Simplified Head-to-Tail Cyclic Polypeptides as Biomaterial-Associated Antimicrobials with Endotoxin Neutralizing and Anti-Inflammatory Capabilities
Abstract
1. Introduction
2. Results
2.1. Design and Characterization of the Peptides
2.2. Circular Dichroism Spectroscopic Analysis
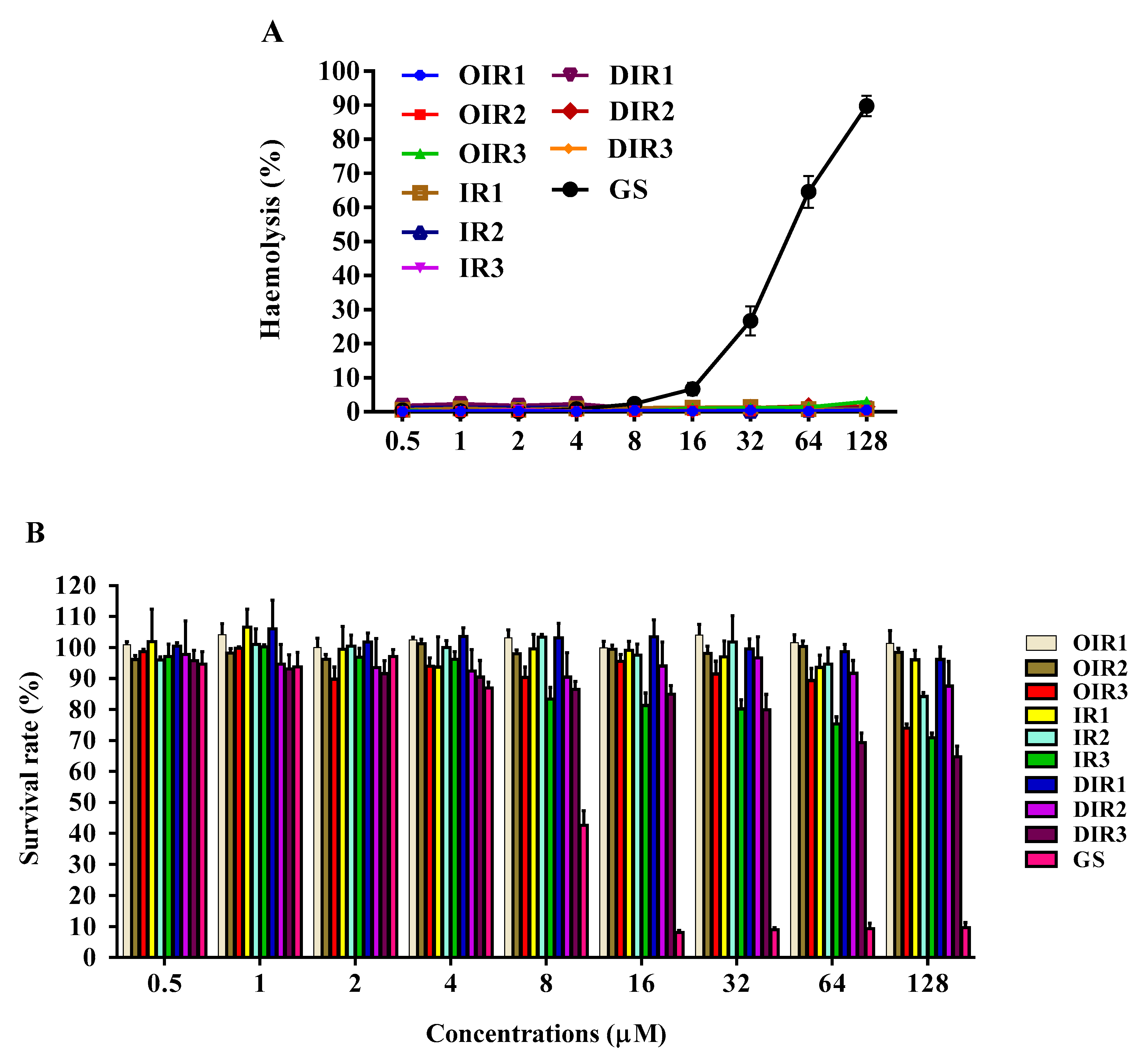
2.3. Antimicrobial and Hemolytic