iTRAQ-Based Quantitative Proteomic Analysis of Digestive Juice across the First 48 Hours of the Fifth Instar in Silkworm Larvae
Abstract
1. Introduction
2. Results
2.1. Protein Profiling
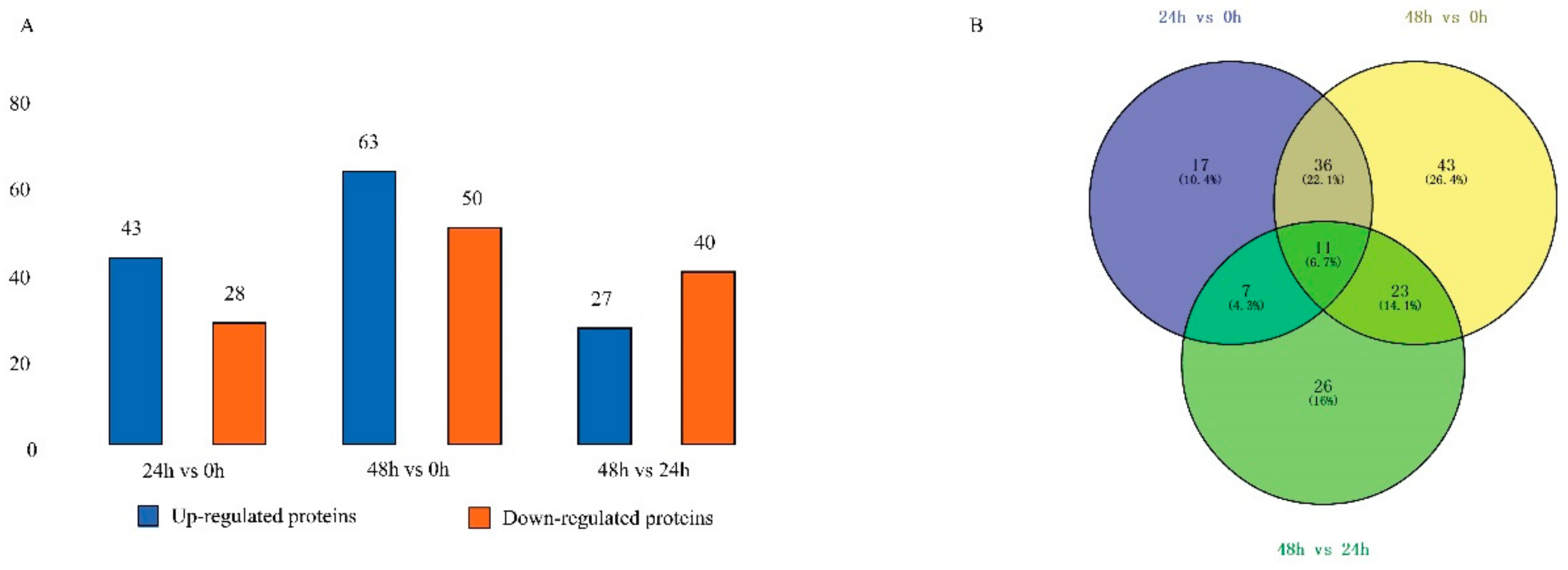
2.2. Identification of Differentially Expressed Proteins
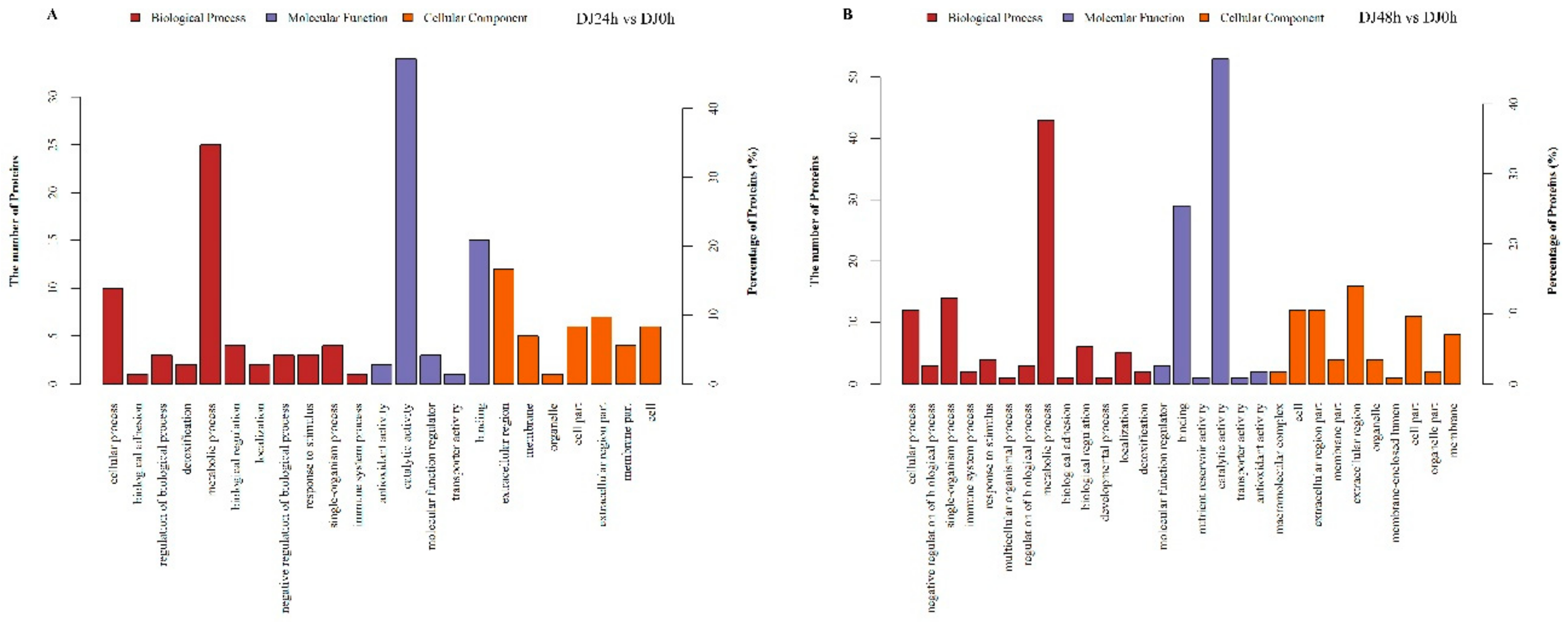
2.3. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis of DEPs
2.4. Protein–Protein Interaction Network Analysis of DEPs
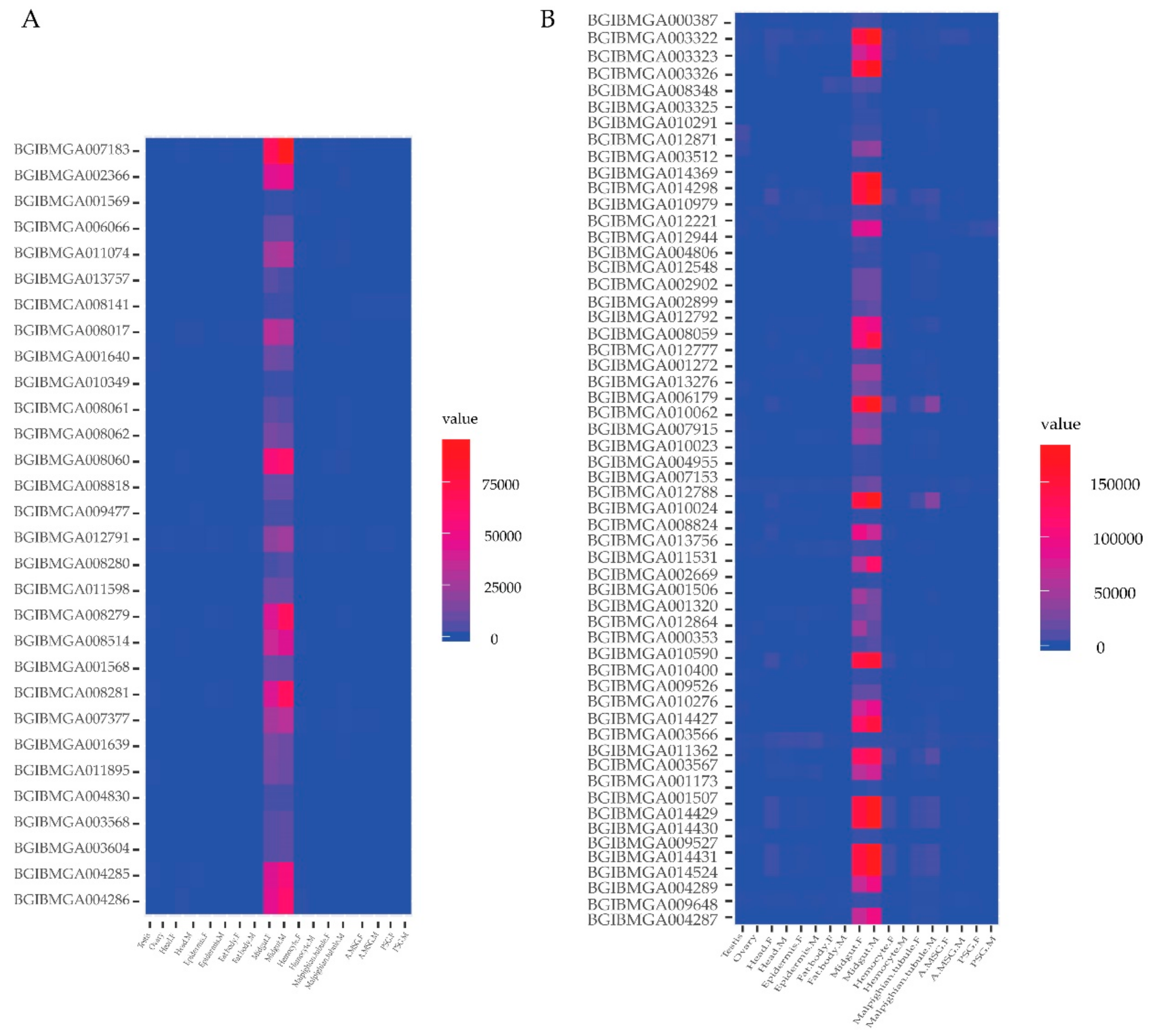
2.5. Spatial Expression Profile of Genes of the Identified Proteins
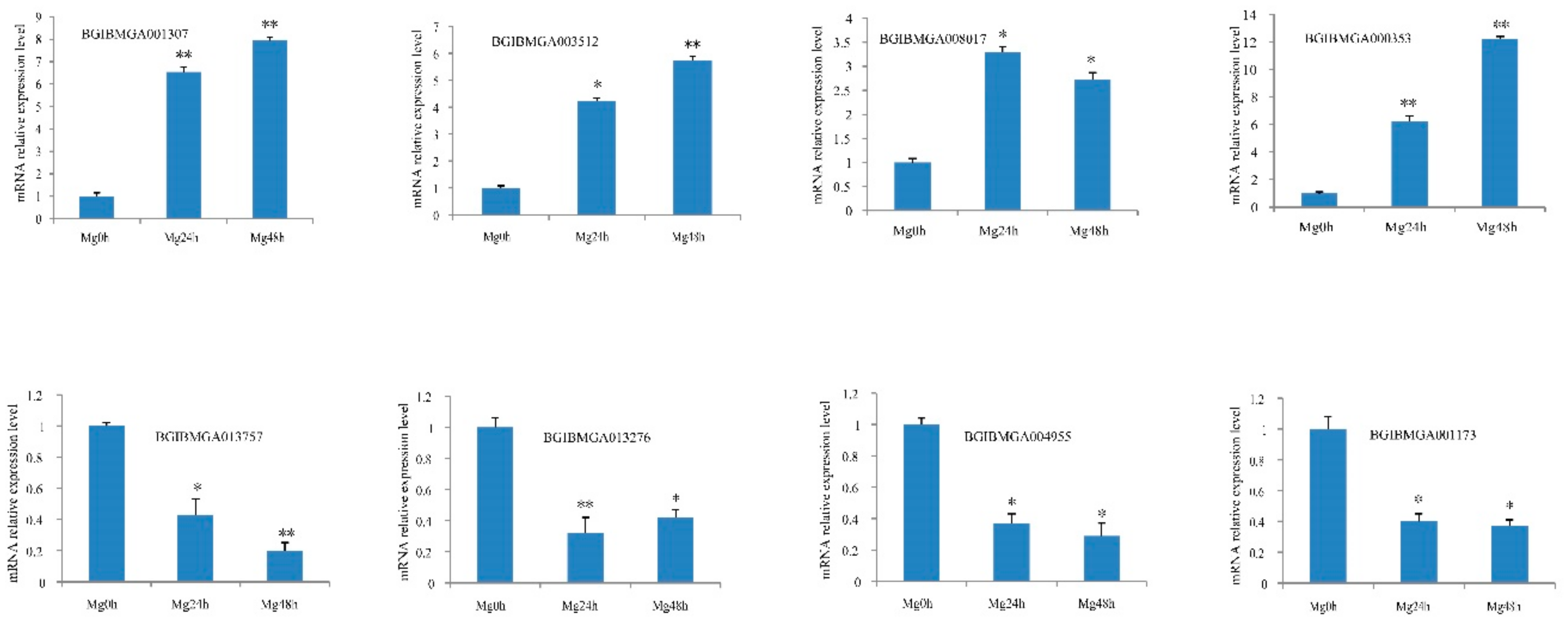
2.6. Expression Analysis of Genes Corresponding to DEPs
3. Discussion
4. Materials and Methods
4.1. Collection of Samples and Protein Preparation
4.2. Protein Digestion and iTRAQ Labeling
4.3. Strong Cation Exchange Fractionation and NanoLC-MS/MS Analysis
4.4. Protein Identification and Annotation
4.5. Tissue Expression Patterns Based on Microarray Data
4.6. RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx Mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Cheng, D.; Duan, J.; Wang, G.; Cheng, T.; Zha, X.; Liu, C.; Zhao, P.; Dai, F.; Zhang, Z.; et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx Mori. Genome Biol. 2007, 8, R162. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, H.; Atiwetin, P.; Nagaoka, S.; Miyata, S.; Kitajima, S.; Sugimura, Y. Absorption of mulberry root urease to the hemolymph of the silkworm, Bombyx mori. J. Insect Physiol. 2005, 51, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, X.; He, Y.; Shuai, J.; Chen, X.; Ling, E. Expression of antimicrobial peptide genes in Bombyx mori gut modulated by oral bacterial infection and development. Dev. Comp. Immunol. 2010, 34, 1191–1198. [Google Scholar] [CrossRef]
- Kataoka, N.; Miyake, S.; Azuma, M. Aquaporin and aquaglyceroporin in silkworms, differently expressed in the hindgut and midgut of Bombyx mori. Insect Mol. Biol. 2009, 18, 303–314. [Google Scholar] [CrossRef]
- Shao, Q.M.; Yang, B.; Xu, Q.Y.; Li, X.Q.; Lu, Z.Q.; Wang, C.S.; Huang, Y.P.; Soderhall, K.; Ling, E.J. Hindgut Innate Immunity and Regulation of Fecal Microbiota through Melanization in Insects. J. Biol. Chem. 2012, 287, 14270–14279. [Google Scholar] [CrossRef]
- Baggio, M.P.; Vessaro-Silva, S.A.; Ribeiro, L.F.; Brancalhao, R.M. Morphology of the Pylorus of Bombyx mori (Linnaeus) (Lepidoptera: Bombycidae). Neotrop. Entomol. 2014, 43, 344–349. [Google Scholar] [CrossRef]
- Vallet-Gely, I.; Lemaitre, B.; Boccard, F. Bacterial strategies to overcome insect defences. Nat. Rev. Microbiol. 2008, 6, 302–313. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, M.; Wang, S.; Zhu, L.; Xue, R.; Cao, G.; Gong, C. Proteomics analysis of digestive juice from silkworm during Bombyx mori nucleopolyhedrovirus infection. Proteomics 2015, 15, 2691–2700. [Google Scholar] [CrossRef]
- Holtof, M.; Lenaerts, C.; Cullen, D.; Vanden Broeck, J. Extracellular nutrient digestion and absorption in the insect gut. Cell Tissue Res. 2019, 377, 397–414. [Google Scholar] [CrossRef]
- Wang, D.D.; Dong, Z.M.; Zhang, Y.; Guo, K.Y.; Guo, P.C.; Zhao, P.; Xia, Q.Y. Proteomics Provides Insight into the Interaction between Mulberry and Silkworm. J. Proteome Res. 2017, 16, 2472–2480. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.N.; Datta, R.K. Hierarchical clustering of 54 races and strains of the mulberry silkworm, Bombyx mori L.: Significance of biochemical parameters. Theor. Appl. Genet. 1992, 85, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.N.; Rao, C.G.; Chatterjee, G.K.; Ashwath, S.K.; Patnaik, A.K. Correlation between yield and biochemical parameters in the mulberry silkworm, Bombyx mori L. Theor. Appl. Genet. 1993, 87, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, G.K.; Sengupta, A.K.; Verma, A.K.; Sen, S.K.; Saratchandra, B. Esterase isozyme polymorphism, specific and nonspecific esterase, syngenic lines development and natural occurrence of a thermostable esterase in the tropical silkworm Bombyx mori L. Insect Biochem. Mol. Biol. 2001, 31, 1191–1199. [Google Scholar] [CrossRef]
