Lipidomic Profiling Reveals Significant Perturbations of Intracellular Lipid Homeostasis in Enterovirus-Infected Cells
Abstract
1. Introduction
2. Results
2.1. Analytical Method Validation
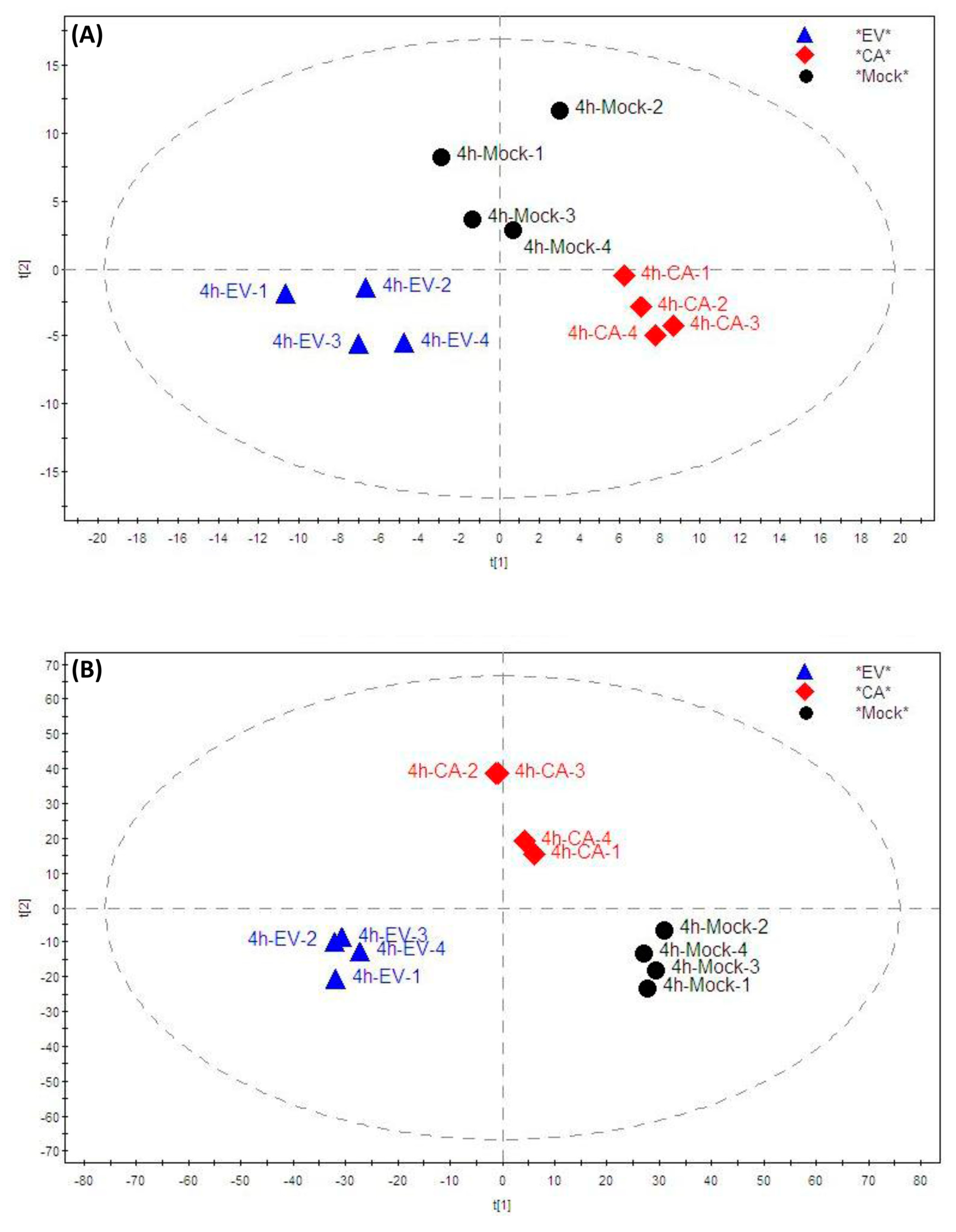
2.2. Lipidomic Profile Analysis
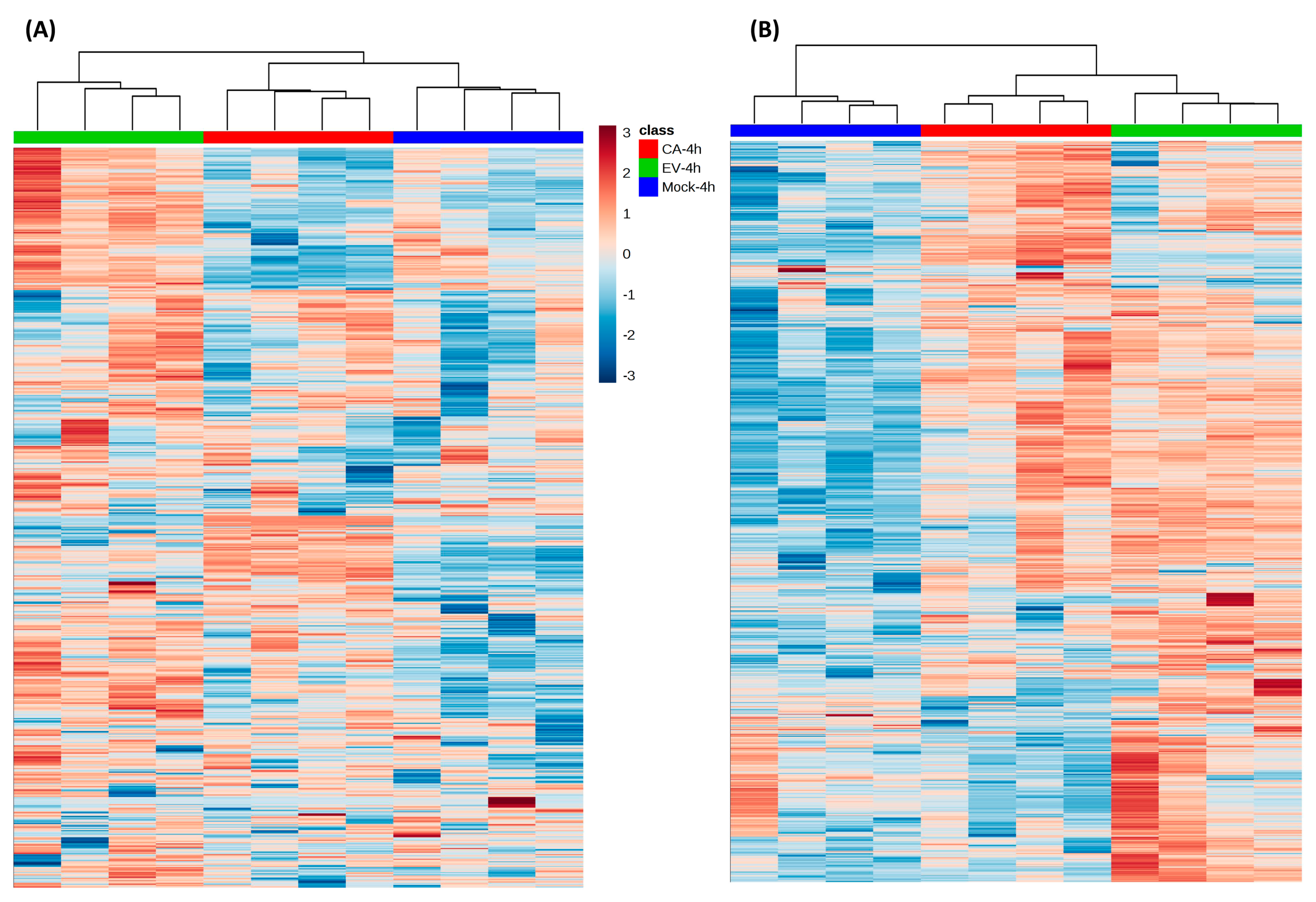
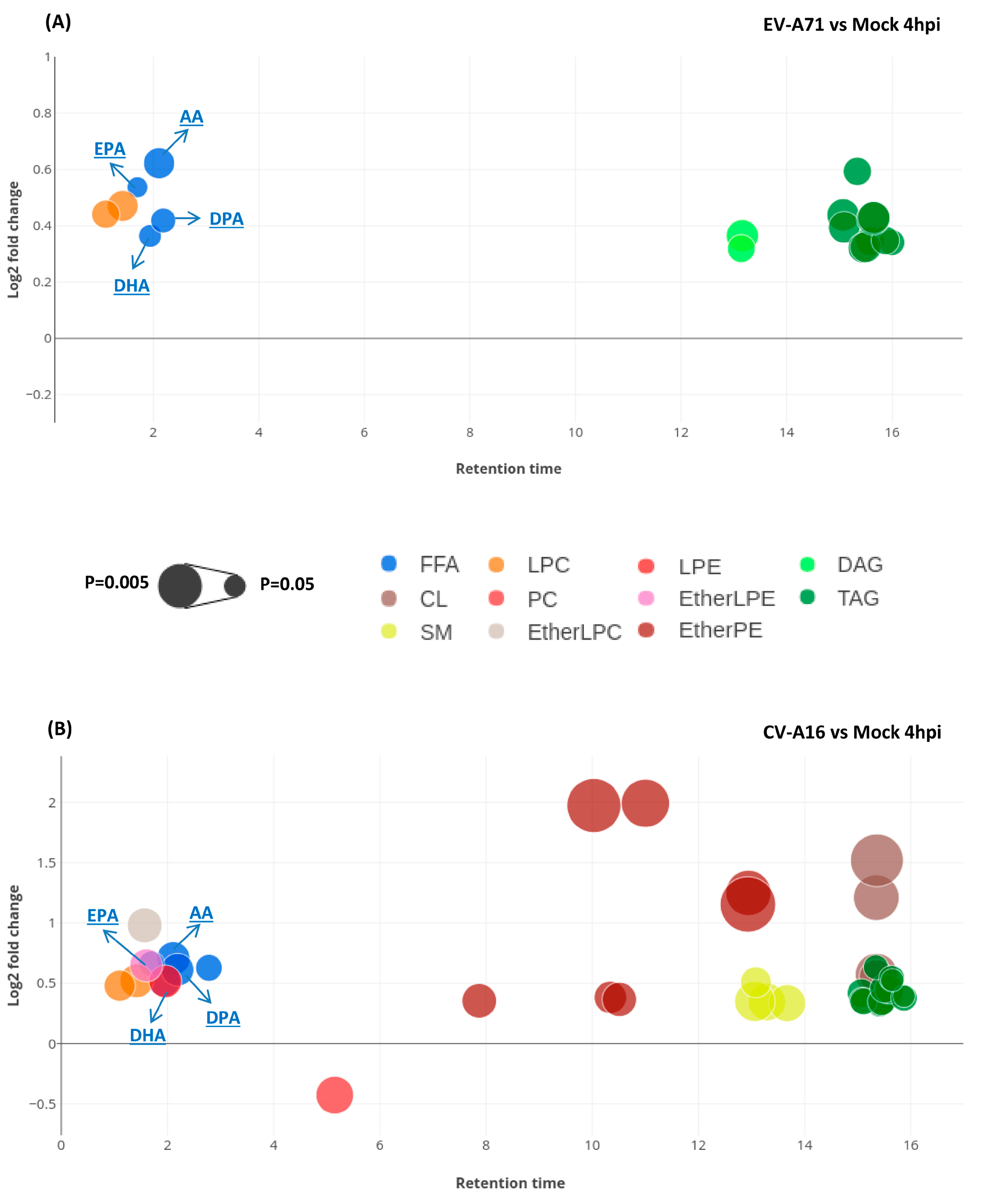
2.3. Significantly Perturbed Lipid Features in Enterovirus Infection
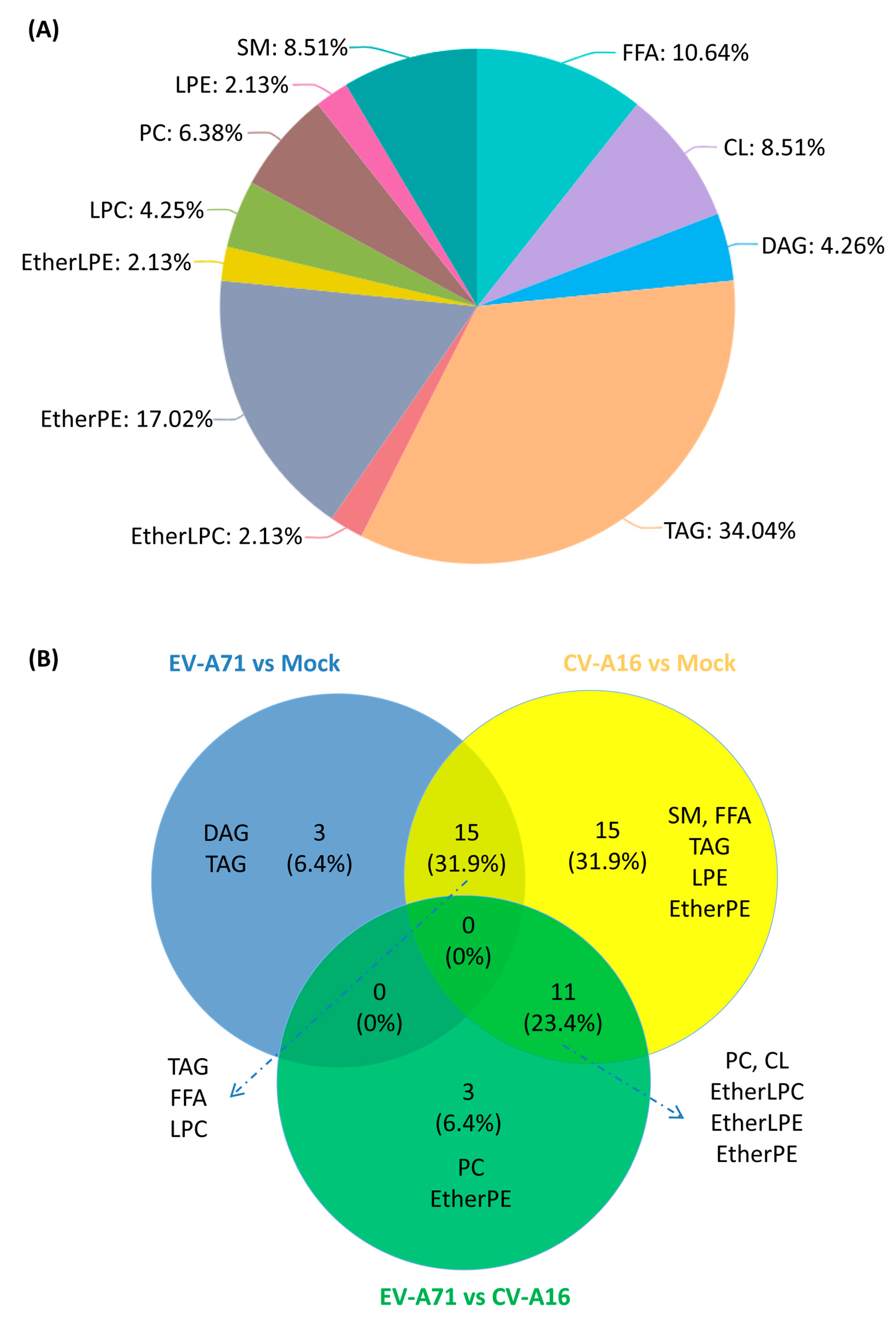
2.4. Lipid Changes Specific to EV-A71 and CV-A16 Infection
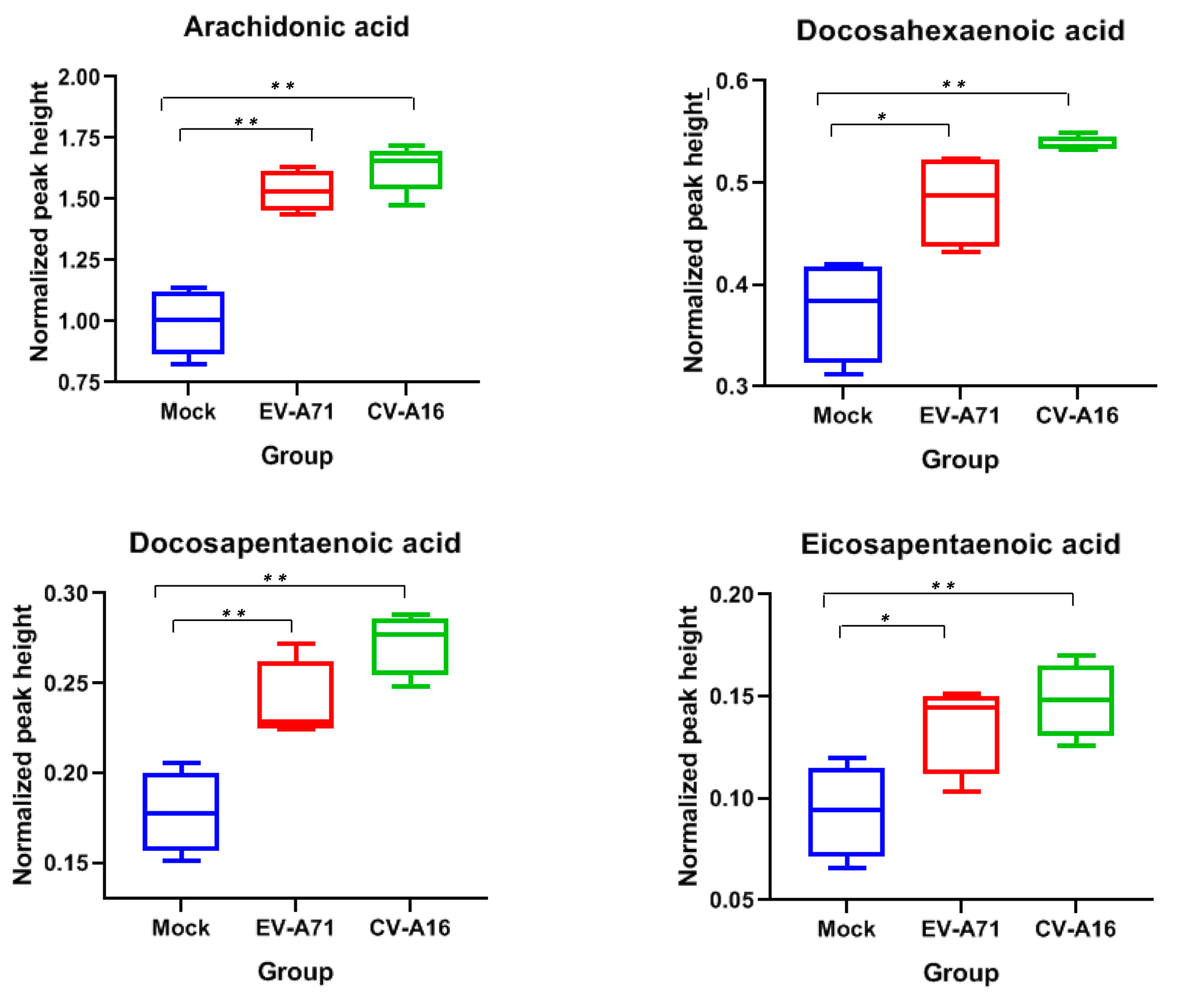
2.5. Characterization of the Specific Lipid Changes in EV-A71 and CV-A16 Infection
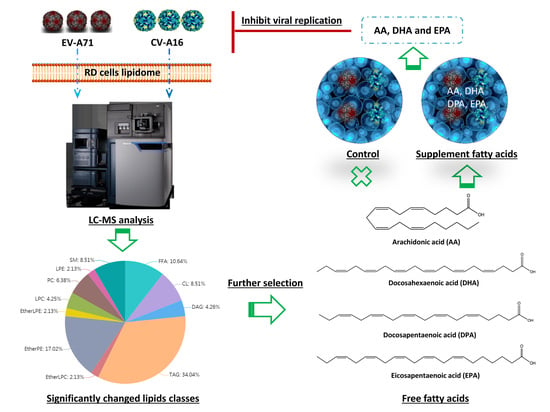
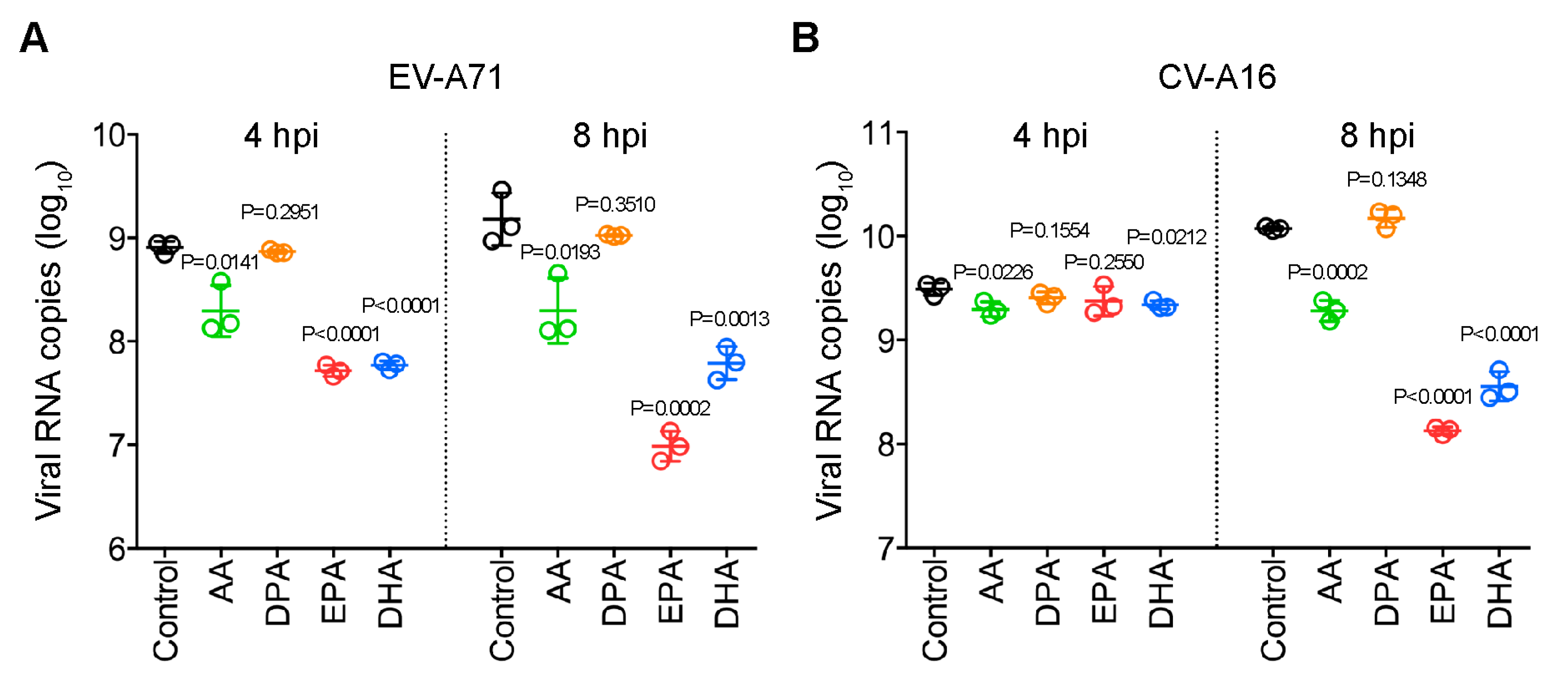
2.6. Antiviral Activites of the Identified Fatty Acids Against EV-A71 and CV-A16
3. Discussion
4. Materials and Methods
4.1. Viruses and Cells
4.2. Chemical Reagents and Standards
4.3. Lipid Treatment of EV-A71-Infected and CV-A16-Infected RD Cells
4.4. Lipid Extraction for Lipidomics Profiling
4.5. Ultra-High Performance Liquid Chromatography-Electrospray Ionization-Quadrupole-Time of Flight-Mass Spectrometry (UPLC-ESI-Q-TOF-MS) Analysis
4.6. Analytical Method Assessment
4.7. Data Processing and Statistical Analysis
4.8. Lipids Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wong, S.S.; Yip, C.C.; Lau, S.K.; Yuen, K.Y. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol Infect. 2010, 138, 1071–1089. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Liao, Q.; Viboud, C.; Zhang, J.; Sun, J.; Wu, J.T.; Chang, Z.; Liu, F.; Fang, V.J.; Zheng, Y.; et al. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect. Dis 2014, 14, 308–318. [Google Scholar] [CrossRef]
- Chang, L.Y.; Lin, T.Y.; Hsu, K.H.; Huang, Y.C.; Lin, K.L.; Hsueh, C.; Shih, S.R.; Ning, H.C.; Hwang, M.S.; Wang, H.S.; et al. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 1999, 354, 1682–1686. [Google Scholar] [CrossRef]
- Yip, C.C.; Lau, S.K.; Woo, P.C.; Yuen, K.Y. Human enterovirus 71 epidemics: what’s next? Emerg Health Threats J. 2013, 6, 19780. [Google Scholar] [CrossRef] [PubMed]
