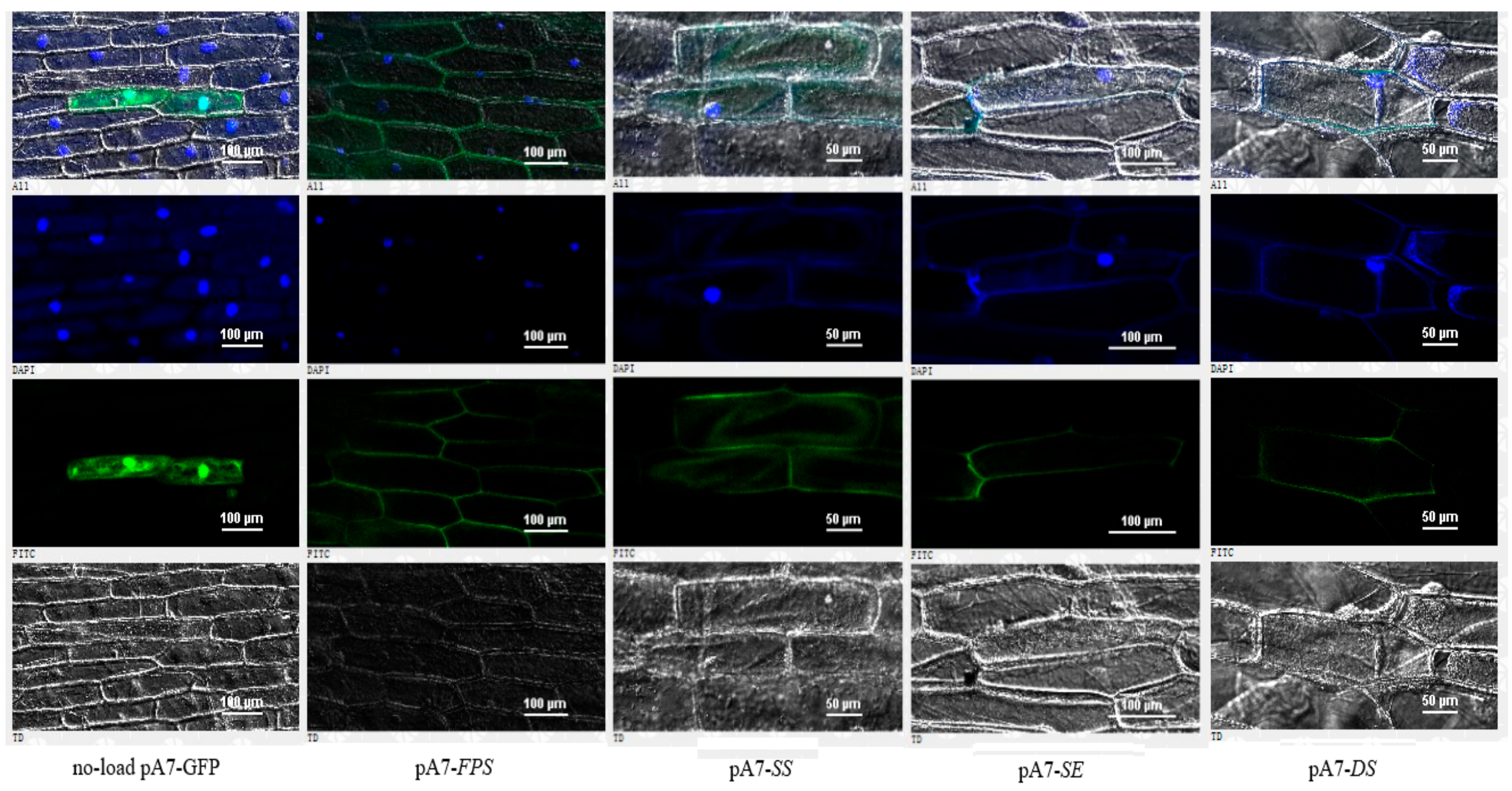
2.4. Subcellular Localizations
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. RNA and DNA Isolation
4.3. Genes Cloning
4.4. Bioinformatics Analysis
4.5. Phylogenetic and Genetic Evolutionary Analysis
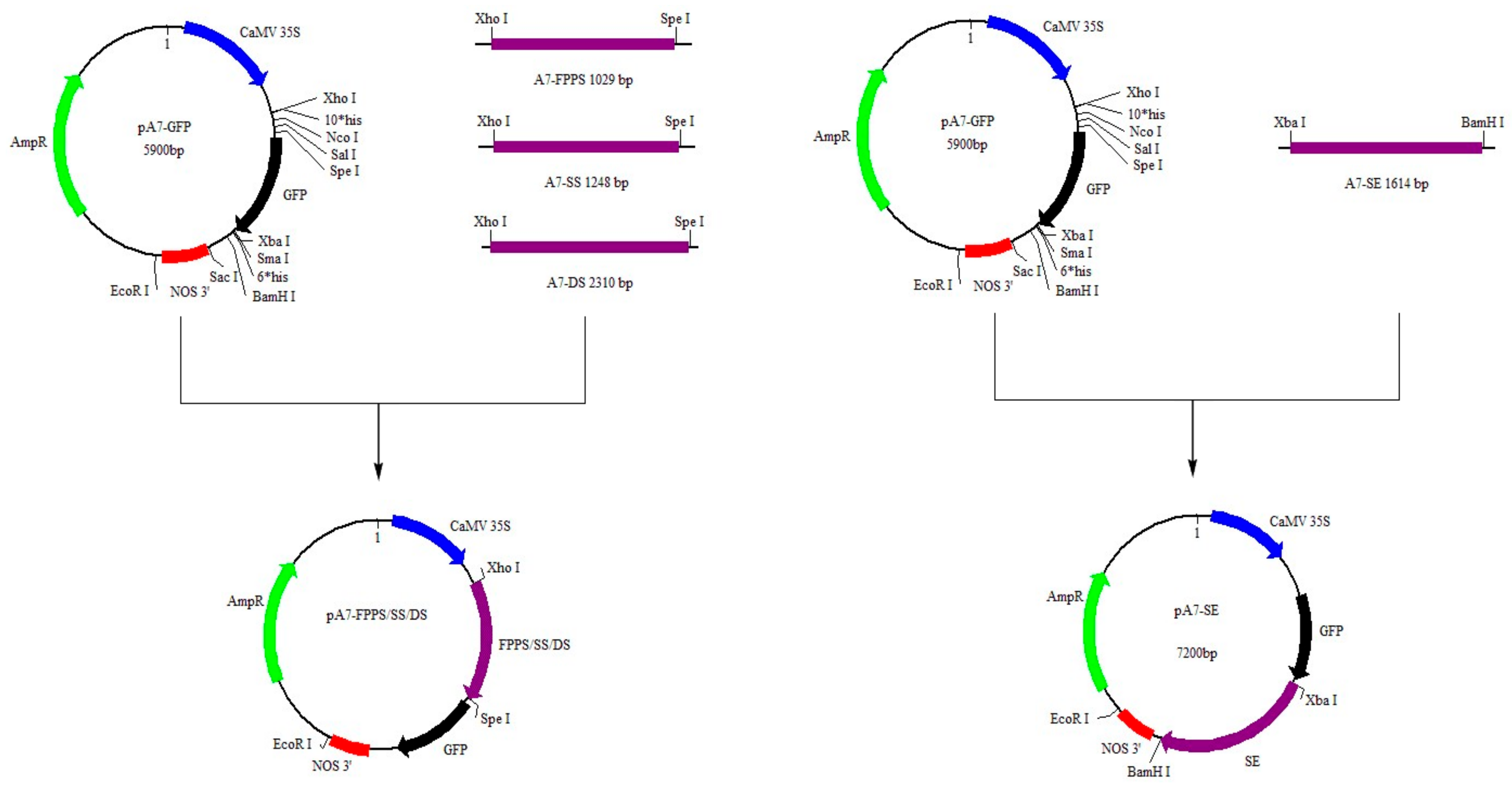
4.6. Construction of GFP Expression Vectors
4.7. Transformation of Onion Epidermal Cells by Gene Guns
4.8. Nuclear Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ng, T.B. Pharmacological activity of sanchi ginseng (Panax notoginseng). J. Pharm. Pharmacol. 2010, 58, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.G.; Zhang, S.C.; Liang, Z.S.; Qi, Z.H. Research history and overview of chemical constituents of Panax notoginseng. Chin. Tradit. Herb. Drugs 2014, 45, 2564–2570. [Google Scholar]
- Liu, Y.; Xie, M.X.; Kang, J.; Zheng, D. Studies on the interaction of total saponins of Panax notoginseng and human serum albumin by fourier transform infrared spectroscopy. Sectrochim. Acta Part A 2003, 59, 2747–2758. [Google Scholar] [CrossRef]
- Nah, S.Y.; Park, H.J.; McCleskey, E.W. A trace component of ginseng that inhibit Ca2+ channels through a pertussis toxin sensitive G protein. Proc. Natl. Acad. Sci. USA 1995, 92, 8739–8743. [Google Scholar] [CrossRef] [PubMed]
- Bo, G.R.; Lu, S.M.; Li, P.; Liu, C.J.; Guo, S.Q.; Chen, F. Study on protective effects of Panax notoginseng saponins Rg1 to mitochondrial injuries of intestinal epithelial cells induced by hemorrhagic shock rats. Chin. Pharm. J. 2003, 38, 665–667. [Google Scholar]
- Zhou, Y.; Wei, A.P. Exprssion of neuronal nitric oxide synthase in rat ischemic brain modulated by notoglnsenoside A comprehensive methods for the study of immrmopharmacology. J. Brain Nerv. Dis. 2000, 8, 196–200. [Google Scholar]
- Zhang, H.G.; Li, X.H.; Yang, Z.C. Effects of Panax notoginseng on myocardial Gsα mRNA expression and ATPase activity after severe cald in rats. Burns 2003, 29, 541–546. [Google Scholar] [CrossRef]
- Zhang, L.B.; Yang, G.Z. The antagonistic effect of notoginsenoside-Rg1 injected in striatum on immunoin flammatory injury in rat model of Parkinson’s disease. Chin. J. Immunol. 2004, 20, 121–123. [Google Scholar]