Activities and Selectivities
2.4. Cytotoxicity
2.5. Salt Sensitivity and Serum Stability
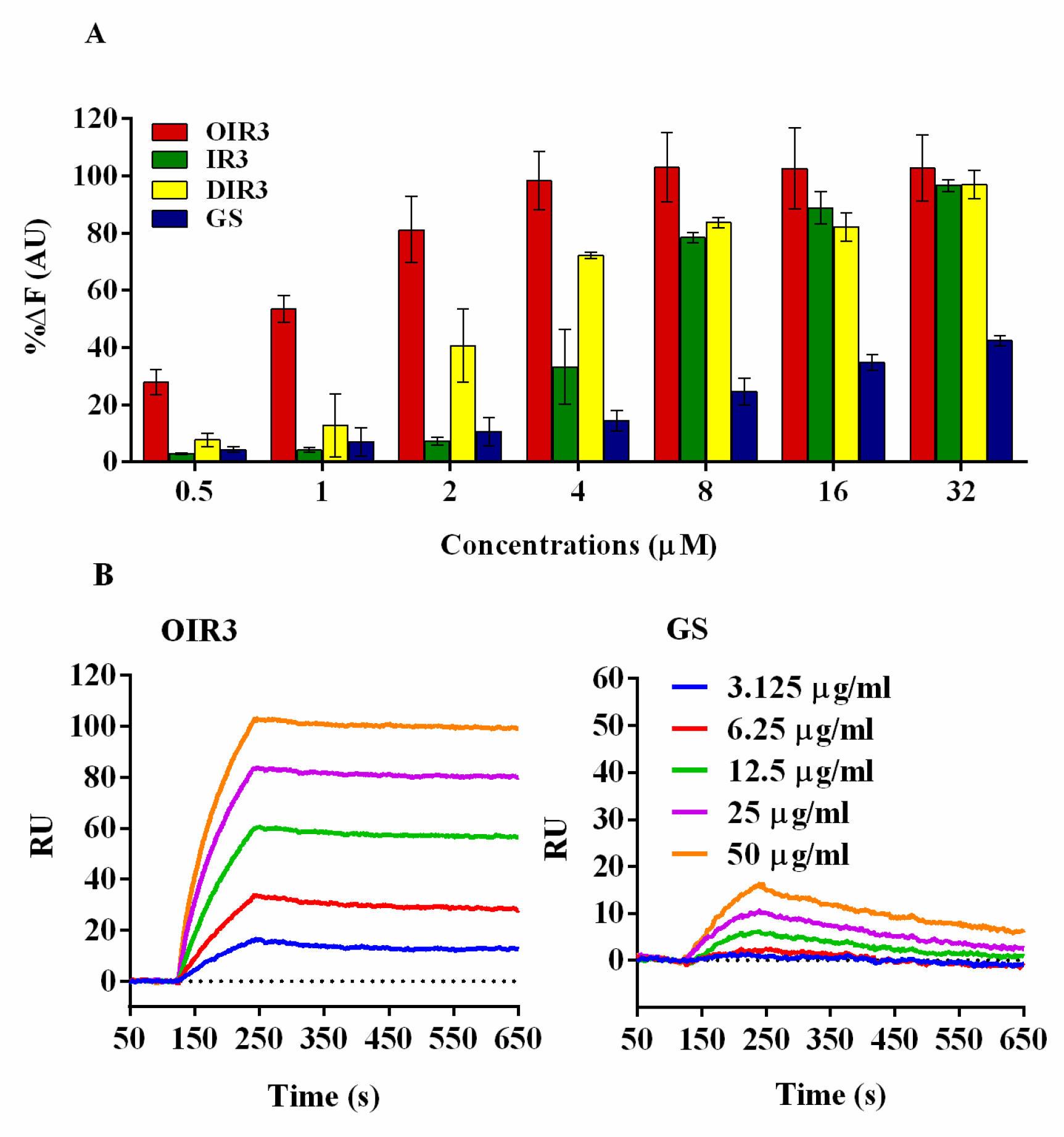