- Srinivasan, A.; Giri, A.P.; Gupta, V.S. Structural and functional diversities in lepidopteran serine proteases. Cell Mol. Biol. Lett. 2006, 11, 132–154. [Google Scholar] [CrossRef]
- Ponnuvel, K.M.; Nithya, K.; Sirigineedi, S.; Awasthi, A.K.; Yamakawa, M. In vitro antiviral activity of an alkaline trypsin from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Arch. Insect Biochem. Physiol. 2012, 81, 90–104. [Google Scholar] [CrossRef]
- Manjunatha, G.K.S.; Peter, A.; Naika, M.B.N.; Niranjana, P.; Shamprasad, P. Identification of In-Vitro Red Fluorescent Protein with Antipathogenic Activity from the Midgut of the Silkworm (Bombyx Mori L.). Protein Pept. Lett. 2018, 25, 302–313. [Google Scholar] [CrossRef]
- Sunagar, S.G.; Lakkappan, V.J.; Ingalhalli, S.S.; Savanurmath, C.J.; Hinchigeri, S.B. Characterization of the photochromic pigments in red fluorescent proteins purified from the gut juice of the silkworm Bombyx mori L. Photochem. Photobiol. 2008, 84, 1440–1444. [Google Scholar] [CrossRef]
- Matti, K.M.; Singh, S.S.; Savanurmath, C.J.; Hinchigeri, S.B. A unique red fluorescent protein of silkworm bearing two photochromic moieties. Photochem. Photobiol. Sci. 2009, 8, 1364–1372. [Google Scholar] [CrossRef]
- Matti, K.M.; Savanurmath, C.J.; Hinchigeri, S.B. A promising broad spectrum antimicrobial red fluorescent protein present in silkworm excreta. Biol. Pharm. Bull. 2010, 33, 1143–1147. [Google Scholar] [CrossRef][Green Version]
- Sunagar, S.G.; Savanurmath, C.J.; Hinchigeri, S.B. The profiles of red fluorescent proteins with antinucleopolyhedrovirus activity in races of the silkworm Bombyx mori. J. Insect Physiol. 2011, 57, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Ponnuvel, K.M.; Nakazawa, H.; Furukawa, S.; Asaoka, A.; Ishibashi, J.; Tanaka, H.; Yamakawa, M. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J. Virol. 2003, 77, 10725–10729. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, H.; Tsuneishi, E.; Ponnuvel, K.M.; Furukawa, S.; Asaoka, A.; Tanaka, H.; Ishibashi, J.; Yamakawa, M. Antiviral activity of a serine protease from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Virology 2004, 321, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Wickham, S.; West, M.B.; Cook, P.F.; Hanigan, M.H. Gamma-glutamyl compounds: Substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal. Biochem. 2011, 414, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, A.; Liu, F.Y.; Illes, K.; Nagar, B. Crystal structure of the human alkaline sphingomyelinase provides insights into substrate recognition. J. Biol. Chem. 2017, 292, 7087–7094. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, A.; Heinz, L.X.; Illes, K.; Superti-Furga, G.; Nagar, B. Crystal Structure of the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3B Provides Insights into Determinants of Substrate Specificity. J. Biol. Chem. 2016, 291, 24054–24064. [Google Scholar] [CrossRef]
- Whitcomb, D.C.; Lowe, M.E. Human pancreatic digestive enzymes. Dig. Dis. Sci. 2007, 52, 1–17. [Google Scholar] [CrossRef]
- Raghavendra, R.; Neelagund, S.; Kuluvar, G.; Bhanuprakash, V.; Revanaiah, Y. Protective effect of partially purified 35 kDa protein from silk worm (Bombyx mori) fecal matter against carbon tetrachloride induced hepatotoxicity and in vitro anti-viral properties. Pharm. Biol. 2010, 48, 1426–1431. [Google Scholar] [CrossRef][Green Version]
- Lee, T.V.; Ding, T.; Chen, Z.; Rajendran, V.; Scherr, H.; Lackey, M.; Bolduc, C.; Bergmann, A. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development 2008, 135, 43–52. [Google Scholar] [CrossRef]
- Fujii, T.; Banno, Y. Identification of a novel function of the silkworm integument in nitrogen metabolism: Uric acid is synthesized within the epidermal cells in B. mori. Insect Biochem. Mol. Biol. 2019, 105, 43–50. [Google Scholar] [CrossRef]