- Ooi, M.H.; Wong, S.C.; Lewthwaite, P.; Cardosa, M.J.; Solomon, T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol 2010, 9, 1097–1105. [Google Scholar] [CrossRef]
- Taube, S.; Jiang, M.X.; Wobus, C.E. Glycosphingolipids as Receptors for Non-Enveloped Viruses. Viruses-Basel 2010, 2, 1011–1049. [Google Scholar] [CrossRef]
- Chazal, N.; Gerlier, D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol. Biol Rev. 2003, 67, 226–237, table of contents. [Google Scholar] [CrossRef]
- Nagy, P.D.; Strating, J.R.P.M.; van Kuppeveld, F.J.M. Building Viral Replication Organelles: Close Encounters of the Membrane Types. Plos Pathogens 2016, 12. [Google Scholar] [CrossRef]
- Hsu, N.Y.; Ilnytska, O.; Belov, G.; Santiana, M.; Chen, Y.H.; Takvorian, P.M.; Pau, C.; van der Schaar, H.; Kaushik-Basu, N.; Balla, T.; et al. Viral Reorganization of the Secretory Pathway Generates Distinct Organelles for RNA Replication. Cell 2010, 141, 799–811. [Google Scholar] [CrossRef]
- Ono, A.; Ablan, S.D.; Lockett, S.J.; Nagashima, K.; Freed, E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl Acad Sci USA 2004, 101, 14889–14894. [Google Scholar] [CrossRef]
- Zhang, J.; Pekosz, A.; Lamb, R.A. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol 2000, 74, 4634–4644. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Hardt, M.; Schwudke, D.; Neuman, B.W.; Pleschka, S.; Ziebuhr, J. Inhibition of Cytosolic Phospholipase A2alpha Impairs an Early Step of Coronavirus Replication in Cell Culture. J. Virol 2018, 92. [Google Scholar] [CrossRef]
- Yan, B.; Chu, H.; Yang, D.; Sze, K.H.; Lai, P.M.; Yuan, S.; Shuai, H.; Wang, Y.; Kao, R.Y.; Chan, J.F.; et al. Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication. Viruses 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Pauling, J.K.; Hermansson, M.; Hartler, J.; Christiansen, K.; Gallego, S.F.; Peng, B.; Ahrends, R.; Ejsing, C.S. Proposal for a common nomenclature for fragment ions in mass spectra of lipids. PLoS ONE 2017, 12, e0188394. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Deng, Y.; Hou, J.; Bi, Q.; Yang, M.; Jiang, B.; Liu, X.; Wu, W.; Guo, D. UHPLC-LTQ-Orbitrap MS combined with spike-in method for plasma metabonomics analysis of acute myocardial ischemia rats and pretreatment effect of Danqi Tongmai tablet. Mol. Biosyst 2015, 11, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim Biophys Acta 2011, 1811, 637–647. [Google Scholar] [CrossRef]