2.6. Endotoxin Neutralization Activities
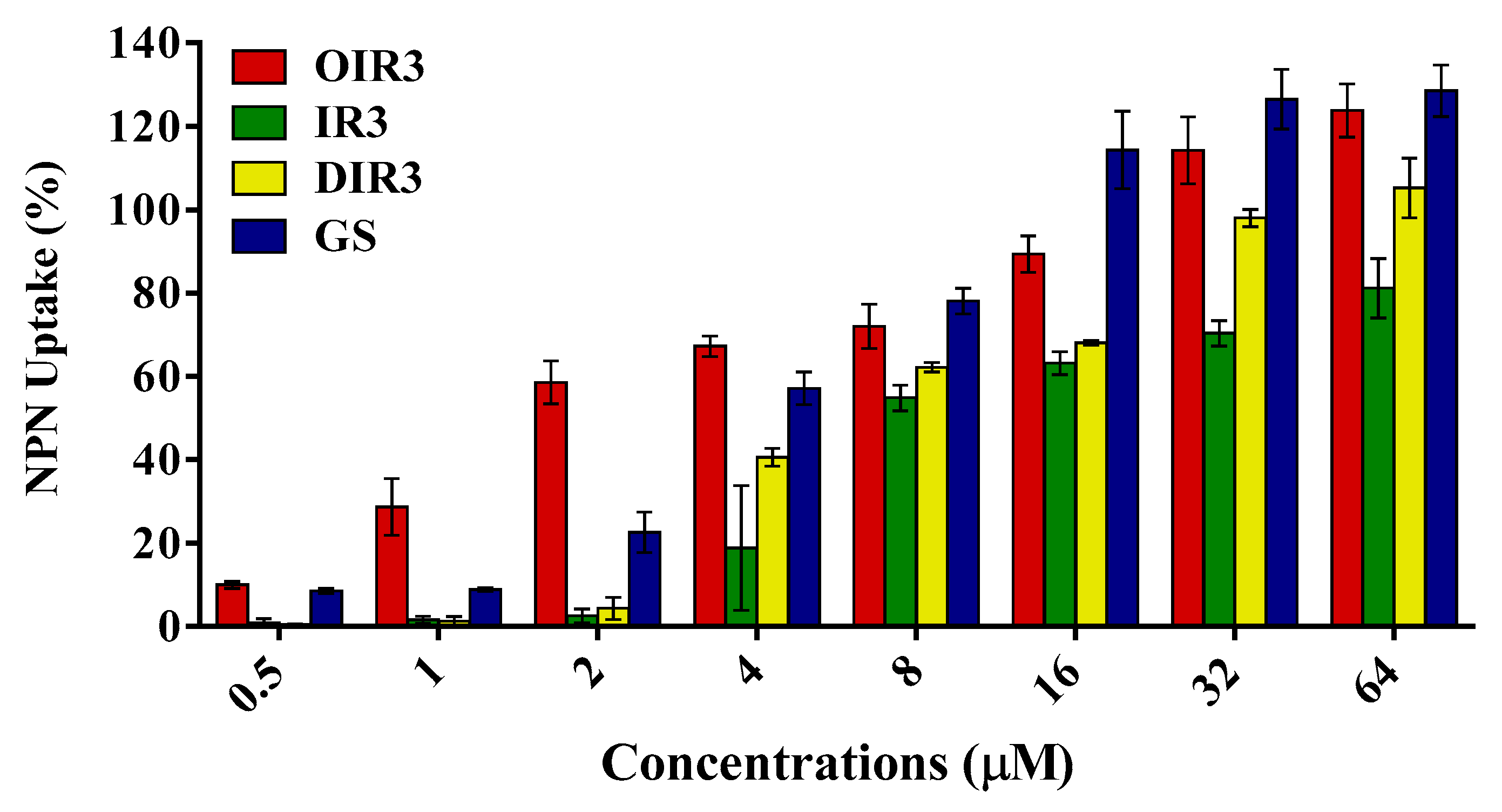
2.7. Outer Membrane Permeability Assay