- Ozawa, R.; Matsumoto, S. Intracellular signal transduction of PBAN action in the silkworm, Bombyx mori: Involvement of acyl CoA reductase. Insect Biochem. Mol. Biol. 1996, 26, 259–265. [Google Scholar] [CrossRef]
- Yamamoto, K.; Wilson, D.K. Identification, characterization, and crystal structure of an aldo-keto reductase (AKR2E4) from the silkworm Bombyx mori. Arch. Biochem. Biophys. 2013, 538, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Kanaya, T.; Kobayashi, J. Purification and characterization of an insect haemolymph protein promoting in vitro replication of the Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 2000, 81, 1135–1141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iatrou, K.; Swevers, L. Transformed lepidopteran cells expressing a protein of the silkmoth fat body display enhanced susceptibility to baculovirus infection and produce high titers of budded virus in serum-free media. J. Biotechnol. 2005, 120, 237–250. [Google Scholar] [CrossRef]
- Okada, T.; Ishiyama, S.; Sezutsu, H.; Usami, A.; Tamura, T.; Mita, K.; Fujiyama, K.; Seki, T. Molecular cloning and expression of two novel beta-N-acetylglucosaminidases from silkworm Bombyx mori. Biosci. Biotechnol. Biochem. 2007, 71, 1626–1635. [Google Scholar] [CrossRef]
- Kimura, S. Genetics of insect hemolymph alpha-mannosidase in the silkworm, Bombyx mori. Biochem. Genet. 1983, 21, 713–724. [Google Scholar] [CrossRef]
- Yamamoto, K.; Higashiura, A.; Suzuki, M.; Shiotsuki, T.; Sugahara, R.; Fujii, T.; Nakagawa, A. Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori. Biochem. Biophys. Res. Commun. 2016, 474, 104–110. [Google Scholar] [CrossRef]
- Uno, T.; Ueno, M.; Kikuchi, M.; Aizono, Y. Purification and characterization of nucleoside diphosphate kinase from the brain of Bombyx mori. Arch. Insect Biochem. Physiol. 2002, 50, 147–155. [Google Scholar] [CrossRef]
- Isobe, M.; Kai, H.; Kurahashi, T.; Suwan, S.; Pitchayawasin-Thapphasaraphong, S.; Franz, T.; Tani, N.; Higashi, K.; Nishida, H. The molecular mechanism of the termination of insect diapause, part 1: A timer protein, TIME-EA4, in the diapause eggs of the silkworm Bombyx mori is a metallo-glycoprotein. Chembiochem A Eur. J. Chem. Biol. 2006, 7, 1590–1598. [Google Scholar] [CrossRef]
- Xu, L.; Liang, H.; Niu, Y.; Wang, Y.; Sima, Y.; Xu, S. Differential expression of the Bombyx mori diapause-termination timer gene Ea4 in diapause-inducing temperature and photoperiod. Arch. Insect Biochem. Physiol. 2012, 79, 182–194. [Google Scholar] [CrossRef]
- Ngernyuang, N.; Kobayashi, I.; Promboon, A.; Ratanapo, S.; Tamura, T.; Ngernsiri, L. Cloning and expression analysis of the Bombyx mori alpha-amylase gene (Amy) from the indigenous Thai silkworm strain, Nanglai. J. Insect Sci. 2011, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Mitsumasu, K.; Azuma, M.; Niimi, T.; Yamashita, O.; Yaginuma, T. Membrane-penetrating trehalase from silkworm Bombyx mori. Molecular cloning and localization in larval midgut. Insect Mol. Biol. 2005, 14, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Cheng, T.; Jin, S.; Jiang, L.; Wang, C.; Xia, Q. Structural, evolutionary and functional analysis of APN genes in the Lepidoptera Bombyx mori. Gene 2014, 535, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M.; Teh, E.M.; Keeling, P.J. Novel ubiquitin fusion proteins: Ribosomal protein P1 and actin. J. Mol. Biol. 2003, 328, 771–778. [Google Scholar] [CrossRef]
- Catic, A.; Ploegh, H.L. Ubiquitin—Conserved protein or selfish gene? Trends Biochem. Sci. 2005, 30, 600–604. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Hao, F.; Li, N.; Liu, X.; Wang, G.; Wang, Y.; Tang, H. Developmental Changes for the Hemolymph Metabolome of Silkworm (Bombyx mori L.). J. Proteome Res. 2015, 14, 2331–2347. [Google Scholar] [CrossRef]
- Grzelak, K. Control of expression of silk protein genes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1995, 110, 671–681. [Google Scholar] [CrossRef]