- Gao, W.Q.; Jia, L.; Zhao, Y.Q. Advance in study on antitumor effect of Panax ginseng and its active mechanism. Drug Eval. Res. 2011, 34, 53–58. [Google Scholar]
- Niu, Y.Y.; Zhu, X.X.; Luo, H.M.; Sun, C.; Yang, T.J.; Dong, L.L.; Huang, L.F.; Chen, S.L. Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng. Gene 2014, 53, 295–303. [Google Scholar] [CrossRef]
- Reumann, S.; Babujee, L.; Ma, C.; Wienkoop, S.; Siemsen, T.; Antonicelli, G.E.; Rasche, N.; Luder, F.; Weckwerth, W.; Jahn, O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 2007, 19, 3170–3193. [Google Scholar] [CrossRef] [PubMed]
- Sapir-Mir, M.; Mett, A.; Belausov, E.; Tal-Meshulam, S.; Frydman, A.; Gidoni, D.; Eyal, Y. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol. 2008, 148, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Guirimand, G.; Papon, N.; Courdavault, V.; Thabet, I.; Ginis, O.; Bouzid, S.; Giglioli-Guivarc’h, N.; Clastre, M. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 2011, 234, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Cunillera, N.; Arro, M.; Delourme, D.; Karst, F.; Boronat, A.; Ferrer, A. Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J. Biol. Chem. 1996, 271, 7774–7780. [Google Scholar] [CrossRef]
- Jarstfer, M.B.; Zhang, D.L.; Poulter, C.D. Recombinant squalene synthase. Synthesis of non-head-to-tail isoprenoids in the absence of NADPH. J. Am. Chem. Soc. 2002, 124, 8834–8845. [Google Scholar] [CrossRef]
- Laden, B.P.; Tang, Y.; Porter, T. Cloning, heterologous expression, and enzymological characterization of human squalene monooxygenase. Arch. Biochem. Biophys. 2000, 374, 381–388. [Google Scholar] [CrossRef]
- Kim, T.D.; Han, J.Y.; Huh, G.H.; Choi, Y.E. Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol. 2011, 52, 125–137. [Google Scholar] [CrossRef]
- Han, J.Y.; Hwang, H.S.; Choi, S.W.; Kim, H.J.; Choi, Y.E. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012, 53, 1535–1545. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, D.Q.; Ge, F.; Chen, C.Y. Effect of over-expressing farnesyl pyrophosphate synthase (FPS) gene of Panax notoginseng cell on saponin synthesis. Mod. Food Sci. Technol. 2015, 31, 59–64. [Google Scholar]
- Hu, F.X.; Zhong, J.J. Jasmonic acid mediates gene transcription of ginsenoside biosynthesis in cell cultures of Panax notoginseng treated with chemically synthesized 2-hydroxyethyl jasmonate. Process. Biochem. 2008, 43, 113–118. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeong, J.H.; Seo, J.W.; Shin, C.G.; Kim, Y.S.; In, J.G.; Yang, D.C.; Yi, J.S.; Choi, Y.E. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing Squalene synthase gene. Plant Cell Physiol. 2004, 45, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.M.; Sun, C.; Sun, Y.Z.; Wu, Q.; Li, Y.; Song, J.Y.; Niu, Y.Y.; Cheng, X.L.; Xu, H.X.; Li, C.Y.; et al. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genom. 2011, 12, S5. [Google Scholar] [CrossRef] [PubMed]
- Tansakul, P.; Shibuya, M.; Kushiro, T.; Ebizuka, Y. Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 2006, 580, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Z.; Wang, Z.H.; Zhang, M.P.; Wang, Y.; Sun, Y.C.; Jiang, S.C. Gene cloning of SQS and SE enzyme and construction of expression vector in ginseng. Mod. Agric. Sci. Technol. 2010, 4, 20–22. [Google Scholar]
- Han, J.Y.; In, J.G.; Kwon, Y.S.; Choi, Y.E. Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry 2010, 71, 36–46. [Google Scholar] [CrossRef]
- Han, J.Y.; Kwon, Y.S.; Yang, D.C.; Jung, Y.R.; Choi, Y.E. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 2006, 47, 1653–1662. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, Y.B.; Uddin, M.R.; Lee, S.; Kim, S.U.; Sang, U.P. Enhanced triterpene accumulation in Panax ginseng hairy roots overexpressing Mevalonate-5-pyrophosphate decarboxylase and farnesyl pyrophosphate synthase. ACS Synth. Biol. 2014, 3, 773–779. [Google Scholar] [CrossRef]
- Liu, C.J.; Meng, Y.L.; Hou, S.S.; Chen, X.Y. Cloning and sequencing of a cDNA encoding farnesyl pyrophosphate synthase from, Gossypium arboreum and its expression pattern in the developing seeds of, Gossypium hirsutum cv. “Sumian-6”. Acta Bot. Sin. 1998, 40, 703–710. [Google Scholar]
- Gao, F.; Luo, X.P.; Tao, L.; Li, C.L.; Ding, C.B.; Chen, H.; Wu, Q. Molecular cloning of squalene synthase gene form Paris polyphylla and its expression in Escherichia coli. Chin. J. Chin. Mater. Med. 2013, 38, 2086–2091. [Google Scholar]