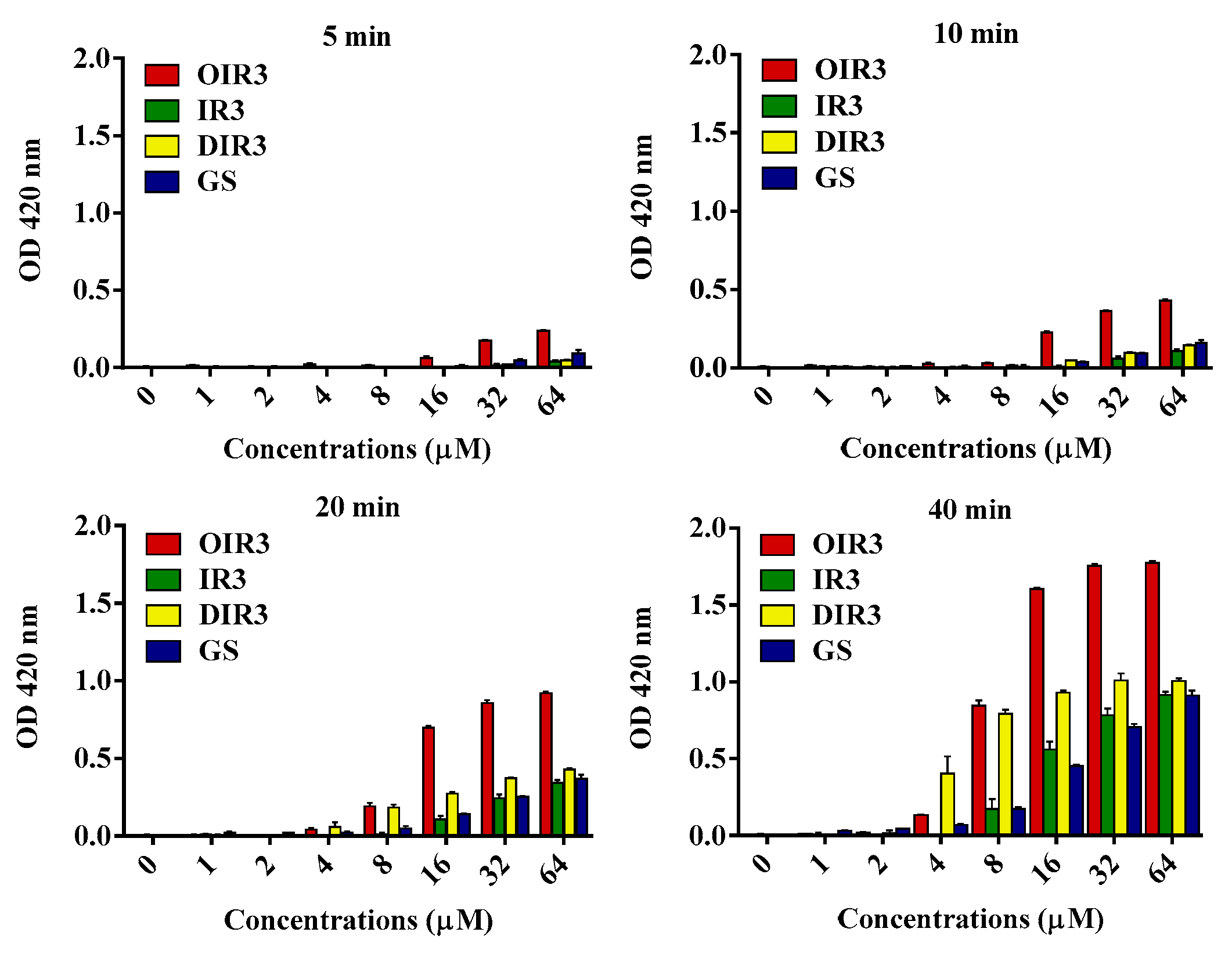
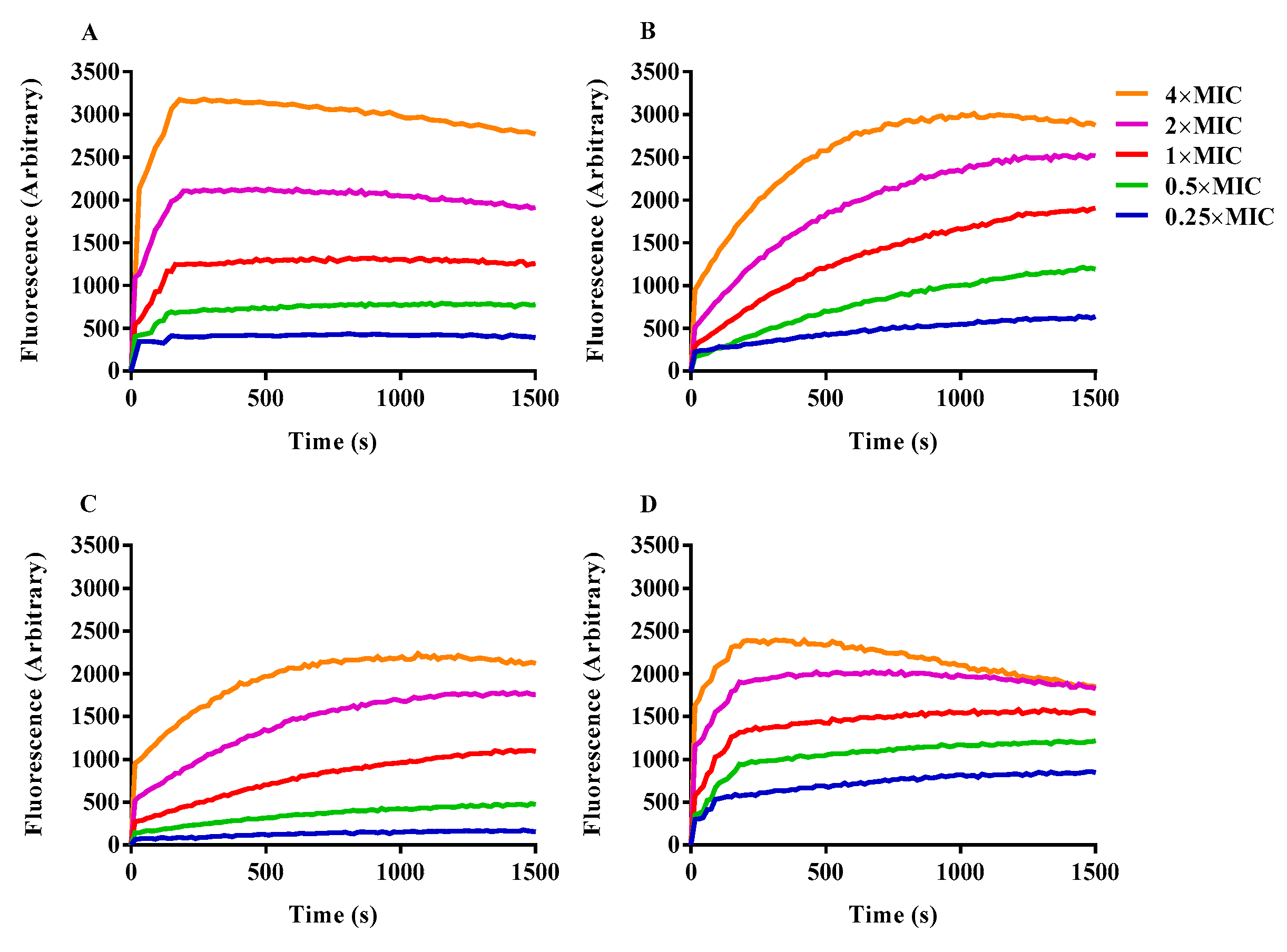
2.8. Inner Membrane