- Liebisch, G.; Vizcaino, J.A.; Kofeler, H.; Trotzmuller, M.; Griffiths, W.J.; Schmitz, G.; Spener, F.; Wakelam, M.J. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013, 54, 1523–1530. [Google Scholar] [CrossRef]
- Thormar, H.; Isaacs, C.E.; Brown, H.R.; Barshatzky, M.R.; Pessolano, T. Inactivation of enveloped viruses and killing of cells by fatty acids and monoglycerides. Antimicrob Agents Chemother 1987, 31, 27–31. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, Y.J.; Wu, K.; Yan, H.; Hao, X.A.; Wu, Y.F. Application of fatty acids as antiviral agents against tobacco mosaic virus. Pesticide Biochemistry and Physiology 2017, 139, 87–91. [Google Scholar] [CrossRef]
- Bienz, K.; Egger, D.; Pfister, T.; Troxler, M. Structural and functional characterization of the poliovirus replication complex. J. Virol 1992, 66, 2740–2747. [Google Scholar] [PubMed]
- Schlegel, A.; Giddings, T.H., Jr.; Ladinsky, M.S.; Kirkegaard, K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol 1996, 70, 6576–6588. [Google Scholar] [PubMed]
- Suhy, D.A.; Giddings, T.H., Jr.; Kirkegaard, K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol 2000, 74, 8953–8965. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Guedan, A.; Mousnier, A.; Swieboda, D.; Zhang, Q.; Horkai, D.; Le Novere, N.; Solari, R.; Wakelam, M.J.O. Host lipidome analysis during rhinovirus replication in HBECs identifies potential therapeutic targets. J. Lipid Res. 2018, 59, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, W.M.; Galli, C. Fat and fatty acid terminology, methods of analysis and fat digestion and metabolism: a background review paper. Ann. Nutr. Metab. 2009, 55, 8–43. [Google Scholar] [CrossRef]
