Proteome and Ubiquitome Changes during Rose Petal Senescence
Abstract
:1. Introduction
2. Results and Discussion
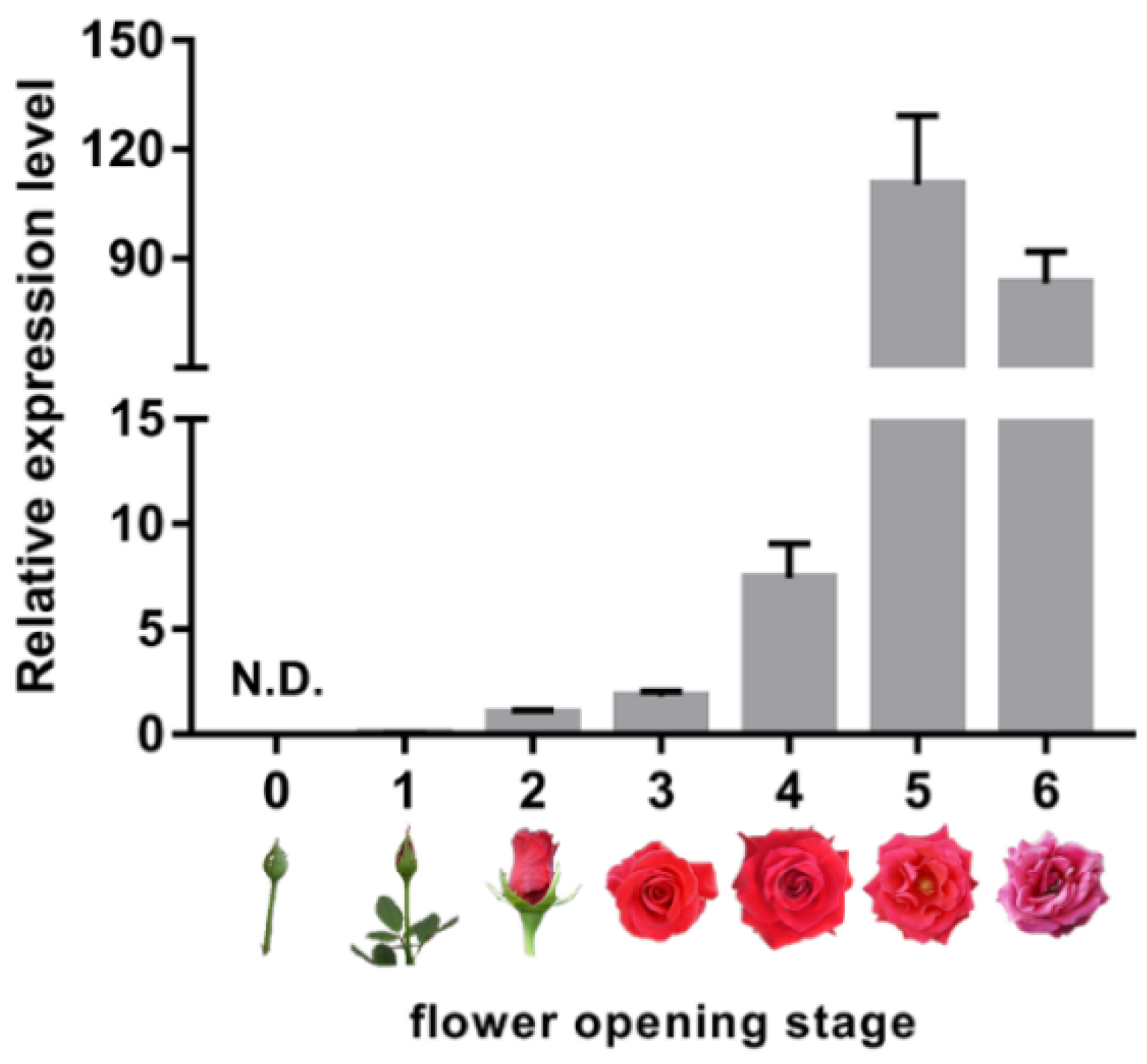
2.1. Expression of Senescence Marker RhSAG12 is Dramatically Increased from Flower Opening Stage 3 to Stage 5
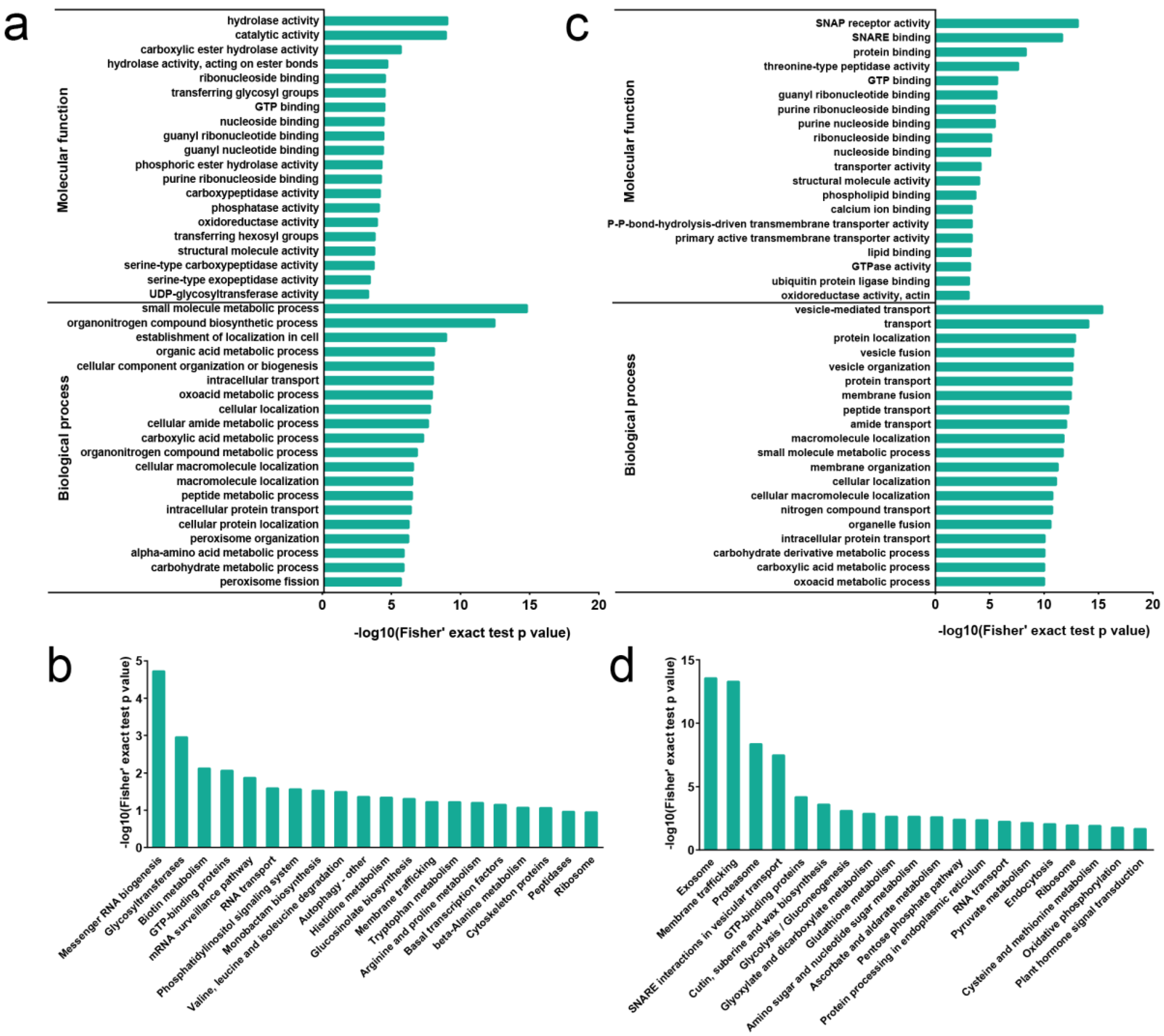
2.2. Proteome Profile in Rose Senesced Petal
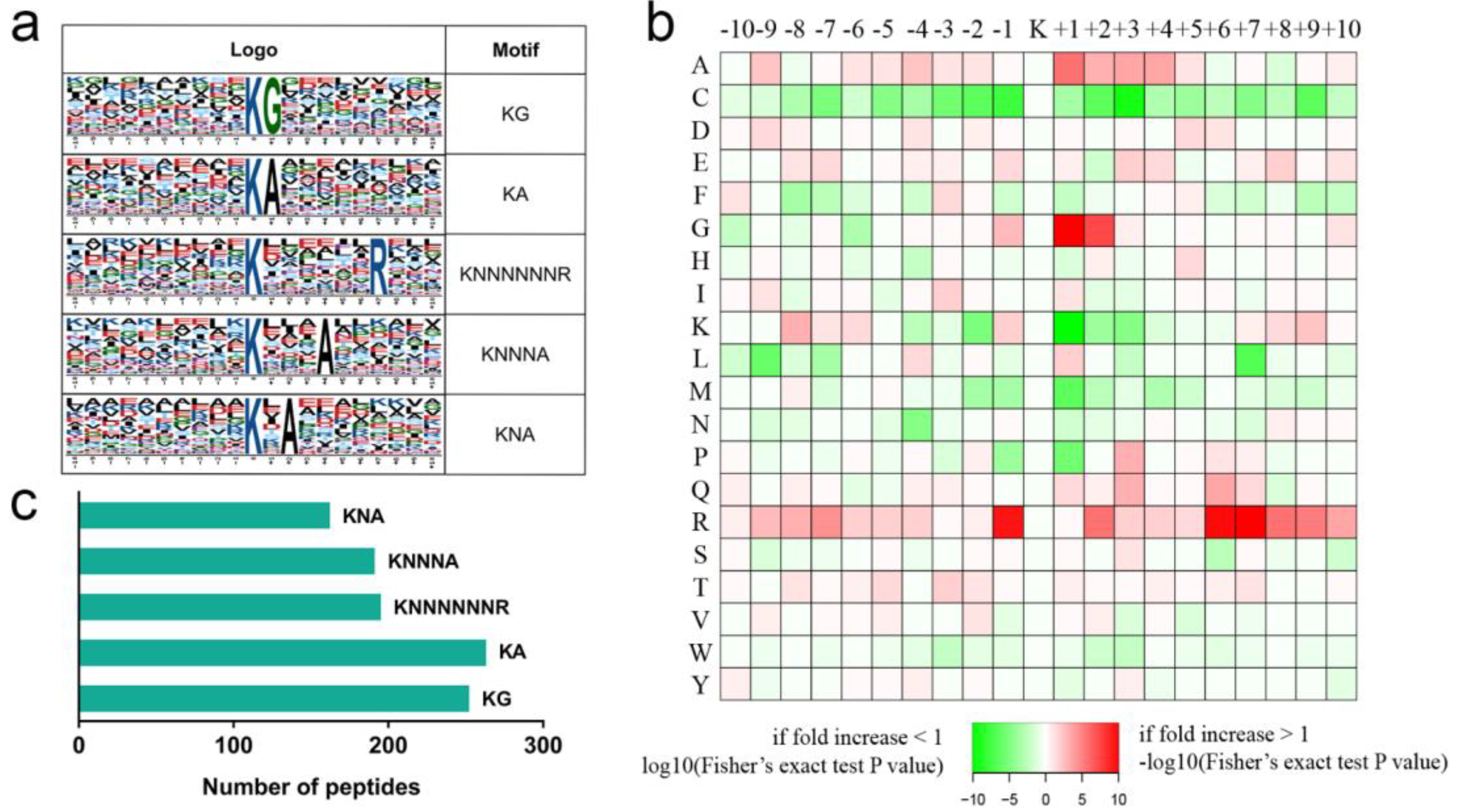
2.3. Ubiquitome Profile in Rose Senesced Petals
2.4. Association between the Global Proteome and Ubiquitome
2.5. Transcription Factors and Protein Kinases
2.6. Proteasome and Non-Proteasome Pathways
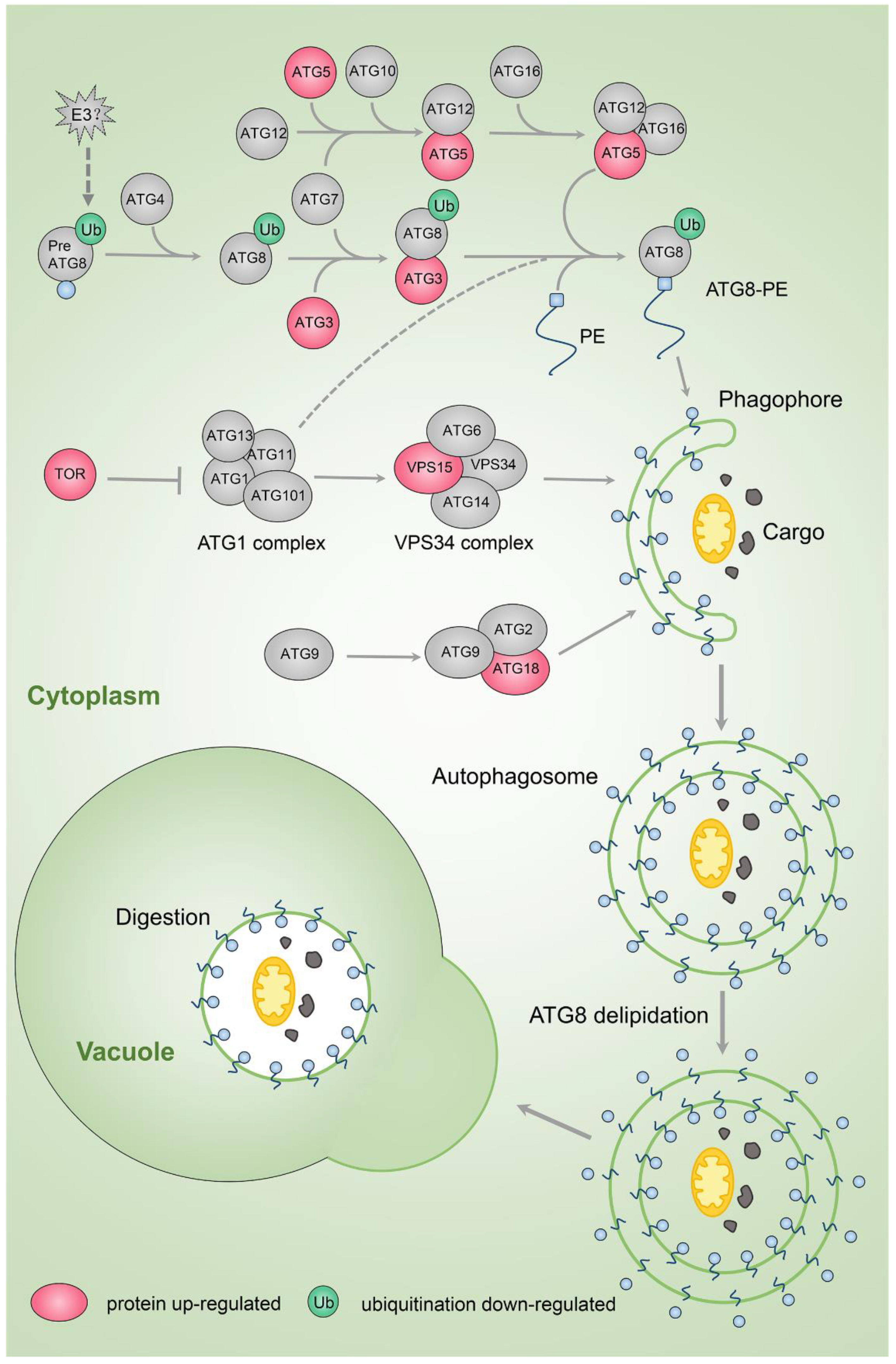
2.7. Autophagy Pathway
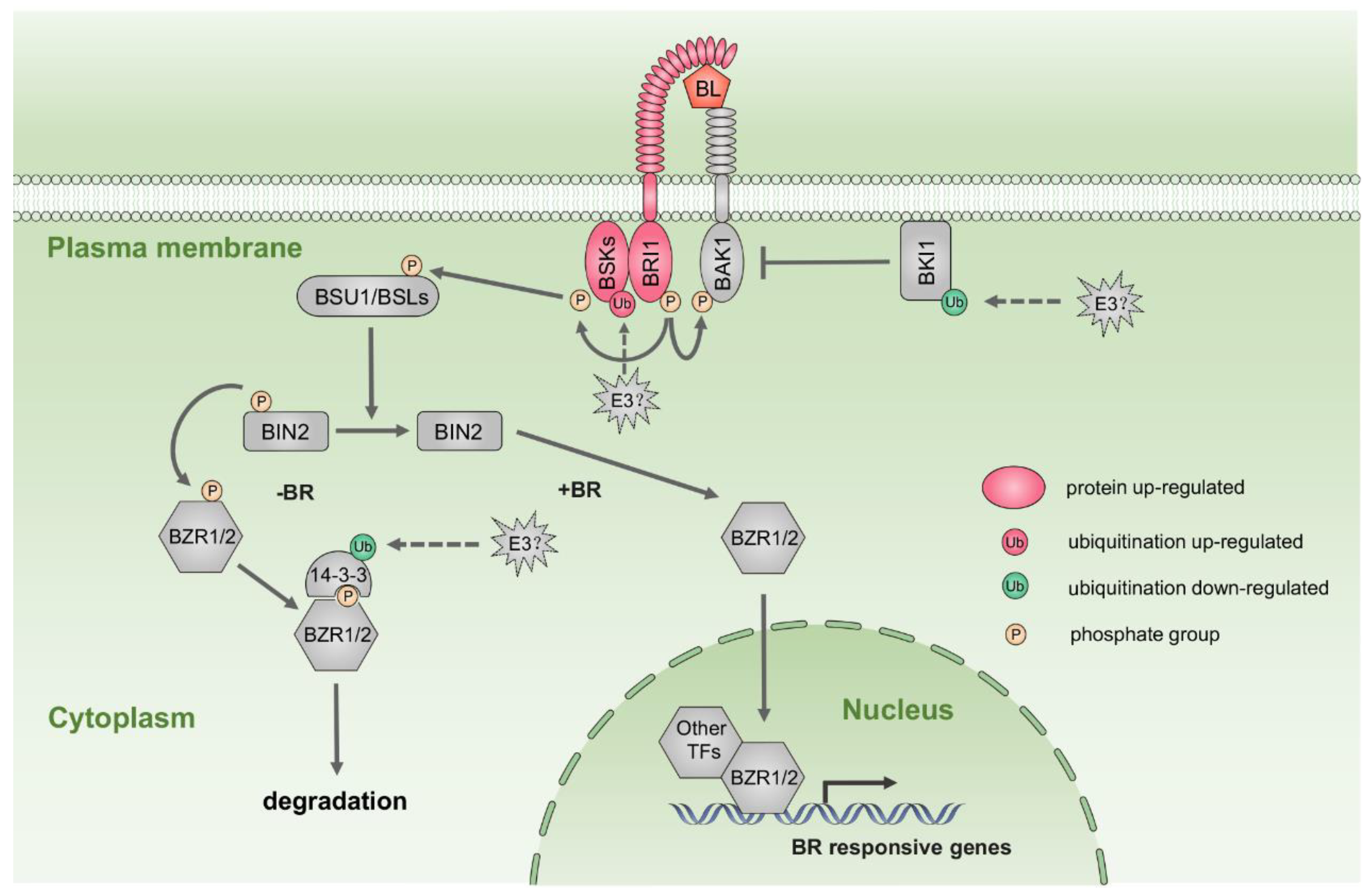
2.8. Hormone Biosynthesis and Signaling Pathways
2.9. Transporter Activity
3. Conclusions
4. Materials and Methods
4.1. Plant Material
4.2. Total RNA Extraction and Quantitative RT-PCR
4.3. Protein Extraction
4.4. Trypsin Digestion
4.5. HPLC Fractionation
4.6. Affinity Enrichment
4.7. LC-MS/MS Analysis
4.8. Database Search
4.9. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van Doorn, W.G. Categories of petal senescence and abscission: A re-evaluation. Ann. Bot. 2001, 87, 447–456. [Google Scholar] [CrossRef]
- Van Doorn, W.G.; Woltering, E.J. Physiology and molecular biology of petal senescence. J. Exp. Bot. 2008, 59, 453–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; Ma, C.; Liu, Y.; Shahid, M.O.; Wang, C.P.; Gao, J.P. Petal senescence: A hormone view. J. Exp. Bot. 2018, 69, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Shahri, W.; Tahir, I. Flower senescence: Some molecular aspects. Planta 2014, 239, 277–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.L.; Chaffin, G.S.; Eason, J.R.; Clark, D.G. Ethylene-sensitivity regulates proteolytic activity and cysteine protease gene expression in petunia corollas. J. Exp. Bot. 2005, 56, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, P.; Rubinstein, B. Characterization of proteolytic activity during senescence in daylilies. Physiol. Plant 1998, 104, 463–473. [Google Scholar] [CrossRef]
- Pak, C.; van Doorn, W.G. Delay of Iris flower senescence by protease inhibitors. New Phytol. 2005, 165, 473–480. [Google Scholar] [CrossRef]
- Borochov, A.; Halevy, A.H.; Shinitzky, M. Senescence and the fluidity of rose petal membranes: Relationship to phospholipid metabolism. Plant Physiol. 1982, 69, 296–299. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.E.; Froese, C.D.; Madey, E.; Smith, M.D.; Hong, Y.W. Lipid metabolism during plant senescence. Prog. Lipid Res. 1998, 37, 119–141. [Google Scholar] [CrossRef]
- Suttle, J.C.; Kende, H. Ethylene action and loss of membrane integrity during petal senescence in Tradescantia. Plant Physiol. 1980, 65, 1067–1072. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.L. Mineral nutrient remobilization during corolla senescence in ethylene-sensitive and -insensitive flowers. AoB Plants 2013, 5, plt023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Woo, H.R.; Nam, H.G. Toward systems understanding of leaf senescence: An integrated multi-omics perspective on leaf senescence research. Mol. Plant 2016, 9, 813–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panavas, T.; Pikula, A.; Reid, P.D.; Rubinstein, B.; Walker, E.L. Identification of senescence-associated genes from daylily petals. Plant Mol. Biol. 1999, 40, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G.; Balk, P.; Van Houwelingen, A.; Hoeberichts, F.; Hall, R.; Vorst, O.; Van Der Schoot, C.; Van Wordragen, M. Gene expression during anthesis and senescence in Iris flowers. Plant Mol. Biol. 2003, 53, 845–863. [Google Scholar] [CrossRef]
- Hoeberichts, F.A.; Van Doorn, W.G.; Vorst, O.; Hall, R.D.; Van Wordragen, M.F. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J. Exp. Bot. 2007, 58, 2873–2885. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Chang, X.X.; Lin, J.; Chang, Y.H.; Chen, J.C.; Reid, M.S.; Jiang, C.Z. Transcriptome profiling reveals regulatory mechanisms underlying corolla senescence in petunia. Hortic. Res. 2018, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef]
- Sullivan, J.A.; Shirasu, K.; Deng, X.W. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet. 2003, 4, 948–958. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.D. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 2000, 11, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007, 315, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.H.; Liu, J.X.; Wei, Q.; Wang, R.M.; Yang, W.Y.; Ma, Y.Y.; Chen, G.J.; Yu, Y.X. Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiol. 2017, 173, 668–687. [Google Scholar] [CrossRef]
- Ma, N.; Cai, L.; Lu, W.J.; Tan, H.; Gao, J.P. Exogenous ethylene influences flower opening of cut roses (Rosa hybrida) by regulating the genes encoding ethylene biosynthesis enzymes. Sci. China Ser. C: Life Sci. 2005, 48, 434–444. [Google Scholar] [CrossRef]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Kang, H.X.; Liu, W.D.; Wang, G.L. Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 2015, 14, 2017–2025. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, L.R.; Shi, C.N.; Tian, Q.Z.; Lv, G.G.; Wang, Y.; Cui, D.Q.; Chen, F. Comprehensive profiling of lysine ubiquitome reveals diverse functions of lysine ubiquitination in common wheat. Sci. Rep. 2017, 7, 13601. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.L.; Xie, X.; Wu, L.Y.; Liu, C.Y.; Zeng, L.R.; Zhou, X.P.; Luo, F.; Wang, G.L.; Liu, W.D. Proteomic analysis of ubiquitinated proteins in Rice (Oryza sativa) after treatment with pathogen-associated molecular pattern (PAMP) elicitors. Front. Plant Sci. 2018, 9, 1064. [Google Scholar] [CrossRef] [Green Version]
- Shabek, N.; Zheng, N. Plant ubiquitin ligases as signaling hubs. Nat. Struct. Mol. Biol. 2014, 21, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Schippers, J.H. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Chang, X.X.; Kasuga, T.; Bui, M.; Reid, M.S.; Jiang, C.Z. A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Hortic. Res. 2015, 2, 15059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prinsi, B.; Negri, A.S.; Quattrocchio, F.M.; Koes, R.E.; Espen, L. Proteomics of red and white corolla limbs in petunia reveals a novel function of the anthocyanin regulator ANTHOCYANIN1 in determining flower longevity. J. Proteom. 2016, 131, 38–47. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Liang, G.; Yang, S.Z.; Yu, D.Q. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, K.; Shimizu, K.; Niki, T.; Ichimura, K. Identification of a NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J. 2014, 79, 1044–1051. [Google Scholar] [CrossRef]
- Lü, P.T.; Zhang, C.Q.; Liu, J.T.; Liu, X.W.; Jiang, G.M.; Jiang, X.Q.; Khan, M.A.; Wang, L.S.; Hong, B.; Gao, J.P. RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J. 2014, 78, 578–590. [Google Scholar] [CrossRef]
- Wu, L.; Ma, N.; Jia, Y.C.; Zhang, Y.; Feng, M.; Jiang, C.Z.; Ma, C.; Gao, J.P. An Ethylene-Induced Regulatory Module Delays Flower Senescence by Regulating Cytokinin Content. Plant Physiol. 2017, 173, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Chang, X.x.; Donnelly, L.; Sun, D.Y.; Rao, J.P.; Reid, M.S.; Jiang, C.z. A petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence. PLoS ONE 2014, 9, e88320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, B.S.; Yao, T.; Seo, J.S.; Wong, E.C.C.; Mitsuda, N.; Huang, C.H.; Chua, N.H. Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 2018, 4, 898–903. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 2018. [Google Scholar] [CrossRef]
- Xiao, D.; Cui, Y.J.; Xu, F.; Xu, X.X.; Gao, G.X.; Wang, Y.X.; Guo, Z.X.; Wang, D.; Wang, N.N. SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE directly interacts with the cytoplasmic domain of SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE and negatively regulates leaf senescence in Arabidopsis. Plant Physiol. 2015, 169, 1275–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Meng, T.; Li, P.L.; Yu, Y.Q.; Cui, Y.J.; Wang, Y.X.; Gong, Q.Q.; Wang, N.N. A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 2011, 157, 2131–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.C.; Hong, S.W.; Whang, S.s.; Lim, P.O.; Nam, H.G.; Koo, J.C. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.J.; Wuriyanghan, H.; Zhang, Y.Q.; Duan, K.X.; Chen, H.W.; Li, Q.T.; Lu, X.; He, S.J.; Ma, B.; Zhang, W.K.; et al. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.J.; Cai, Z.H.; Guo, Y.F.; Gan, S.S. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009, 150, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chang, Y.; Zhao, C.C.; Yang, H.L.; Ren, D.T. Expression of the inactive ZmMEK1 induces salicylic acid accumulation and salicylic acid-dependent leaf senescence. J. Integr. Plant. Biol. 2016, 58, 724–736. [Google Scholar] [CrossRef] [Green Version]
- Schippers, J.H.; Schmidt, R.; Wagstaff, C.; Jing, H.C. Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol. 2015, 169, 914–930. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.J.; Jiang, C.z.; Donnelly, L.; Reid, M.S. Functional analysis of a RING domain ankyrin repeat protein that is highly expressed during flower senescence. J. Exp. Bot. 2007, 58, 3623–3630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, T.; Ichimura, K.; Kanekatsu, M.; van Doorn, W.G. Gene expression in opening and senescing petals of morning glory (Ipomoea nil) flowers. Plant Cell Rep. 2007, 26, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.Y.; Willard, B.; Chapin, L.J.; Kinter, M.T.; Francis, D.M.; Stead, A.D.; Jones, M.L. Proteomic analysis of pollination-induced corolla senescence in petunia. J. Exp. Bot. 2010, 61, 1089–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eason, J.R.; Ryan, D.J.; Pinkney, T.T.; O’Donoghue, E.M. Programmed cell death during flower senescence: Isolation and characterization of cysteine proteinases from Sandersonia aurantiaca. Funct. Plant Biol. 2002, 29, 1055–1064. [Google Scholar] [CrossRef]
- Jones, M.L.; Larsen, P.B.; Woodson, W.R. Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Mol. Biol. 1995, 28, 505–512. [Google Scholar] [CrossRef]
- Breeze, E.; Wagstaff, C.; Harrison, E.; Bramke, I.; Rogers, H.; Stead, A.; Thomas, B.; Buchanan-Wollaston, V. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnol. J. 2004, 2, 155–168. [Google Scholar] [CrossRef]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.X.; Zhang, X.G.; Otegui, M.S. Plant autophagy: New flavors on the menu. Curr. Opin. Plant Biol. 2018, 46, 113–121. [Google Scholar] [CrossRef]
- Chiba, A.; Ishida, H.; Nishizawa, N.K.; Makino, A.; Mae, T. Exclusion of ribulose-1, 5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol. 2003, 44, 914–921. [Google Scholar] [CrossRef] [Green Version]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.-S.; Penfold, C.A.; Jenkins, D. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 873–894. [Google Scholar] [CrossRef] [Green Version]
- Yamada, T.; Ichimura, K.; Kanekatsu, M.; van Doorn, W.G. Homologs of genes associated with programmed cell death in animal cells are differentially expressed during senescence of Ipomoea nil petals. Plant Cell Physiol. 2009, 50, 610–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibuya, K.; Shimizu, K.; Yamada, T.; Ichimura, K. Expression of autophagy-associated ATG8 genes during petal senescence in Japanese morning glory. J. Jpn. Soc. Hortic. Sci. 2011, 80, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, K.; Niki, T.; Ichimura, K. Pollination induces autophagy in petunia petals via ethylene. J. Exp. Bot. 2013, 64, 1111–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.W.; Wang, Z.Y. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 2010, 61, 681–704. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.H.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.M.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Clouse, S.D.; Sasse, J.M. Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef] [Green Version]
- He, Y.J.; Xu, R.J.; Zhao, Y.J. Enhancement of senescence by epibrassinolide in leaves of mung bean seedling. Acta Phytophysiol. Sin. 1996, 22, 58–62. [Google Scholar]
- de Assis-Gomes, M.D.M.; Pinheiro, D.T.; Bressan-Smith, R.; Campostrini, E. Exogenous brassinosteroid application delays senescence and promotes hyponasty in Carica papaya L. leaves. Theor. Exp. Plant Physiol. 2018, 30, 193–201. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [Green Version]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Dave, A.; Graham, I.A. Oxylipin signaling: A distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front. Plant Sci. 2012, 3, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef] [PubMed]
- He, Y.H.; Fukushige, H.; Hildebrand, D.F.; Gan, S.S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, S.; Dai, L.Y.; Liu, F.Q.; Wang, Z.L.; Peng, W.; Xie, D.X. COS1: An Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 2004, 16, 1132–1142. [Google Scholar] [CrossRef] [Green Version]
- Van Doorn, W.G.; Celikel, F.G.; Pak, C.; Harkema, H. Delay of iris flower senescence by cytokinins and jasmonates. Physiol. Plant 2013, 148, 105–120. [Google Scholar] [CrossRef]
- Winkenbach, F. Zum Stoffwechsel der aufblühenden und welkenden Korolle der Prunkwinde Ipomoea purpurea. I. Beziehungen zwischen Gestaltwandel, Stofftransport, Atmung und Invertaseaktivität. Ber. Schweiz. Bot. Ges. 1970, 80, 374–390. [Google Scholar]
- Verlinden, S. Changes in mineral nutrient concentrations in petunia corollas during development and senescence. HortScience 2003, 38, 71–74. [Google Scholar] [CrossRef]
- Chapin, L.J.; Jones, M.L. Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during petunia corolla senescence. J. Exp. Bot. 2009, 60, 2179–2190. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.T.; Fan, Y.W.; Zou, J.; Fang, Y.Q.; Wang, L.H.; Wang, M.; Jiang, X.Q.; Liu, Y.Q.; Gao, J.P.; Zhang, C.Q. A RhABF2/Ferritin module affects rose (Rosa hybrida) petal dehydration tolerance and senescence by modulating iron levels. Plant J. 2017, 92, 1157–1169. [Google Scholar] [CrossRef] [Green Version]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Xue, J.Q.; Li, Y.H.; Liu, X.J.; Dai, F.W.; Jia, W.S.; Luo, Y.B.; Gao, J.P. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol. 2008, 148, 894–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Y.; Li, N.; Tian, J.; Gao, J.P.; Zhang, C.Q. Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci. Hortic. 2013, 158, 16–21. [Google Scholar] [CrossRef] [Green Version]

| Protein Accession | Protein Description | Mol. Weight (kDa) |
|---|---|---|
| XP_024163091.1 | Copper transporter 5.1 | 15.9 |
| XP_024180604.1 | Potassium transporter 4 | 87.8 |
| XP_024185106.1 | Magnesium transporter MRS2-1 | 50.6 |
| XP_024169378.1 | Molybdate-anion transporter-like | 50.6 |
| XP_024165719.1 | Sugar transporter ERD6-like 7 | 36.8 |
| XP_024166942.1 | Auxin transporter-like protein 3 LAX3 | 52.9 |
| XP_024170862.1 | Oligopeptide transporter 7 | 84.5 |
| XP_024173227.1 | GDP-mannose transporter GONST3 | 59.0 |
| XP_024183764.1 | Oligopeptide transporter 4-like | 83.0 |
| XP_024165085.1 | Lysine histidine transporter-like 8 | 58.6 |
| XP_024182501.1 | Lysine histidine transporter-like 8 | 60.4 |
| XP_024179150.1 | ABC transporter I family member 1 | 25.7 |
| XP_024160504.1 | ABC transporter B family member 25 | 79.3 |
| XP_024158019.1 | ABC transporter A family member 2-like | 106.4 |
| XP_024167415.1 | ABC transporter C family member 5 | 170.6 |
| XP_024189449.1 | Putative ABC transporter C family member 15 isoform X1 | 169.6 |
| XP_024190413.1 | ABC transporter G family member 15-like | 75.4 |
| XP_024191990.1 | ABC transporter I family member 17 isoform X2 | 29.5 |
| Protein Accession | Position | Protein Description | Modified Sequence | Regulation Type |
|---|---|---|---|---|
| XP_024166523.1 | 1271 | ABC transporter C family member 14-like | APPTNWPTHGNVELK(1)DLQVR | Up |
| XP_024167390.1 | 215 | Sugar transporter ERD6-like 7 | HK(1)EFEVALQK | Up |
| XP_024196922.1 | 254 | Equilibrative nucleotide transporter 1 | ELK(1)GPLTGSVLR | Up |
| XP_024158774.1 | 1314 | ABC transporter C family member 2-like | ILIDGCDIGK(1)FGLEDLRK | Up |
| XP_024176720.1 | 876 | ABC transporter C family member 10-like | LK(1)GNKGDQLIK | Up |
| XP_024176720.1 | 879 | ABC transporter C family member 10-like | GNK(1)GDQLIK | Up |
| XP_024163367.1 | 603 | Sulfate transporter 3.1-like isoform X3 | FMLHTTK(0.997)SDPVK(0.003)EEPGTWNNV | Up |
| XP_024186868.1 | 547 | ABC transporter B family member 1 | VANAHSFIVK(1)LPDGFDTQVGER | Up |
| XP_024173043.1 | 1356 | ABC transporter C family member 10-like | K(0.986)AIEEK(0.014)EEGLDTFVVQDGTNWSTGQR | Up |
| XP_024176720.1 | 632 | ABC transporter C family member 10-like | SASFSWDNLSK(1)ATLR | Up |
| XP_024189449.1 | 725 | Putative ABC transporter C family member 15 isoform X1 | ENILFGNAYDK(1)AK | Up |
| XP_024165536.1 | 194 | Bidirectional sugar transporter SWEET1-like isoform X2 | QQPSAEDNVELGLEK(1)PHQSK | Down |
| XP_024191501.1 | 1219 | ABC transporter D family member 1 isoform X2 | LYQEGGK(1)FDDSTNILDMR | Down |
| XP_024181861.1 | 1066 | ABC transporter B family member 19 | FYDPIVGK(1)VMIDGK | Down |
| XP_024158774.1 | 1478 | ABC transporter C family member 2-like | SAFSK(1)MVQSTGAANAQYLR | Down |
| XP_024194123.1 | 478 | Ammonium transporter 1 member 1 | HGGFAYVYHDEDDAGK(1)PAGIQLR | Down |
| XP_024165085.1 | 332 | Lysine histidine transporter-like 8 | GHNLILEIQATMPSSEK(1)HPSR | Down |
| XP_024176551.1 | 64 | Copper transport protein ATX1-like | K(1)TAYWEAEAPAEPEAK | Down |
| XP_024169378.1 | 441 | Molybdate-anion transporter-like | LFVITDGHK(1)SK | Down |
| XP_024194192.1 | 4 | Aluminum-activated malate transporter 4 | AAK(1)IGSFR | Down |
| XP_024163410.1 | 64 | ABC transporter E family member 2 | K(1)CPFEAIQIINLPK | Down |
| XP_024165658.1 | 153 | Zinc transporter 4, chloroplastic-like | K(1)QGLARPTEDQVR | Down |
| XP_024166097.1 | 99 | Amino acid transporter AVT3B-like | KLESPDAPTK(1)IASFGDLGFR | Down |
| XP_024176551.1 | 46 | Copper transport protein ATX1-like | VTVK(1)GNVPPETVLQTVTK | Down |
| XP_024176711.1 | 886 | ABC transporter C family member 10-like | GDQLIK(1)LEER | Down |
| XP_024180604.1 | 145 | Potassium transporter 4 | YGPSSQVAASSPLK(1)R | Down |
| XP_024180746.1 | 363 | ABC transporter G family member 11-like | SYK(1)SSENYHQLQR | Down |
| XP_024181557.1 | 442 | Copper transport protein CCH-like | DTIPSNFQK(1)R | Down |
| XP_024186868.1 | 1208 | ABC transporter B family member 1 | FVSALPDGYK(1)TFVGER | Down |
| XP_024186868.1 | 731 | ABC transporter B family member 1 | LEK(1)LAFK | Down |
| XP_024186868.1 | 66 | ABC transporter B family member 1 | K(0.824)ESNDSGGGEK(0.176)PEAVPSIGFGEVFR | Down |
| XP_024186868.1 | 76 | ABC transporter B family member 1 | ESNDSGGGEK(1)PEAVPSIGFGEVFR | Down |
| XP_024192899.1 | 4 | Sucrose transport protein SUC3 | AGK(1)TDSVSIR | Down |
| XP_024193193.1 | 198 | Putative potassium transporter 12 | LKLPTPELK(1)R | Down |
| XP_024194123.1 | 486 | Ammonium transporter 1 member 1 | K(1)VEPNSSTPNSV | Down |
| XP_024198608.1 | 337 | ABC transporter F family member 1 | FGHGSAK(1)LAR | Down |
| XP_024198608.1 | 192 | ABC transporter F family member 1 | LEALDAATAEK(1)R | Down |
| XP_024198608.1 | 325 | ABC transporter F family member 1 | WEQEQIANMK(1)EYIAR | Down |
| XP_024177255.1 | 253 | Probable aquaporin PIP-type 7a | SLGAAIIFNK(1)DR | Down |
| XP_024177255.1 | 14 | Probable aquaporin PIP-type 7a | MQAK(1)EEDVSLGANK(1)FPER | Down |
| XP_024175740.1 | 14 | Aquaporin PIP1-3 | LGANK(1)FSER | Down |
| XP_024166688.1 | 269 | Aquaporin PIP2-7 | AAAIK(1)ALGSFR | Down |
| XP_024166688.1 | 3 | Aquaporin PIP2-7 | TK(1)EVSEEPQAHHHK | Down |
| XP_024193331.1 | 204 | Auxin efflux carrier component 3-like PIN3-like | DFLETDAEIGDDGK(1)LHVK | Down |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Xu, Y.; Fan, Y.; Wang, Y.; Zhang, G.; Liang, Y.; Jiang, C.; Hong, B.; Gao, J.; Ma, C. Proteome and Ubiquitome Changes during Rose Petal Senescence. Int. J. Mol. Sci. 2019, 20, 6108. https://doi.org/10.3390/ijms20246108
Lu J, Xu Y, Fan Y, Wang Y, Zhang G, Liang Y, Jiang C, Hong B, Gao J, Ma C. Proteome and Ubiquitome Changes during Rose Petal Senescence. International Journal of Molecular Sciences. 2019; 20(24):6108. https://doi.org/10.3390/ijms20246108
Chicago/Turabian StyleLu, Jingyun, Yanjie Xu, Youwei Fan, Yaru Wang, Guifang Zhang, Yue Liang, Chuyan Jiang, Bo Hong, Junping Gao, and Chao Ma. 2019. "Proteome and Ubiquitome Changes during Rose Petal Senescence" International Journal of Molecular Sciences 20, no. 24: 6108. https://doi.org/10.3390/ijms20246108
APA StyleLu, J., Xu, Y., Fan, Y., Wang, Y., Zhang, G., Liang, Y., Jiang, C., Hong, B., Gao, J., & Ma, C. (2019). Proteome and Ubiquitome Changes during Rose Petal Senescence. International Journal of Molecular Sciences, 20(24), 6108. https://doi.org/10.3390/ijms20246108