Permeability Assay
2.9. Cytoplasmic Membrane Electrical Potential Measurement
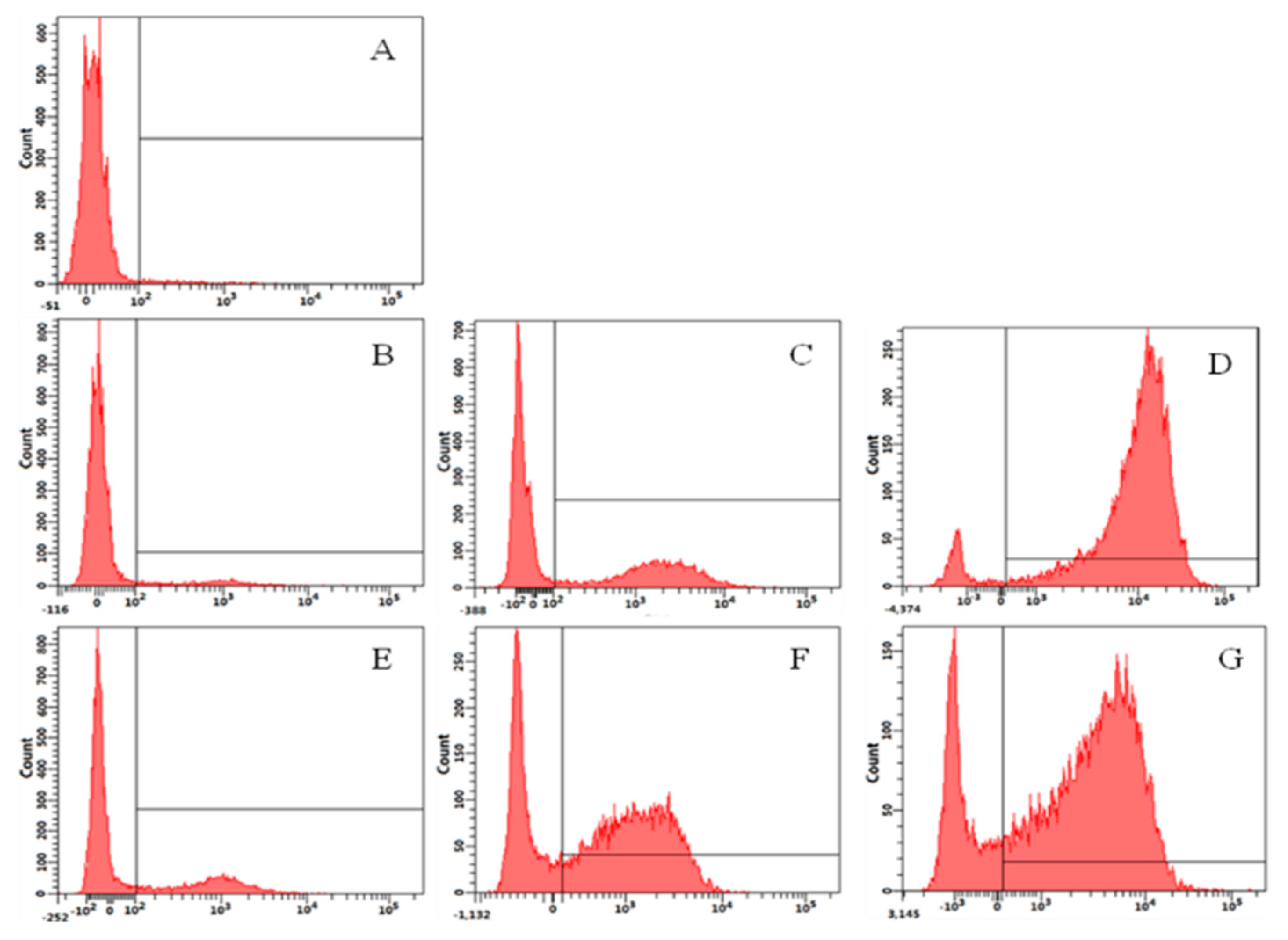
2.10. Flow Cytometry Analysis
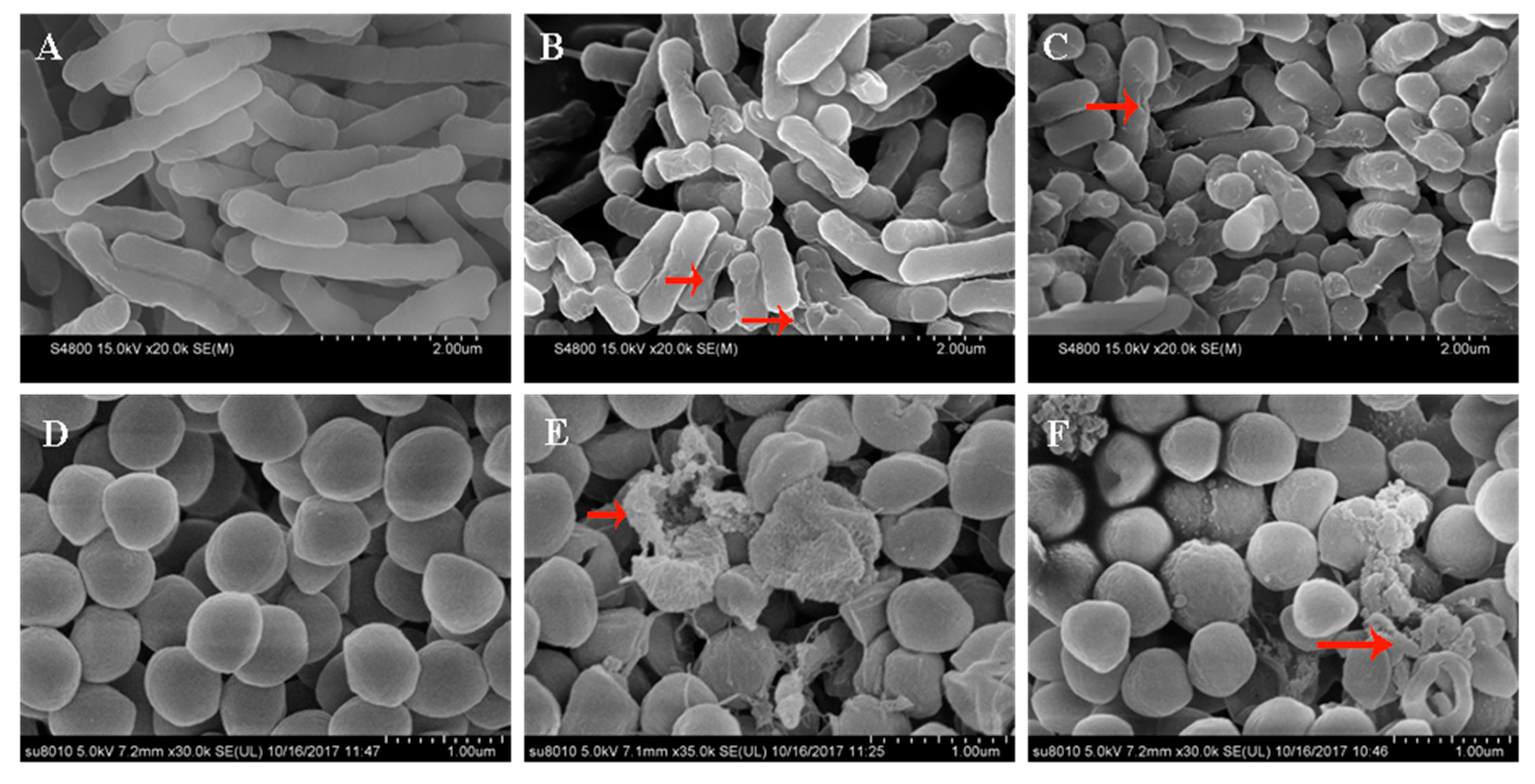
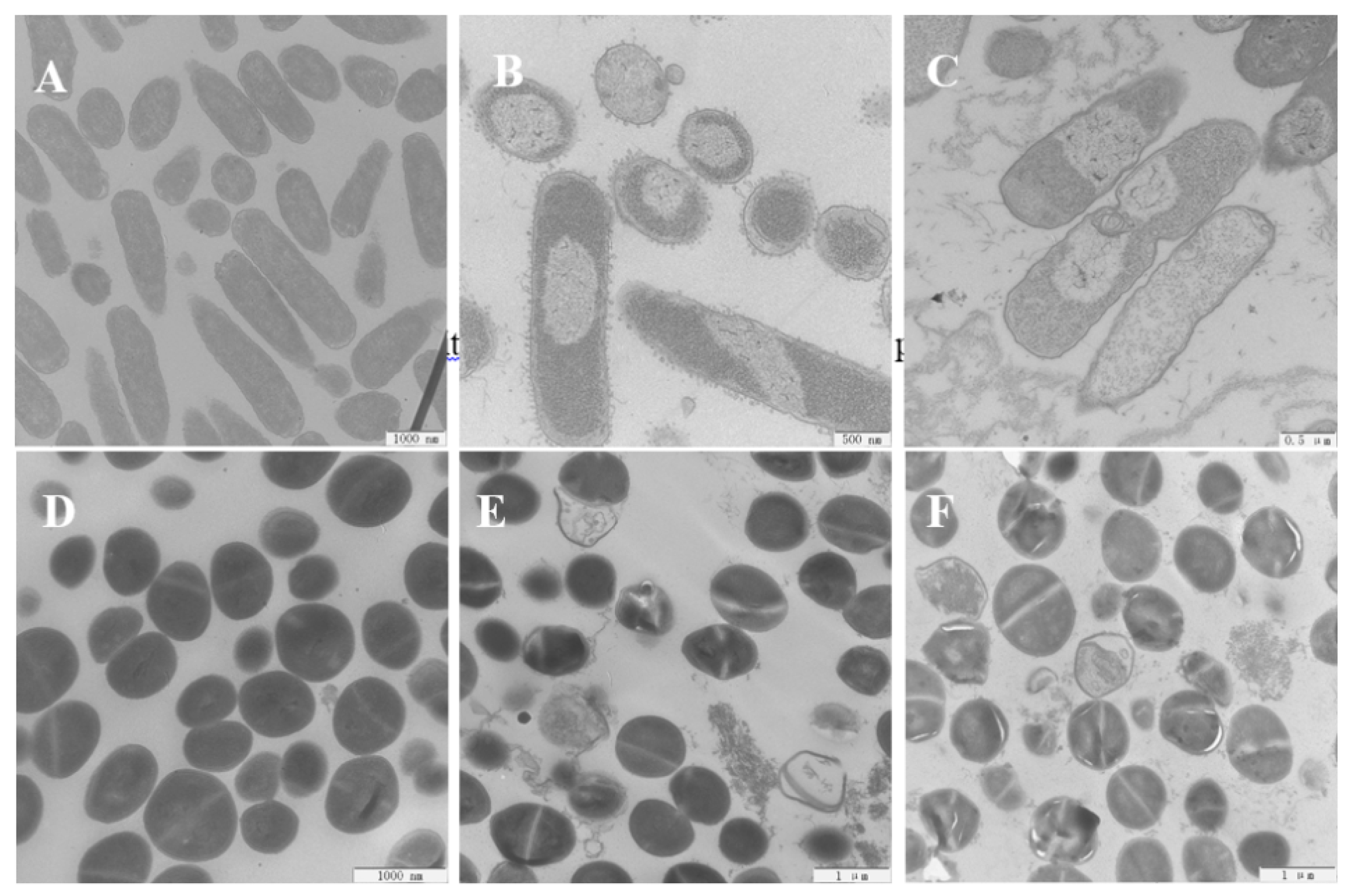
2.11. