Genetic and Functional Diversity of Nitrilases in Agaricomycotina
Abstract
1. Introduction
2. Results
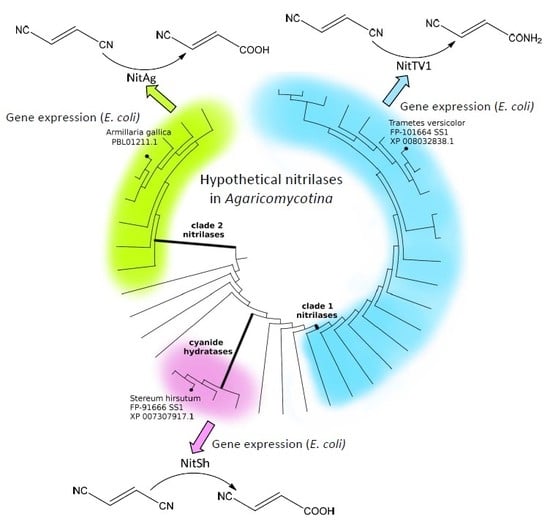
2.1. Occurrence of Putative Nitrilases (NLases) in Agaricomycotina
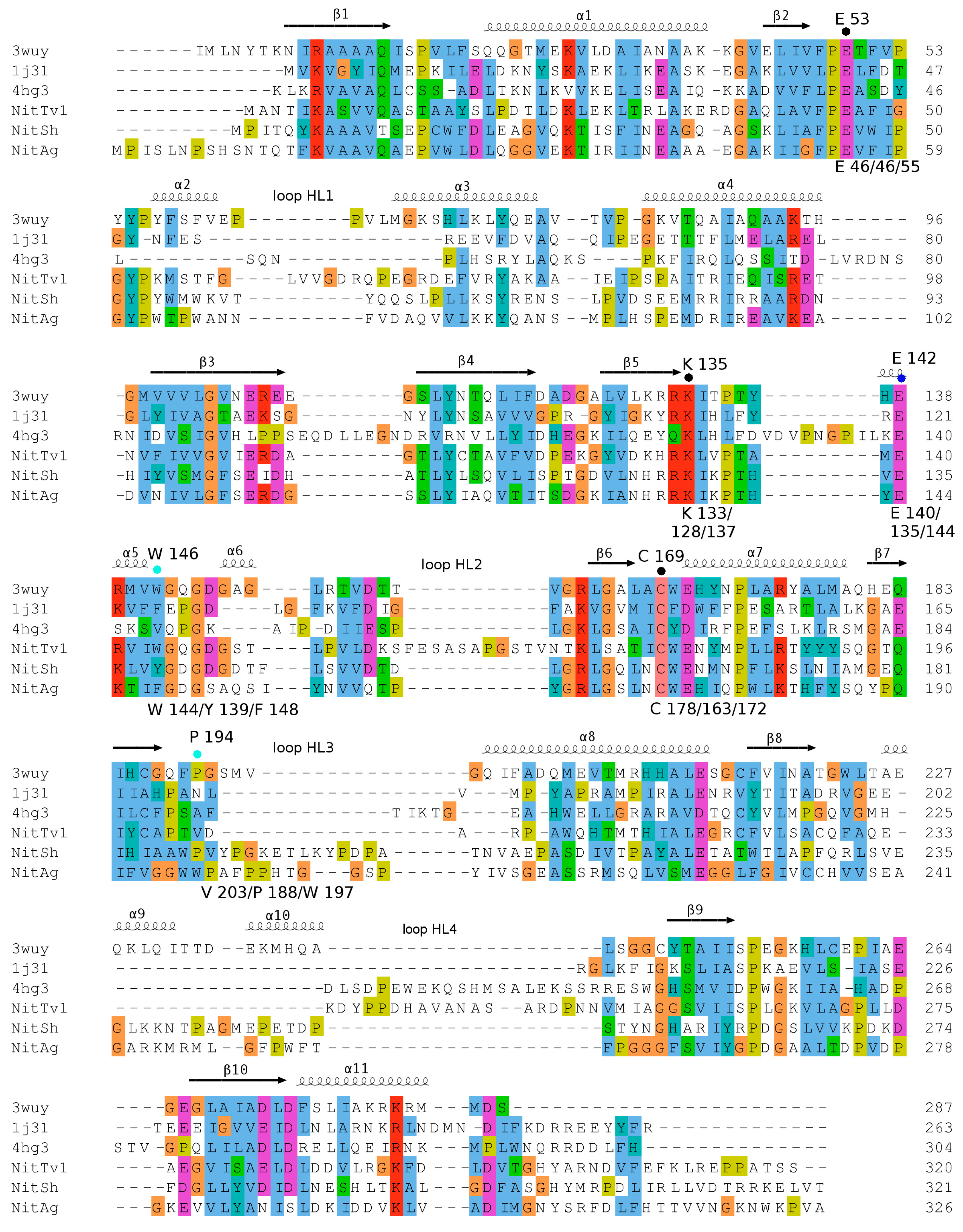
2.2. Selection of Nitrilases for Overproduction, Multiple Alignment and Substrate Specificity Predictions
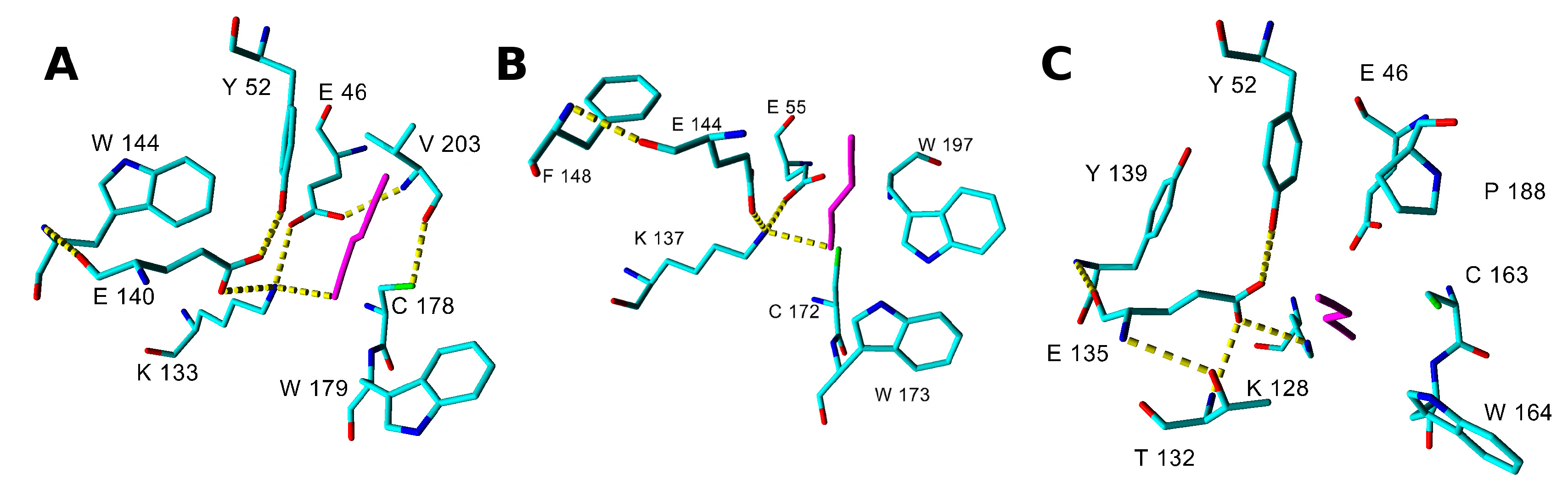
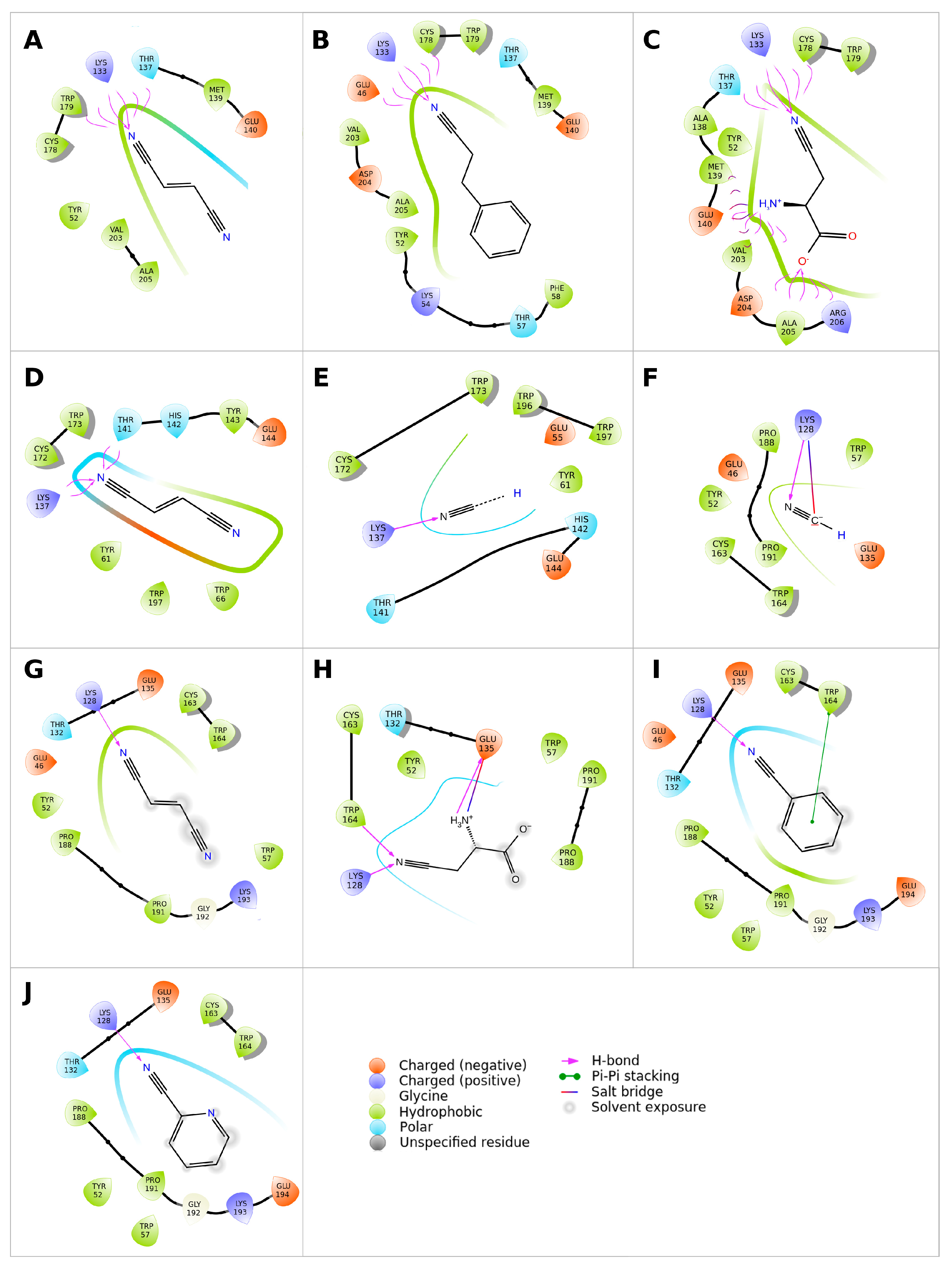
2.3. Modeling Substrate Interaction with the Active Site Residues
2.4. Determination of Nitrilase Activities in E. coli Cells
2.5. Phylogenetic Distribution of NLases in Basidiomycota
3. Discussion
4. Materials and Methods
4.1. Sequence Analysis
4.2. Nitrilase Modeling and Substrate Docking
4.3. Nitrilase Overproduction
4.4. Nitrilase Activity Assays
4.5. Analytical High-Performance Liquid Chromatography (HPLC)
4.6. Liquid Chromatography–Mass Spectrometry (LC–MS) Analysis
4.7. Nuclear Magnetic Resonance (NMR) Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACN | Acetonitrile |
| BN | Benzonitrile |
| β-CA | β-Cyano-L-alanine |
| 2CP | 2-Cyanopyridine |
| 4CP | 4-Cyanopyridine |
| CynH | Cyanide hydratase |
| CynD | Cyanide dihydratase |
| DL | Desolvation line |
| DMSO-d6 | Deuterated dimethylsulfoxide |
| ESI | Electrospray ionization |
| FN | Fumaronitrile |
| GDT | Global Distance Test |
| HB | Hydrogen bond |
| HGT | Horizontal gene transfer |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| MN | Mandelonitrile |
| MUSCLE | Multiple Sequence Comparison by Log-Expectation |
| NitAg | Nitrilase from Armillaria gallica |
| NitSh | Nitrilase from Stereum hirsutum |
| NitTv1 | Nitrilase from Trametes versicolor |
| NLase | Nitrilase |
| PAN | Phenylacetonitrile |