- Ma, Y.M.; Yuan, L.C.; Zhang, L.S.; Hou, X.M.; Lu, S.F. Cloning and identification of two squalene synthase genes from Salvia miltiorrhiza. Chin. Tradit. Herb. Drugs 2014, 45, 1307–1312. [Google Scholar]
- Tao, C.C.; Ma, C.T.; Wu, Y.S.; Zhou, Q.N.; Su, H.L.; Chao, N.X.; Luo, Y. Cloning and sequence analysis of squalene synthase gene from Trichosanthes rubriflos. Chin. Tradit. Herb. Drugs 2015, 46, 1034–1041. [Google Scholar]
- Zhang, G.; Tang, Z.S.; Zhou, L.Y.; Liu, Q.; Li, Y.M.; Zhang, X.F.; Lu, X. Characterization of a squalene synthase gene in Dendrobium officinale Kimura et Migo. J. Northwest A F Univ. 2013, 41, 151–158. [Google Scholar]
- Hu, W.; Liu, N.; Tian, Y.H.; Li, Y.T.; Zhang, L.X. Cloning, expression of squalene epoxidase from Panax ginseng. J. Northwest A F Univ. 2012, 40, 207–212. [Google Scholar]
- Xing, Z.B.; Cao, L.; Chen, L.; He, S.; Li, B.C.; Zhu, J.L. Cloning and sequence analysis on cDNA of squalene epoxidase gene in Eleutherococcus senticosus. Chin. J. Chin. Mater. Med. 2012, 37, 172–175. [Google Scholar]

| The Source Species | Accession No. | The Source Species | Accession No. |
|---|---|---|---|
| P. ginseng | AAY87903 | B. falcatum | AY964186.1 |
| C. microcarpa | AF390138 | S. nigrum | JX984610.1 |
| C. asiatica | AY787627 | G. hirsutum | EF688567 |
| B. chinense | EU400219 | S. miltiorrhiza | JQ974834.1 |
| C. sinensis | FS949364 | W. somnifera | GU181386.1 |
| P. quinquefolium | GQ401664 | P. ginseng | AB122078.1 |
| A. elata | HM219226 | P. vietnamensis | KJ946469.1 |
| A. ageratoides | JX424562 | E. senticosus | JN228206.1 |
| E. ulmoides | KC468536 | P. quinquefolium | KC524469.1 |
| S. album | KF011939 | A. elata | GU354314.1 |
| P. ginseng | AB010148 | T. cacao | XM_007047548.1 |
| S. tuberosum | JF802612.1 | C. clementina | XM_006426079.1 |
| C. asiatica | AY787628.1 | P. trichocarpa | XM_006386402.1 |
| B. chinense | GQ889266.1 | P. persica | XM_007208396.1 |
| A. indica | JQ327160.1 | Z. mays | BT056068.1 |
| P. quinquefolium | GU997681 | P. ginseng | GU183405.1 |
| A. elata | GU354313 | P. quinquefolium | KC316048.1 |
| T. cacao | XM_007041985 | P. vietnamensis | KF306328.1 |
| B. platyphylla | KP723830.1 | O. europaea | AB291240.1 |
| C. oleifera | JX914592.1 | E. senticosus | JF818131.1 |
| Primer | Primer Sequences (5′-3′) |
|---|---|
| FPSF | ATGAGCGATCTGAAGACGAGATT |
| FPSR | TTACTTTTGCCGCTTATATATCTTTCC |
| SSF | ATGGGAAGTTTGGGGGCAAT |
| SSR | TCACTGTTTGTTCGGTAGTAGGTTT |
| SEF | ATGAATTCATCTTCTTCTACTAGTAC |
| SER | TTAGTGAATGGGGGGAGCTCT |
| DSF | ATGTGGAAGCTGAAGGTTGCT |
| DSR | TTAAATTTTGAGCTGCTGGTGC |
| Primer | Primer Sequences (5′-3′) | Restriction Sites |
|---|---|---|
| A7-FPPSF | GGGGCTCGAGATGAGCGATCTGAAGA | XhoI |
| A7-FPPSR | GGGGGACTAGTGCCTTTTGCCGCTTATAT | BcuI (SpeI) |
| A7-SSF | TTTTTTTTCTCGAGATGGGAAGTTTGGGGG | XhoI |
| A7-SSR | GGGACTAGTGCCTGTTTGTTCGGTAGT | BcuI (SpeI) |
| A7-SEF | GGGGGGGTCTAGACATGAATTCATCTTCTT | XbaI |
| A7-SER | TTTGGATCCTTAGTGAATGGGGGGAGCT | BamHI |
| A7-DSF | TTTTTTCTCGAGATGTGGAAGCTGAAGG | XhoI |
| A7-DSR | TTGGACTAGTGCAATTTTGAGCTGCTG | BcuI (SpeI) |
| Ingredient | Dosage (μL) | Ingredient | Dosage (μL) |
|---|---|---|---|
| 10×Buffer Tango | 2.5 | 10×Buffer Tango | 2.5 |
| XhoI | 2.0 | XbaI | 1.0 |
| BcuI (SpeI) | 0.5 | BamHI | 1.0 |
| FPS/SS/DS/pA7-GFP plasmid | ≤1.0 μg | SE/pA7-GFP plasmid | ≤1.0 μg |
| RNase Free H2O | up to 25 | RNase Free H2O | up to 25 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, P.; Zheng, Y.; Liang, Z. Structure and Location Studies on Key Enzymes in Saponins Biosynthesis of Panax notoginseng. Int. J. Mol. Sci. 2019, 20, 6121. https://doi.org/10.3390/ijms20246121
Xia P, Zheng Y, Liang Z. Structure and Location Studies on Key Enzymes in Saponins Biosynthesis of Panax notoginseng. International Journal of Molecular Sciences. 2019; 20(24):6121. https://doi.org/10.3390/ijms20246121
Chicago/Turabian StyleXia, Pengguo, Yujie Zheng, and Zongsuo Liang. 2019. "Structure and Location Studies on Key Enzymes in Saponins Biosynthesis of Panax notoginseng" International Journal of Molecular Sciences 20, no. 24: 6121. https://doi.org/10.3390/ijms20246121
APA StyleXia, P., Zheng, Y., & Liang, Z. (2019). Structure and Location Studies on Key Enzymes in Saponins Biosynthesis of Panax notoginseng. International Journal of Molecular Sciences, 20(24), 6121. https://doi.org/10.3390/ijms20246121