- Hou, Y.; Zou, Y.; Wang, F.; Gong, J.; Zhong, X.; Xia, Q.; Zhao, P. Comparative analysis of proteome maps of silkworm hemolymph during different developmental stages. Proteome Sci. 2010, 8, 45. [Google Scholar] [CrossRef]
- Ye, J.; Li, Y.; Liu, H.W.; Li, J.; Dong, Z.; Xia, Q.; Zhao, P. Genome-Wide Identification and Characterization of Carboxypeptidase Genes in Silkworm (Bombyx mori). Int. J. Mol. Sci. 2016, 17, 1203. [Google Scholar] [CrossRef]
- Konno, K.; Ono, H.; Nakamura, M.; Tateishi, K.; Hirayama, C.; Tamura, Y.; Hattori, M.; Koyama, A.; Kohno, K. Mulberry latex rich in antidiabetic sugar-mimic alkaloids forces dieting on caterpillars. Proc. Natl. Acad. Sci. USA 2006, 103, 1337–1341. [Google Scholar] [CrossRef]
- Gan, Q.; Zhang, X.W.; Zhang, D.B.; Shi, L.; Zhou, Y.; Sun, T.T.; Jiang, S.; Gao, J.S.; Meng, Y. BmSUC1 is essential for glycometabolism modulation in the silkworm, Bombyx mori. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, M.; Bandani, A.R.; Mehrabadi, R.; Alizadeh, H. Inhibitory activity of proteinaceous alpha-amylase inhibitors from Triticale seeds against Eurygaster integriceps salivary alpha-amylases: Interaction of the inhibitors and the insect digestive enzymes. Pestic. Biochem. Physiol. 2012, 102, 220–228. [Google Scholar] [CrossRef]
- Da Lage, J.L. The Amylases of Insects. Int. J. Insect Sci. 2018, 10, 1179543318804783. [Google Scholar] [CrossRef] [PubMed]
- Asano, N.; Yamashita, T.; Yasuda, K.; Ikeda, K.; Kizu, H.; Kameda, Y.; Kato, A.; Nash, R.J.; Lee, H.S.; Ryu, K.S. Polyhydroxylated alkaloids isolated from mulberry trees (Morusalba L.) and silkworms (Bombyx mori L.). J. Agric. Food Chem. 2001, 49, 4208–4213. [Google Scholar] [CrossRef]
- Li, X.T.; Shi, L.G.; Dai, X.P.; Chen, Y.J.; Xie, H.Q.; Feng, M.; Chen, Y.Y.; Wang, H.B. Expression plasticity and evolutionary changes extensively shape the sugar-mimic alkaloid adaptation of nondigestive glucosidase in lepidopteran mulberry-specialist insects. Mol. Ecol. 2018, 27, 2858–2870. [Google Scholar] [CrossRef]
- Daimon, T.; Taguchi, T.; Meng, Y.; Katsuma, S.; Mita, K.; Shimada, T. Beta-fructofuranosidase genes of the silkworm, Bombyx mori: Insights into enzymatic adaptation of B. mori to toxic alkaloids in mulberry latex. J. Biol. Chem. 2008, 283, 15271–15279. [Google Scholar] [CrossRef]
- Horne, I.; Haritos, V.S.; Oakeshott, J.G. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem. Mol. Biol. 2009, 39, 547–567. [Google Scholar] [CrossRef]
- Van der Horst, D.J. Insect adipokinetic hormones: Release and integration of flight energy metabolism. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 136, 217–226. [Google Scholar] [CrossRef]
- Fruttero, L.L.; Leyria, J.; Canavoso, L.E. Lipids in Insect Oocytes: From the Storage Pathways to Their Multiple Functions. Results Probl. Cell Differ. 2017, 63, 403–434. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Liu, Q.; Tan, X.; Xie, X.; Xia, Q.; Zhao, P. GC/MS-based metabolomics analysis reveals active fatty acids biosynthesis in the Filippi’s gland of the silkworm, Bombyx mori, during silk spinning. Insect Biochem. Mol. Biol. 2019, 105, 1–9. [Google Scholar] [CrossRef]
- Arimatsu, Y.; Kotani, E.; Sugimura, Y.; Furusawa, T. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2007, 37, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Swevers, L.; Iatrou, K.; Huvenne, H.; Smagghe, G. Bombyx mori DNA/RNA non-specific nuclease: Expression of isoforms in insect culture cells, subcellular localization and functional assays. J. Insect Physiol. 2012, 58, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, N.; Imamura, M.; Kadotani, T.; Yaoi, K.; Iwahana, H.; Sato, R. The lipopolysaccharide-binding protein participating in hemocyte nodule formation in the silkworm Bombyx mori is a novel member of the C-type lectin superfamily with two different tandem carbohydrate-recognition domains. FEBS Lett. 1999, 443, 139–143. [Google Scholar] [CrossRef]
- Gao, K.; Deng, X.Y.; Qian, H.Y.; Qin, G.X.; Hou, C.X.; Guo, X.J. Cloning and expression analysis of a peptidoglycan recognition protein in silkworm related to virus infection. Gene 2014, 552, 24–31. [Google Scholar] [CrossRef]
- Ao, J.Q.; Ling, E.; Yu, X.Q. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol. Immunol. 2008, 45, 543–552. [Google Scholar] [CrossRef]
- Takahasi, K.; Ochiai, M.; Horiuchi, M.; Kumeta, H.; Ogura, K.; Ashida, M.; Inagaki, F. Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition. Proc. Natl. Acad. Sci. USA 2009, 106, 11679–11684. [Google Scholar] [CrossRef]
- Zou, Z.; Evans, J.D.; Lu, Z.; Zhao, P.; Williams, M.; Sumathipala, N.; Hetru, C.; Hultmark, D.; Jiang, H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007, 8, R177. [Google Scholar] [CrossRef]
- Zheng, X.; Xia, Y. beta-1, 3-Glucan recognition protein (betaGRP) is essential for resistance against fungal pathogen and opportunistic pathogenic gut bacteria in Locusta migratoria manilensis. Dev. Comp. Immunol. 2012, 36, 602–609. [Google Scholar] [CrossRef]
- Pauchet, Y.; Freitak, D.; Heidel-Fischer, H.M.; Heckel, D.G.; Vogel, H. Immunity or digestion: Glucanase activity in a glucan-binding protein family from Lepidoptera. J. Biol. Chem. 2009, 284, 2214–2224. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, P.; Cheng, T.C.; Sun, Q.; Peng, Z.W.; Dang, Y.H.; Wu, X.W.; Wang, G.H.; Jin, S.K.; Lin, P.; et al. A transgenic animal with antiviral properties that might inhibit multiple stages of infection. Antivir. Res. 2013, 98, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Selot, R.; Kumar, V.; Sekhar, S.C.; Kumar, P.G. Molecular characterization and expression analysis of BmNOX in two strains of Bombyx mori with contrasting viral resistance phenotype. Arch. Insect Biochem. Physiol. 2010, 73, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Yu, H.; Wang, X.; Xu, J.; Ma, Y.; Zhang, S.; Yu, D.; Fei, D.; Muhammad, A. iTRAQ-based quantitative proteomics analysis of molecular mechanisms associated with Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic strains. J. Proteom. 2017, 165, 35–50. [Google Scholar] [CrossRef]
- Sandberg, A.; Lindell, G.; Kallstrom, B.N.; Branca, R.M.; Danielsson, K.G.; Dahlberg, M.; Larson, B.; Forshed, J.; Lehtio, J. Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes. Mol. Cell Proteom. 2012, 11, M112-016998. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, G.H.; Dong, Z.M.; Duan, J.; Xu, P.Z.; Cheng, T.C.; Xiang, Z.H.; Xia, Q.Y. Genome-wide identification and expression analysis of serine proteases and homologs in the silkworm Bombyx mori. BMC Genom. 2010, 11, 405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.R.; Qin, S.; Xu, P.Z.; Zhang, G.Z. Identifying potential maternal genes of Bombyx mori using digital gene expression profiling. PLoS ONE 2018, 13, e0192745. [Google Scholar] [CrossRef] [PubMed]

| Time (h) | Number | Accession | SilkDB Accession | Fold Change | Function |
|---|---|---|---|---|---|
| DJ24/0 h | 1 | H9J8I4 | BGIBMGA005826 | 2.11 | Alcohol dehydrogenase activity |
| 2 | H9IW77 | BGIBMGA001508 | 2.19 | Carboxylic ester hydrolase activity | |
| 3 | Q58I81 | — | 4.18 | Ester bonds hydrolase activity | |
| 4 | H9JTK9 | BGIBMGA012871 | 2.02 | Ester bonds hydrolase activity | |
| 5 | H9ISX5 | BGIBMGA000353 | 3.24 | Innate immune | |
| 6 | Q86D78 | BGIBMGA003512 | 2.72 | Beta-glucosidase | |
| 7 | H9JCF2 | BGIBMGA007153 | 2.73 | Glycosyl hydrolase activity | |
| 8 | H9JL39 | BGIBMGA010240 | 3.96 | Chitinase activity | |
| 9 | H9JSN7 | BGIBMGA012548 | 3.21 | Sphingomyelin phosphodiesterase activity | |
| 10 | B1Q138 | — | 2.37 | Carboxylesterase activity | |
| 11 | H9JTY2 | BGIBMGA012994 | 3.01 | CN_hydrolase activity | |
| 12 | B1Q137 | — | 3.08 | Carboxylesterase activity | |
| 13 | H9IWD7 | BGIBMGA001568 | 2.57 | Maltase-glucoamylase activity | |
| 14 | B2ZZX0 | BGIBMGA008818 | 4.87 | Phosphatase activity | |
| 15 | H9JT78 | BGIBMGA012740 | 2.82 | Peroxidase activity | |
| 16 | H9JBZ7 | BGIBMGA007042 | 2.92 | Peroxidase activity | |
| 17 | P10831 | — | 3.33 | Peptidase regulator | |
| 18 | Q03383 | — | 2.02 | Antichymotrypsin | |
| 19 | C0J8G5 | BGIBMGA003292 | 3.63 | Serine protease inhibitor | |
| 20 | H9IXK0 | BGIBMGA001983 | 10.24 | Serine protease inhibitor | |
| 21 | H9JH30 | BGIBMGA008827 | 13.22 | Serine protease inhibitor | |
| 22 | I3VR74 | BGIBMGA008061 | 2.53 | AMP deaminase activity | |
| 23 | B5TZ28 | BGIBMGA007915 | 4.92 | Metallopeptidase activity | |
| 24 | H9J232 | BGIBMGA003569 | 2.96 | Serine protease activity | |
| 25 | H9J267 | BGIBMGA003604 | 3.19 | Serine protease activity | |
| 26 | H9JL75 | BGIBMGA010276 | 3.15 | Serine protease activity | |
| 27 | H9JJ25 | BGIBMGA009526 | 4.40 | Serine protease activity | |
| 28 | H9JJ26 | BGIBMGA009527 | 3.72 | Serine protease activity | |
| 29 | H9IWJ8 | BGIBMGA001630 | 2.32 | Cholinesterase activity | |
| 30 | H9JDE9 | BGIBMGA007546 | 2.19 | Carboxylesterase activity | |
| 31 | H9J067 | BGIBMGA002902 | 2.84 | Cholinesterase activity | |
| 32 | H9IWK8 | BGIBMGA001640 | 2.89 | Aminopeptidase activity | |
| 33 | H9JJ19 | BGIBMGA009520 | 6.95 | WD repeat domain phosphoinositide-interacting | |
| 34 | Q1HPY8 | — | 4.25 | Guanine nucleotide binding | |
| 35 | H9J1D5 | BGIBMGA003322 | 2.28 | Juvenile hormone binding | |
| 36 | H9JK60 | BGIBMGA009911 | 2.05 | Ubiquitin-activating | |
| 37 | H9IVY4 | BGIBMGA001415 | 2.08 | Ubiquitin-mediated protein binding | |
| 38 | C1K001 | BGIBMGA004287 | 5.34 | N/A | |
| 39 | H9JN76 | BGIBMGA010979 | 2.20 | N/A | |
| 40 | H9J1D8 | BGIBMGA003325 | 3.80 | N/A | |
| 41 | H9JXD9 | BGIBMGA014204 | 2.36 | N/A | |
| 42 | H9JPZ3 | BGIBMGA011598 | 2.39 | N/A | |
| 43 | H9J1D9 | BGIBMGA003326 | 2.33 | N/A | |
| DJ48/0 h | 1 | H9IW76 | BGIBMGA001507 | 5.27 | Lipase |
| 2 | C7EPE2 | BGIBMGA000158 | 5.42 | Glucose-methanol-choline (GMC) oxidoreductase activity | |
| 3 | H9JFH1 | BGIBMGA008268 | 3.06 | Aldehyde oxidase activity | |
| 4 | H9J8I4 | BGIBMGA005826 | 3.90 | Alcohol dehydrogenase activity | |
| 5 | H9JTY7 | BGIBMGA012999 | 2.94 | Glucose dehydrogenase activity | |
| 6 | H9ISL2 | BGIBMGA000239 | 3.87 | Peroxidase activity | |
| 7 | H9ISL1 | BGIBMGA000238 | 7.03 | Peroxidase activity | |
| 8 | H9JT78 | BGIBMGA012740 | 3.46 | Peroxidase activity | |
| 9 | H9IX93 | BGIBMGA001876 | 2.78 | Alpha-amylase activity | |
| 10 | Q86D78 | BGIBMGA003512 | 3.44 | Beta-glucosidase | |
| 11 | B2DD57 | BGIBMGA005696 | 2.81 | Glycosyl hydrolase activity | |
| 12 | H9JCF2 | BGIBMGA007153 | 3.18 | Glycosyl hydrolase activity | |
| 13 | A0A077JI83 | BGIBMGA006066 | 3.90 | O-Glycosyl hydrolase activity | |
| 14 | H9ISX5 | BGIBMGA000353 | 8.99 | Innate immune | |
| 15 | H9JPS6 | BGIBMGA011531 | 2.85 | Phospholipase C activity | |
| 16 | H9IVS0 | BGIBMGA001351 | 3.11 | Oxidoreductase activity | |
| 17 | B2ZZX0 | BGIBMGA008818 | 2.74 | Phosphatase activity | |
| 18 | Q9NGS0 | — | 3.05 | N/A | |
| 19 | P81902 | — | 3.16 | Trypsin inhibitor | |
| 20 | Q03383 | — | 3.14 | Antichymotrypsin | |
| 21 | C0J8G5 | BGIBMGA003292 | 7.33 | Serine protease inhibitor | |
| 22 | C0J8H1 | — | 12.35 | N/A | |
| 23 | C4B489 | BGIBMGA004445 | 2.19 | Serine protease activity | |
| 24 | H9JFI3 | BGIBMGA008280 | 3.17 | Serine protease activity | |
| 25 | H9J229 | BGIBMGA003566 | 2.04 | Serine protease activity | |
| 26 | H9JKH3 | BGIBMGA010024 | 2.37 | Serine protease activity | |
| 27 | H9JG67 | BGIBMGA008514 | 2.02 | Serine protease activity | |
| 28 | H9JY09 | BGIBMGA014427 | 2.44 | Serine protease activity | |
| 29 | H9J231 | BGIBMGA003568 | 2.72 | Serine protease activity | |
| 30 | H9JJ26 | BGIBMGA009527 | 2.64 | Serine protease activity | |
| 31 | H9JIY5 | BGIBMGA009486 | 2.33 | Carboxypeptidase activity | |
| 32 | H9JES0 | BGIBMGA008017 | 2.37 | AMP deaminase activity | |
| 33 | H9JEW2 | BGIBMGA008059 | 4.47 | Aminopeptidase activity | |
| 34 | H9JDE9 | BGIBMGA007546 | 3.86 | Carboxylesterase activity | |
| 35 | H9JE19 | BGIBMGA007766 | 2.35 | Phosphoric diester hydrolase activity | |
| 36 | B5TZ28 | BGIBMGA007915 | 3.13 | Metallopeptidase activity | |
| 37 | H9IWJ8 | BGIBMGA001630 | 2.01 | Cholinesterase activity | |
| 38 | H9JSJ8 | BGIBMGA012509 | 3.51 | Glucosinolate sulphatase activity | |
| 39 | H9J064 | BGIBMGA002899 | 2.18 | Carboxylesterase activity | |
| 40 | H9J067 | BGIBMGA002902 | 3.26 | Cholinesterase activity | |
| 41 | B2ZDZ0 | BGIBMGA009544 | 2.04 | Carboxylesterase activity | |
| 42 | C0SQ80 | BGIBMGA008354 | 2.00 | Odorant binding | |
| 43 | A1YQ87 | BGIBMGA005493 | 3.08 | Phosphopyruvate hydratase activity | |
| 44 | H9JP12 | BGIBMGA011266 | 3.08 | Insect hexamerins | |
| 45 | H9J128 | BGIBMGA003215 | 4.77 | RNA binding | |
| 46 | H9JLC5 | BGIBMGA010326 | 3.09 | Mitochondrial carriers | |
| 47 | H9IVY4 | BGIBMGA001415 | 3.89 | Ubiquitin-mediated protein binding | |
| 48 | H9IT95 | BGIBMGA000475 | 5.80 | Cation binding | |
| 49 | Q69FX2 | BGIBMGA008221 | 2.34 | Innate immunity and lipid metabolism | |
| 50 | S5M110 | BGIBMGA005577 | 2.25 | Carbohydrate derivative binding | |
| 51 | C1K001 | BGIBMGA004287 | 3.16 | N/A | |
| 52 | H9JYG4 | BGIBMGA009648 | 5.55 | DNA binding | |
| 53 | H9IYN2 | BGIBMGA002366 | 2.16 | N/A | |
| 54 | Q2F645 | BGIBMGA014211 | 2.03 | Transketolase activity | |
| 55 | H9J1D5 | BGIBMGA003322 | 4.40 | Juvenile hormone binding | |
| 56 | H9J5L9 | BGIBMGA004809 | 3.95 | Lyase activity | |
| 57 | H9J1D8 | BGIBMGA003325 | 4.85 | N/A | |
| 58 | H9JT75 | BGIBMGA012737 | 2.58 | Peroxidase activity | |
| 59 | H9JXD9 | BGIBMGA014204 | 2.02 | N/A | |
| 60 | H9JPZ3 | BGIBMGA011598 | 3.59 | N/A | |
| 61 | H9J3M9 | BGIBMGA004116 | 2.02 | Transferase activity | |
| 62 | H9J1D9 | BGIBMGA003326 | 8.57 | N/A | |
| 63 | H9JXN1 | BGIBMGA014298 | 12.23 | N/A | |
| DJ48/24 h | |||||
| 1 | H9IW76 | BGIBMGA001507 | 2.87 | Lipase | |
| 2 | C7EPE2 | BGIBMGA000158 | 4.05 | Glucose-methanol-choline (GMC) oxidoreductase activity | |
| 3 | H9JFH1 | BGIBMGA008268 | 2.12 | Aldehyde oxidase activity | |
| 4 | H9ISL2 | BGIBMGA000239 | 2.30 | Peroxidase activity | |
| 5 | H9ISL1 | BGIBMGA000238 | 4.67 | Peroxidase activity | |
| 6 | H9IX93 | BGIBMGA001876 | 2.52 | Alpha-amylase activity | |
| 7 | B2DD57 | BGIBMGA005696 | 2.01 | Glycosyl hydrolase activity | |
| 8 | A0A077JI83 | BGIBMGA006066 | 3.67 | O-Glycosyl hydrolase activity | |
| 9 | A1YQ87 | BGIBMGA005493 | 3.06 | Phosphopyruvate hydratase activity | |
| 10 | P81902 | — | 2.03 | Trypsin inhibitor | |
| 11 | C0J8H1 | — | 2.02 | N/A | |
| 12 | H9JKL1 | BGIBMGA010062 | 3.09 | Serine-type endopeptidase activity | |
| 13 | H9J5K7 | BGIBMGA004797 | 2.14 | Metallocarboxypeptidase activity | |
| 14 | H9JG67 | BGIBMGA008514 | 2.28 | Serine protease activity | |
| 15 | H9J5P0 | BGIBMGA004830 | 2.12 | Metallocarboxypeptidase activity | |
| 16 | H9JIY5 | BGIBMGA009486 | 2.41 | Carboxypeptidase activity | |
| 17 | H9JG68 | BGIBMGA008515 | 4.20 | Serine protease activity | |
| 18 | H9JHZ0 | BGIBMGA009138 | 2.00 | Aminopeptidase activity | |
| 19 | A7LIK7 | BGIBMGA004403 | 2.14 | 30K protein | |
| 20 | H9J128 | BGIBMGA003215 | 2.72 | RNA binding | |
| 21 | H9JLC5 | BGIBMGA010326 | 3.06 | Mitochondrial carriers | |
| 22 | H9IVY4 | BGIBMGA001415 | 2.24 | Ubiquitin-mediated protein binding | |
| 23 | H9JUE4 | BGIBMGA013157 | 3.54 | Ribosomal protein | |
| 24 | Q8N0P2 | BGIBMGA002381 | 3.43 | 70 kilodalton heat shock protein | |
| 25 | H9J5L9 | BGIBMGA004809 | 2.80 | Lyase activity | |
| 26 | H9J1D9 | BGIBMGA003326 | 3.69 | N/A | |
| 27 | H9JXN1 | BGIBMGA014298 | 6.68 | N/A |
| Number | Map_Name | Map_ID | Protein_ID | Definition | Fold Change |
|---|---|---|---|---|---|
| 1 | Galactose metabolism | map00480 | F8V3L0 | gamma-glutamyltranspeptidase [24] | 0.47 |
| 2 | Lysosome | map04142 | H9JSN7 | sphingomyelin phosphodiesterase [25] | 3.21 |
| 3 | Sphingolipid metabolism | map00600 | H9JSN7 | sphingomyelin phosphodiesterase [26] | 3.21 |
| 4 | Arachidonic acid metabolism | map00590 | F8V3L0 | gamma-glutamyltranspeptidase [24] | 0.47 |
| 5 | Taurine and hypotaurine metabolism | map00430 | F8V3L0 | gamma-glutamyltranspeptidase [24] | 0.47 |
| 6 | Neuroactive ligand-receptor interaction | map04080 | H9JL75 | trypsin [27] | 3.15 |
| 7 | Folate biosynthesis | map00790 | B2ZZX0 | alkaline phosphatase [28] | 4.18 |
| 8 | Thiamine metabolism | map00730 | B2ZZX0 | alkaline phosphatase [28] | 4.18 |
| 9 | Ubiquitin mediated proteolysis | map04120 | H9JK60 | ubiquitin-activating enzyme E1 [29] | 2.05 |
| 10 | Caffeine metabolism | map00232 | H9JFX4 | xanthine dehydrogenase/oxidase [30] | 0.28 |
| Number | Map_Name | Map_ID | Protein_ID | Definition | Fold Change |
|---|---|---|---|---|---|
| 1 | Folate biosynthesis | map00790 | H9IVS0 | aldehyde reductase [31] | 3.11 |
| B2ZZX0 | alkaline phosphatase [28] | 4.18 | |||
| H9JTG9 | aldehyde reductase [32] | 0.50 | |||
| H9IVT6 | aldehyde reductase | 0.06 | |||
| 2 | Lysosome | map04142 | Q69FX2 | niemann-Pick C2 protein [33,34] | 2.34 |
| A4PHN6 | hexosaminidase [35] | 0.50 | |||
| H9IWR3 | lysosomal alpha-mannosidase [36] | 0.47 | |||
| 3 | Galactose metabolism | map00052 | H9IVS0 | aldehyde reductase [37] | 3.11 |
| H9JTG9 | aldehyde reductase [32] | 0.50 | |||
| H9IVT6 | aldehyde reductase | 0.06 | |||
| 4 | Fructose and mannose metabolism | map00051 | H9IVS0 | aldehyde reductase [37] | 3.11 |
| H9JTG9 | aldehyde reductase [32] | 0.50 | |||
| H9IVT6 | aldehyde reductase | 0.06 | |||
| 5 | Pentose and glucuronate interconversions | map00040 | H9IVS0 | aldehyde reductase [37] | 3.11 |
| H9JTG9 | aldehyde reductase [32] | 0.50 | |||
| H9IVT6 | aldehyde reductase | 0.06 | |||
| 6 | Glycerolipid metabolism | map00561 | H9IVS0 | aldehyde reductase [37] | 3.11 |
| H9JTG9 | aldehyde reductase [32] | 0.50 | |||
| H9IVT6 | aldehyde reductase | 0.06 | |||
| 7 | Purine metabolism | map00230 | H9JFH1 | xanthine dehydrogenase/oxidase [30] | 3.06 |
| H9JDV4 | nucleoside-diphosphate kinase [38] | 0.26 | |||
| H9JFX4 | xanthine dehydrogenase/oxidase [30] | 0.38 | |||
| 8 | Peroxisome | map04146 | H9JFH1 | xanthine dehydrogenase/oxidase [30] | 3.06 |
| Q08J22 | superoxide dismutase, Cu-Zn family [39,40] | 0.33 | |||
| H9JFX4 | xanthine dehydrogenase/oxidase [30] | 0.38 | |||
| 9 | Starch and sucrose metabolism | map00500 | H9IX93 | alpha-amylase [41] | 2.78 |
| H9J822 | alpha-trehalase [42] | 0.42 | |||
| 10 | Glutathione metabolism | map00480 | H9JES0 | aminopeptidase N [43] | 2.37 |
| H9JEW2 | aminopeptidase N [43] | 4.47 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, P.; Zhang, M.; Qian, P.; Li, J.; Wang, X.; Wu, Y. iTRAQ-Based Quantitative Proteomic Analysis of Digestive Juice across the First 48 Hours of the Fifth Instar in Silkworm Larvae. Int. J. Mol. Sci. 2019, 20, 6113. https://doi.org/10.3390/ijms20246113
Xu P, Zhang M, Qian P, Li J, Wang X, Wu Y. iTRAQ-Based Quantitative Proteomic Analysis of Digestive Juice across the First 48 Hours of the Fifth Instar in Silkworm Larvae. International Journal of Molecular Sciences. 2019; 20(24):6113. https://doi.org/10.3390/ijms20246113
Chicago/Turabian StyleXu, Pingzhen, Meirong Zhang, Ping Qian, Jiawei Li, Xueyang Wang, and Yangchun Wu. 2019. "iTRAQ-Based Quantitative Proteomic Analysis of Digestive Juice across the First 48 Hours of the Fifth Instar in Silkworm Larvae" International Journal of Molecular Sciences 20, no. 24: 6113. https://doi.org/10.3390/ijms20246113
APA StyleXu, P., Zhang, M., Qian, P., Li, J., Wang, X., & Wu, Y. (2019). iTRAQ-Based Quantitative Proteomic Analysis of Digestive Juice across the First 48 Hours of the Fifth Instar in Silkworm Larvae. International Journal of Molecular Sciences, 20(24), 6113. https://doi.org/10.3390/ijms20246113