| PPN | 3-Phenylpropionitrile |
| RP | Reverse phase |
References
- Pace, H.C.; Brenner, C. The nitrilase superfamily: Classification, structure and function. Genome Biol. 2001, 2, reviews0001-1. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.-S.; Lu, Z.-M.; Li, H.; Shi, J.-S.; Zhou, Z.-M.; Xu, Z.-H. Nitrilases in nitrile biocatalysis: Recent progress and forthcoming research. Microb. Cell Fact. 2012, 11, 142. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Rucká, L.; Nešvera, J.; Pátek, M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J. Microbiol. Biotechnol. 2017, 33, 8. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, T.C.; Kumar, V.; Kumar, V.; Thakur, N.; Savitri. Nitrile metabolizing enzymes in biocatalysis and biotransformation. Appl. Biochem. Biotechnol. 2018, 185, 925–946. [Google Scholar] [CrossRef]
- Martínková, L.; Veselá, A.B.; Rinágelová, A.; Chmátal, M. Cyanide hydratases and cyanide dihydratases: Emerging tools in the biodegradation and biodetection of cyanide. Appl. Microbiol. Biotechnol. 2015, 99, 8875–8882. [Google Scholar] [CrossRef]
- Howden, A.J.M.; Preston, G.M. Nitrilase enzymes and their role in plant-microbe interactions. Microb. Biotechnol. 2009, 2, 441–451. [Google Scholar] [CrossRef]
- Thuku, R.N.; Brady, D.; Benedik, M.J.; Sewell, B.T. Microbial nitrilases: Versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 2009, 106, 703–727. [Google Scholar] [CrossRef]
- Thimann, K.V.; Mahadevan, S.; Nitrilase, I. Occurrence, preparation and general properties of enzyme. Arch. Biochem. Biophys. 1964, 105, 133–141. [Google Scholar] [CrossRef]
- Harper, D.B. Fungal degradation of aromatic nitriles. Enzymology of C–N cleavage by Fusarium solani. Biochem. J. 1977, 167, 685–692. [Google Scholar] [CrossRef]
- Goldlust, A.; Bohak, Z. Induction, purification, and characterization of the nitrilase Fusarium oxysporum f. sp. melonis. Biotechnol. Appl. Biochem. 1989, 11, 581–601. [Google Scholar]
- Brunner, S.; Eppinger, E.; Fischer, S.; Gröning, J.; Stolz, A. Conversion of aliphatic nitriles by the arylacetonitrilase from Pseudomonas fluorescens EBC191. World J. Microbiol. Biotechnol. 2018, 34, 91. [Google Scholar] [CrossRef] [PubMed]
- Vorwerk, S.; Biernacki, S.; Hillebrand, H.; Janzik, I.; Müller, A.; Weiler, E.W.; Piotrowski, M. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 2001, 212, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, M.; Schönfelder, S.; Weiler, E.W. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-L-alanine hydratase/nitrilase. J. Biol. Chem. 2001, 276, 2616–2621. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.-S.; Li, H.; Zhu, X.-Y.; Lu, Z.-M.; Wu, Y.; Shi, J.-S.; Xu, Z.-H. Fungal His-tagged nitrilase from Gibberella intermedia: Gene cloning, heterologous expression and biochemical properties. PLoS ONE 2012, 7, e50622. [Google Scholar] [CrossRef]
- Kaplan, O.; Veselá, A.B.; Petříčková, A.; Pasquarelli, F.; Pičmanová, M.; Rinágelová, A.; Bhalla, T.C.; Pátek, M.; Martínková, L. A comparative study of nitrilases identified by genome mining. Mol. Biotechnol. 2013, 54, 996–1003. [Google Scholar] [CrossRef]
- Veselá, A.B.; Rucká, L.; Kaplan, O.; Pelantová, H.; Nešvera, J.; Pátek, M.; Martínková, L. Bringing nitrilase sequences from databases to life: The search for novel substrate specificities with a focus on dinitriles. Appl. Microbiol. Biotechnol. 2016, 100, 2193–2202. [Google Scholar] [CrossRef]
- Shoresh, M.; Harman, G.E.; Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Ann. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef]
- Kameshwar, A.K.S.; Qin, W. Comparative study of genome-wide plant biomass-degrading CAZymes in white rot, brown rot and soft rot fungi. Mycology 2018, 9, 93–105. [Google Scholar] [CrossRef]
- Hiscox, J.; O´Leary, J.; Boddy, L. Fungus wars: Basidiomycete battles in wood decay. Stud. Mycol. 2018, 89, 117–124. [Google Scholar] [CrossRef]
- Legras, J.L.; Chuzel, G.; Arnaud, A.; Galzy, P. Natural nitriles and their metabolism. World J. Microbiol. Biotechnol. 1990, 6, 83–108. [Google Scholar] [CrossRef]
- Strobel, G.A. The fixation of hydrocyanic acid by a psychrophilic basidiomycete. J. Biol. Chem. 1966, 241, 2618–2621. [Google Scholar]
- Strobel, G.A. 4-Amino-4-cyanobutyric acid as an intermediate in glutamate biosynthesis. J. Biol. Chem. 1967, 242, 3265–3269. [Google Scholar]
- Cabuk, A.; Unal, A.T.; Kolankaya, N. Biodegradation of cyanide by a white rot fungus, Trametes versicolor. Biotechnol. Lett. 2006, 28, 1313–1317. [Google Scholar] [CrossRef]
- Luo, H.; Fan, L.; Chang, Y.; Ma, J.; Yu, H.; Shen, Z. Gene cloning, overexpression, and characterization of the nitrilase from Rhodococcus rhodochrous tg1-A6 in E. coli. Appl. Biochem. Biotechnol. 2010, 160, 393–400. [Google Scholar] [CrossRef]
- Rinágelová, A.; Kaplan, O.; Veselá, A.B.; Chmátal, M.; Křenková, A.; Plíhal, O.; Pasquarelli, F.; Cantarella, M.; Martínková, L. Cyanide hydratase from Aspergillus niger K10: Overproduction in Escherichia coli, purification, characterization and use in continuous cyanide degradation. Proc. Biochem. 2014, 49, 445–450. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; p. 126. [Google Scholar]
- Zhang, L.; Yin, B.; Wang, C.; Jiang, S.; Wang, H.; Yuan, Y.A.; Wei, D. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Synechocystis sp. PCC6803. J. Struct. Biol. 2014, 188, 93–101. [Google Scholar] [CrossRef]
- Raczynska, J.; Vorgias, C.; Antranikian, G.; Rypniewski, W. Crystallographic analysis of a thermoactive nitrilase. J. Struct. Biol. 2010, 173, 294–302. [Google Scholar] [CrossRef]
- Liu, H.; Gao, Y.; Zhang, M.; Qiu, X.; Cooper, A.J.; Niu, L.; Teng, M. Structures of enzyme-intermediate complexes of yeast Nit2: Insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2. Acta Crystallogr. D 2013, 69, 1470–1481. [Google Scholar] [CrossRef]
- Thuku, R.N.; Weber, B.W.; Varsani, A.; Sewell, B.T. Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J. 2007, 274, 2099–2108. [Google Scholar] [CrossRef]
- Fernandes, B.C.M.; Mateo, C.; Kiziak, C.; Chmura, A.; Wacker, J.; van Rantwijk, F.; Stolz, A.; Sheldon, R.A. Nitrile hydratase activity of a recombinant nitrilase. Adv. Synth. Catal. 2006, 348, 2597–2603. [Google Scholar] [CrossRef]
- Yeom, S.J.; Kim, H.J.; Lee, J.K.; Kim, D.E.; Oh, D.K. An amino acid at position 142 in nitrilase from Rhodococcus rhodochrous ATCC 33278 determines the substrate specificity for aliphatic and aromatic nitriles. Biochem. J. 2008, 415, 401–407. [Google Scholar] [CrossRef]
- Martínková, L. Nitrile metabolism in fungi: A review of its key enzymes nitrilases with focus on their biotechnological impact. Fung. Biol. Rev. 2019, 33, 149–157. [Google Scholar] [CrossRef]
- Hibbett, D.S. Agaricomycotina. Jelly Fungi, Yeasts, and Mushrooms. Version 20 April 2007. Available online: http://tolweb.org/Agaricomycotina/20531/2007.04.20 (accessed on 22 October 2019).
- Ghosh, S.; Gupta, S.K.; Jha, G. Identification and functional analysis of AG1-IA specific genes of Rhizoctonia solani. Curr. Genet. 2014, 60, 327–341. [Google Scholar] [CrossRef]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host-multi-pathogen warfare: Pathogen interactions in co-infected plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef]
- Wang, P.; Van Etten, H.D. Cloning and properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochem. Biophys. Res. Commun. 1992, 187, 1048–1054. [Google Scholar] [CrossRef]
- Basile, L.J.; Willson, R.C.; Sewell, B.T.; Benedik, M.J. Genome mining of cyanide degrading nitrilases from filamentous fungi. Appl. Microbiol. Biotechnol. 2008, 80, 427–435. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Clamp, M.; Cuff, J.; Searle, S.M.; Barton, G.J. The Jalview Java alignment editor. Bioinformatics 2004, 20, 426–427. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., III; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: Allatom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, 375–383. [Google Scholar] [CrossRef]
- Schrödinger Release 2017-1: Canvas; Schrödinger LLC: New York, NY, USA, 2017.
- Cluness, M.J.; Turner, P.D.; Clements, E.; Brown, D.T.; O´Reilly, C. Purification and properties of cyanide hydratase from Fusarium lateritium and analysis of the corresponding chy1 gene. J. Gen. Biol. 1993, 139, 1807–1815. [Google Scholar] [CrossRef][Green Version]
- Black, G.W.; Brown, N.L.; Perry, J.J.B.; Randall, D.; Turnbull, G.; Zhang, M. A high-throughput screening method for determining the substrate scope of nitrilases. Chem. Commun. 2015, 51, 2660–2662. [Google Scholar] [CrossRef]
- Olsen, B.A. Hydrophilic interaction chromatography using amino and silica columns for the determination of polar pharmaceuticals and impurities. J. Chromatogr. A 2001, 913, 113–122. [Google Scholar] [CrossRef]



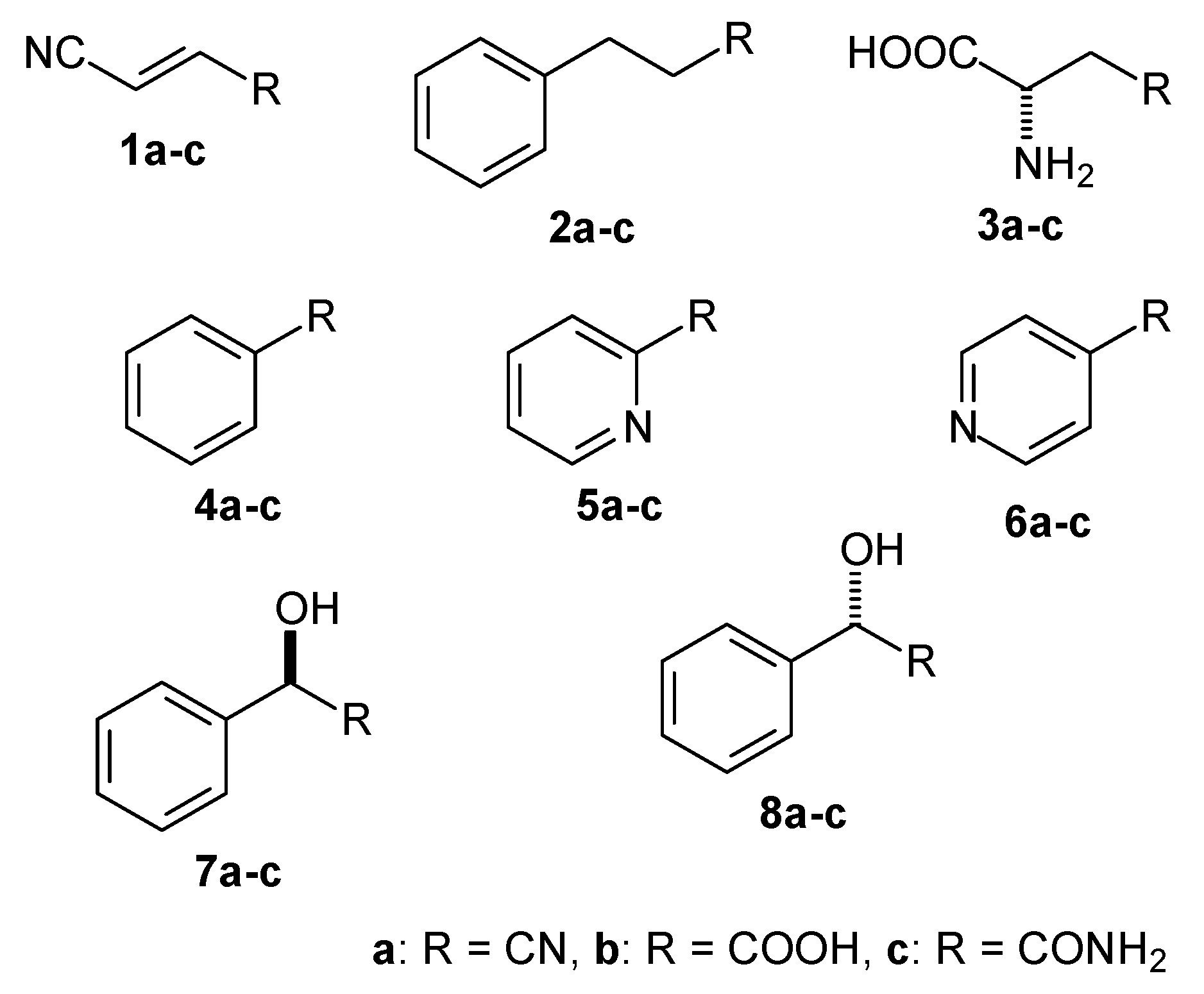
| Proposed Substrate | Docking Glide Score [kcal/mol] | ||
|---|---|---|---|
| NitTv1 | NitAg | NitSh | |
| Fumaronitrile (1a) | −0.15 | −1.0 | −0.77 |
| 3-Phenylpropionitrile (2a) | −3.49 | − 1 | n.i. |
| β-Cyano-L-alanine (3a) | −2.82 | n.i. | −2.60 |
| Benzonitrile (4a) | n.i. | n.i. | −4.56 |
| 2-Cyanopyridine (5a) | n.i. | n.i. | −4.22 |
| 4-Cyanopyridine (6a) | n.i. | n.i. | n.i. |
| (R)-Mandelonitrile (7a) | n.i. | n.i. | −5.18 |
| (S)-Mandelonitrile (8a) | n.i. | n.i. | −4.78 |
| Hydrogen cyanide (HCN) | n.i. | −3.50 | −3.12 |
| Substrate | Relative Activity [%] | ||
|---|---|---|---|
| NitTv1 | NitAg | NitSh | |
| Fumaronitrile (1a) | 100 1 | 0.140 ± 0.003 | 0.199 ± 0.003 |
| 3-Phenylpropionitrile (2a) | 6.26 ± 0.13 | 0 | 0 |
| β-Cyano-L-alanine (3a) | 7.96 ± 1.57 | 0 | traces |
| Benzonitrile (4a) | 0 | 0 | 0.135 ± 0.010 |
| 2-Cyanopyridine (5a) | traces | 0 | 1.21 ± 0.03 |
| 4-Cyanopyridine (6a) | 5.26 ± 0.26 | traces | traces |
| HCN | 0 | 100 2 | 100 3 |
| Order 1 [Number of Sequences] | Genus 1 [Number of Sequences] | ||
|---|---|---|---|
| Clade 1 | Clade 2 | Others | |
| Agaricales (38) | Armillaria (3) Cylindrobasidium (1) Dendrothele (1) Fistulina (1) Gymnopus (1) Hypsizygus (1) Lentinus (1) Moniliophthora (1) Pleurotus (1) Pluteus (1) Schizophyllum (1) Termitomyces (1) | Armillaria (5) Dendrothele (1) Gymnopus (1) Lentinus (1) Moniliophthora (3) Mycena (1) Pleurotus (2) | Agaricus (1) Cylindrobasidium (1) Dendrothele (3) Pluteus (1) Moniliophthora (2) Mycena (2) |
| Amylocorticiales (2) | Plicaturopsis (1) | Plicaturopsis (1) | |
| Atheliales (1) | Fibulanrhizoctonia (1) | ||
| Auriculariales (7) | Auricularia (1) Exidia (1) | Auricularia (4 2) Exidia (1 3) | |
| Cystofilobasidiales (2) | Xanthophylomyces (2) | ||
| Boletales (7) | Coniophora (1) Hydnomerulius (1) Paxillus (1) Rhizopogon (2) Serpula (1) Suillus (1) | ||
| Cantharellales (12) | Botryobasidium (1) Rhizoctonia (4) | Rhizoctonia (7) | |
| Corticiales (1) | Punctularia (1) | ||
| Dacrymycetales (2) | Calocera (1) Dacryopinax (1) | ||
| Geastrales (1) | Sphaerobolus (1) | ||
| Gloeophyllales (2) | Neolentinus (1) Heliocybe (1) | ||
| Hymenochaetales (5) | Fomitiporia (1) Phellinidium (1) Rickenella (1) Sanghuangporus (1) Schizopora (1) | ||
| Jaapiales (1) | Jaapia (1) | ||
| Polyporales (16) | Dichomitus (1) Ganoderma (1) Gelatoporia (1) Grifola (1) Obba (1) Phanerochaete (3) Phlebiopsis (1) Polyporus (2) Steccherinum (1) Trametes (4) | ||
| Russulales (10) | Dentipellis (1) Heterobasidion (1) Peniophora (1) Stereum (1) | Bondarzewia (1) Dentipellis (1) Hericium (2) Heterobasidion (1) | Stereum (1 3) |
| Trechisporales (2) | Sistotremastrum (2) | ||
| Tremellales (21) | Cryptococcus (4) Kockovaella (1) Kwoniella (13) Naematelia (2) Saitozyma (1) | ||
| Trichosporonales (4) | Apiotrichum (2) Cutaneotrichosporon (1) Trichosporon (1) | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rucká, L.; Chmátal, M.; Kulik, N.; Petrásková, L.; Pelantová, H.; Novotný, P.; Příhodová, R.; Pátek, M.; Martínková, L. Genetic and Functional Diversity of Nitrilases in Agaricomycotina. Int. J. Mol. Sci. 2019, 20, 5990. https://doi.org/10.3390/ijms20235990
Rucká L, Chmátal M, Kulik N, Petrásková L, Pelantová H, Novotný P, Příhodová R, Pátek M, Martínková L. Genetic and Functional Diversity of Nitrilases in Agaricomycotina. International Journal of Molecular Sciences. 2019; 20(23):5990. https://doi.org/10.3390/ijms20235990
Chicago/Turabian StyleRucká, Lenka, Martin Chmátal, Natalia Kulik, Lucie Petrásková, Helena Pelantová, Petr Novotný, Romana Příhodová, Miroslav Pátek, and Ludmila Martínková. 2019. "Genetic and Functional Diversity of Nitrilases in Agaricomycotina" International Journal of Molecular Sciences 20, no. 23: 5990. https://doi.org/10.3390/ijms20235990
APA StyleRucká, L., Chmátal, M., Kulik, N., Petrásková, L., Pelantová, H., Novotný, P., Příhodová, R., Pátek, M., & Martínková, L. (2019). Genetic and Functional Diversity of Nitrilases in Agaricomycotina. International Journal of Molecular Sciences, 20(23), 5990. https://doi.org/10.3390/ijms20235990