- Morita, M.; Kuba, K.; Ichikawa, A.; Nakayama, M.; Katahira, J.; Iwamoto, R.; Watanebe, T.; Sakabe, S.; Daidoji, T.; Nakamura, S.; et al. The Lipid Mediator Protectin D1 Inhibits Influenza Virus Replication and Improves Severe Influenza. Cell 2013, 153, 112–125. [Google Scholar] [CrossRef]
- Schmitz, G.; Ecker, J. The opposing effects of n-3 and n-6 fatty acids. Prog Lipid Res. 2008, 47, 147–155. [Google Scholar] [CrossRef]
- Lee, J.Y.; Plakidas, A.; Lee, W.H.; Heikkinen, A.; Chanmugam, P.; Bray, G.; Hwang, D.H. Differential modulation of Toll-like receptors by fatty acids: preferential inhibition by n-3 polyunsaturated fatty acids. J. Lipid Res. 2003, 44, 479–486. [Google Scholar] [CrossRef]
- Ibrahim, A.; Mbodji, K.; Hassan, A.; Aziz, M.; Boukhettala, N.; Coeffier, M.; Savoye, G.; Dechelotte, P.; Marion-Letellier, R. Anti-inflammatory and anti-angiogenic effect of long chain n-3 polyunsaturated fatty acids in intestinal microvascular endothelium. Clin. Nutr 2011, 30, 678–687. [Google Scholar] [CrossRef]
- Arita, M.; Yoshida, M.; Hong, S.; Tjonahen, E.; Glickman, J.N.; Petasis, N.A.; Blumberg, R.S.; Serhan, C.N. Resolvin E1, an endogenous lipid mediator derived from omega-3 eicosapentaenoic acid, protects against 2,4,6-trinitrobenzene sulfonic acid-induced colitis. Proc. Natl Acad Sci USA 2005, 102, 7671–7676. [Google Scholar] [CrossRef]
- Marion-Letellier, R.; Savoye, G.; Ghosh, S. Polyunsaturated fatty acids and inflammation. IUBMB Life 2015, 67, 659–667. [Google Scholar] [CrossRef]
- Hofmann, S.; Krajewski, M.; Scherer, C.; Scholz, V.; Mordhorst, V.; Truschow, P.; Schobel, A.; Reimer, R.; Schwudke, D.; Herker, E. Complex lipid metabolic remodeling is required for efficient hepatitis C virus replication. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids 2018, 1863, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chu, H.; Chan, J.F.; Ye, Z.W.; Wen, L.; Yan, B.; Lai, P.M.; Tee, K.M.; Huang, J.; Chen, D.; et al. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun 2019, 10, 120. [Google Scholar] [CrossRef]
- Burnum-Johnson, K.E.; Kyle, J.E.; Eisfeld, A.J.; Casey, C.P.; Stratton, K.G.; Gonzalez, J.F.; Habyarimana, F.; Negretti, N.M.; Sims, A.C.; Chauhan, S.; et al. MPLEx: a method for simultaneous pathogen inactivation and extraction of samples for multi-omics profiling. Analyst 2017, 142, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Measurement of DNA concentration as a normalization strategy for metabolomic data from adherent cell lines; Silva, L.P., Lorenzi, P.L., Purwaha, P., Yong, V., Hawke, D.H., Weinstein, J.N., Eds.; 2013; Volume 85, pp. 9536–9542. [Google Scholar]
- Damen, C.W.; Isaac, G.; Langridge, J.; Hankemeier, T.; Vreeken, R.J. Enhanced lipid isomer separation in human plasma using reversed-phase UPLC with ion-mobility/high-resolution MS detection. J. Lipid Res. 2014, 55, 1772–1783. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Increasing lipidomic coverage by selecting optimal mobile-phase modifiers in LC–MS of blood plasma. Metabolomics 2016, 12. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Kind, T.; Liu, K.H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J. Chem. 2014, 28, 623–632. [Google Scholar] [CrossRef]
| Significant Lipids | Detection Mode | Retention Time | Accurate Mass in Detection Mode | Adduct Ion Name | Lipid Class | Identification Information | p-Value in EV71 Infection vs Mock | p-Value in CA16 Infection vs Mock | p-Value in CA16 vs EV71 Infection | Fold Change in EV71 Infection vs Mock | Fold Change in CA16 Infection vs Mock | Fold Change in CA16 vs EV71 Infection |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arachidonic acid | negative | 2.11 | 303.2315 | [M-H]− | FFA | STD | 0.0007 | 0.0004 | NS | 1.54 | 1.63 | 1.06 |
| Docosahexaenoic acid | negative | 1.94 | 327.2319 | [M-H]− | FFA | STD | 0.0196 | 0.0007 | NS | 1.29 | 1.43 | 1.11 |
| Docosapentaenoic acid | negative | 2.19 | 329.2466 | [M-H]− | FFA | STD | 0.0095 | 0.0006 | NS | 1.34 | 1.53 | 1.14 |
| Eicosapentaenoic acid | negative | 1.7 | 301.2167 | [M-H]− | FFA | STD | 0.0382 | 0.0096 | NS | 1.45 | 1.58 | 1.09 |
| Adrenic acid | negative | 2.78 | 331.2624 | [M-H]− | FFA | MS/MS | NS | 0.0067 | NS | 1.25 | 1.55 | 1.23 |
| CL (16:0/18:1/16:0/18:1) | negative | 15.35 | 1403.989 | [M-H]− | CL | MS/MS | NS | 0.0000 | 0.0000 | 0.87 | 1.49 | 1.71 |
| CL (70:5) | negative | 15.35 | 1425.9708 | [M-H]− | CL | MS/MS | NS | 0.0003 | 0.0003 | 0.86 | 1.47 | 1.71 |
| CL (16:1/18:0/18:1/18:1) | negative | 15.35 | 1430.004 | [M-H]− | CL | MS/MS | NS | 0.0000 | 0.0000 | 0.91 | 2.31 | 2.54 |
| CL (72:4) | negative | 15.36 | 1456.0192 | [M-H]− | CL | MS/MS | NS | 0.0000 | 0.0000 | 0.67 | 2.87 | 4.30 |
| SM (d14:2/28:2) | positive | 13.08 | 809.6527 | [M+H]+ | SM | MS/MS | NS | 0.0013 | NS | 1.21 | 1.42 | 1.18 |
| SM (d18:1/24:1) | positive | 13.06 | 813.6888 | [M+H]+ | SM | MS/MS | NS | 0.0000 | NS | 1.14 | 1.27 | 1.12 |
| SM (d18:0/24:1) | positive | 13.29 | 815.6979 | [M+H]+ | SM | MS/MS | NS | 0.0001 | NS | 1.10 | 1.27 | 1.16 |
| SM (d18:1/24:0) | positive | 13.67 | 815.7034 | [M+H]+ | SM | MS/MS | NS | 0.0001 | NS | 1.15 | 1.26 | 1.10 |
| PC (O-16:0/0:0) | positive | 1.57 | 482.363 | [M+H]+ | EtherLPC | MS/MS | NS | 0.0002 | 0.0007 | 1.23 | 1.97 | 1.60 |
| PC (16:1/0:0) | positive | 1.1 | 494.3254 | [M+H]+ | LPC | MS/MS | 0.0027 | 0.0011 | NS | 1.36 | 1.39 | 1.03 |
| PC (18:1/0:0) | positive | 1.42 | 522.3575 | [M+H]+ | LPC | MS/MS | 0.0008 | 0.0004 | NS | 1.39 | 1.43 | 1.04 |
| PC (16:0/20:4) | positive | 6.51 | 782.5679 | [M+H]+ | PC | MS/MS | NS | NS | 0.0204 | 1.08 | 0.81 | 0.75 |
| PC (20:4/20:4) | positive | 5.15 | 830.5708 | [M+H]+ | PC | MS/MS | NS | 0.0000 | 0.0000 | 1.12 | 0.74 | 0.66 |
| PC (20:3/20:4) | positive | 5.89 | 832.5861 | [M+H]+ | PC | MS/MS | NS | NS | 0.0000 | 1.09 | 0.85 | 0.78 |
| PE (P-16:0/0:0) | negative | 1.6 | 436.2826 | [M-H]− | EtherLPE | MS/MS | NS | 0.0005 | 0.0003 | 1.10 | 1.57 | 1.43 |
| PE (18:0/0:0) | negative | 1.96 | 480.3081 | [M-H]− | LPE | MS/MS | NS | 0.0006 | NS | 1.23 | 1.43 | 1.17 |
| PE (P-16:0/16:0) | negative | 10.03 | 674.5099 | [M-H]− | EtherPE | MS/MS | NS | 0.0000 | 0.0000 | 0.98 | 3.92 | 4.02 |
| PE (P-16:0/17:0) | negative | 11 | 688.5256 | [M-H]− | EtherPE | MS/MS | NS | 0.0000 | 0.0000 | 0.92 | 3.99 | 4.33 |
| PE (P-18:0/16:0) | negative | 12.93 | 702.543 | [M-H]− | EtherPE | MS/MS | NS | 0.0000 | 0.0000 | 1.04 | 2.22 | 2.13 |
| PE (P-16:0/16:1) | positive | 7.87 | 674.5112 | [M+H]+ | EtherPE | MS/MS | NS | 0.0002 | NS | 1.16 | 1.28 | 1.10 |
| PE (P-16:0/18:1) | negative | 10.09 | 700.5308 | [M-H]− | EtherPE | MS/MS | NS | NS | 0.0000 | 1.14 | 6.49 | 5.72 |
| PE (O-16:0/18:2) | positive | 10.34 | 702.5461 | [M+H]+ | EtherPE | MS/MS | NS | 0.0006 | NS | 1.20 | 1.30 | 1.09 |
| PE (O-18:0/16:1(9Z)) | positive | 12.94 | 704.5615 | [M+H]+ | EtherPE | MS/MS | NS | 0.0000 | 0.0000 | 1.01 | 2.37 | 2.35 |
| PE (P-18:0/18:2) | positive | 10.51 | 728.5601 | [M+H]+ | EtherPE | MS/MS | NS | 0.0003 | NS | 1.17 | 1.29 | 1.10 |
| DG (18:1/18:1/0:0) | positive | 13.16 | 638.5742 | [M+NH4]+ | DAG | MS/MS | 0.0004 | NS | NS | 1.29 | 1.23 | NS |
| DG (38:4) | positive | 13.14 | 662.575 | [M+NH4]+ | DAG | MS/MS | 0.0030 | NS | NS | 1.25 | 1.20 | NS |
| TG (44:1) | positive | 15.07 | 766.692 | [M+NH4]+ | TAG | MS/MS | 0.0004 | 0.0033 | NS | 1.36 | 1.34 | 0.99 |
| TG (46:2) | positive | 15.09 | 792.7092 | [M+NH4]+ | TAG | MS/MS | 0.0006 | 0.0069 | NS | 1.31 | 1.28 | 0.97 |
| TG (14:0/16:0/16:1) | positive | 15.41 | 794.7274 | [M+NH4]+ | TAG | MS/MS | NS | 0.0112 | NS | 1.24 | 1.25 | 1.01 |
| TG (47:1) | positive | 15.57 | 808.7391 | [M+NH4]+ | TAG | MS/MS | NS | 0.0023 | NS | 1.23 | 1.32 | 1.07 |
| TG (14:1/16:1/18:1) | positive | 15.11 | 818.7252 | [M+NH4]+ | TAG | MS/MS | NS | 0.0066 | NS | 1.18 | 1.27 | 1.08 |
| TG (14:0/16:1/18:1) | positive | 15.43 | 820.7425 | [M+NH4]+ | TAG | MS/MS | NS | 0.0064 | NS | 1.23 | 1.26 | 1.02 |
| TG (49:2) | positive | 15.58 | 834.7562 | [M+NH4]+ | TAG | MS/MS | 0.0022 | 0.0026 | NS | 1.27 | 1.36 | 1.07 |
| TG (16:1/16:1/18:1) | positive | 15.45 | 846.7579 | [M+NH4]+ | TAG | MS/MS | 0.0010 | 0.0043 | NS | 1.25 | 1.27 | 1.02 |
| TG (16:1/17:1/18:1) | positive | 15.59 | 860.7723 | [M+NH4]+ | TAG | MS/MS | NS | 0.0031 | NS | 1.27 | 1.37 | 1.08 |
| TG (14:0/18:1/20:4) | positive | 15.34 | 870.757 | [M+NH4]+ | TAG | MS/MS | 0.0025 | 0.0137 | NS | 1.51 | 1.55 | 1.03 |
| TG (16:1/18:1/18:2) | positive | 15.5 | 872.7713 | [M+NH4]+ | TAG | MS/MS | 0.0008 | 0.0031 | NS | 1.25 | 1.37 | 1.10 |
| TG (16:0/18:1/18:1) | positive | 15.99 | 876.7005 | [M+NH4]+ | TAG | MS/MS | 0.0062 | NS | NS | 1.27 | 1.18 | NS |
| TG (16:1/17:1/20:1) | positive | 15.87 | 888.8029 | [M+NH4]+ | TAG | MS/MS | 0.0026 | 0.0077 | NS | 1.27 | 1.30 | 1.02 |
| TG (54:5) | positive | 15.64 | 898.7866 | [M+NH4]+ | TAG | MS/MS | 0.0002 | 0.0100 | NS | 1.34 | 1.46 | 1.08 |
| TG (16:0/18:1/22:5) | positive | 15.65 | 924.8024 | [M+NH4]+ | TAG | MS/MS | 0.0004 | 0.0256 | NS | 1.35 | 1.44 | 1.07 |
| TG (16:0/18:1/22:4) | positive | 15.87 | 926.8177 | [M+NH4]+ | TAG | MS/MS | NS | 0.0326 | NS | 1.24 | 1.31 | 1.06 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, B.; Zou, Z.; Chu, H.; Chan, G.; Tsang, J.O.-L.; Lai, P.-M.; Yuan, S.; Yip, C.C.-Y.; Yin, F.; Kao, R.Y.-T.; et al. Lipidomic Profiling Reveals Significant Perturbations of Intracellular Lipid Homeostasis in Enterovirus-Infected Cells. Int. J. Mol. Sci. 2019, 20, 5952. https://doi.org/10.3390/ijms20235952
Yan B, Zou Z, Chu H, Chan G, Tsang JO-L, Lai P-M, Yuan S, Yip CC-Y, Yin F, Kao RY-T, et al. Lipidomic Profiling Reveals Significant Perturbations of Intracellular Lipid Homeostasis in Enterovirus-Infected Cells. International Journal of Molecular Sciences. 2019; 20(23):5952. https://doi.org/10.3390/ijms20235952
Chicago/Turabian StyleYan, Bingpeng, Zijiao Zou, Hin Chu, Gabriella Chan, Jessica Oi-Ling Tsang, Pok-Man Lai, Shuofeng Yuan, Cyril Chik-Yan Yip, Feifei Yin, Richard Yi-Tsun Kao, and et al. 2019. "Lipidomic Profiling Reveals Significant Perturbations of Intracellular Lipid Homeostasis in Enterovirus-Infected Cells" International Journal of Molecular Sciences 20, no. 23: 5952. https://doi.org/10.3390/ijms20235952
APA StyleYan, B., Zou, Z., Chu, H., Chan, G., Tsang, J. O.-L., Lai, P.-M., Yuan, S., Yip, C. C.-Y., Yin, F., Kao, R. Y.-T., Sze, K.-H., Lau, S. K.-P., Chan, J. F.-W., & Yuen, K.-Y. (2019). Lipidomic Profiling Reveals Significant Perturbations of Intracellular Lipid Homeostasis in Enterovirus-Infected Cells. International Journal of Molecular Sciences, 20(23), 5952. https://doi.org/10.3390/ijms20235952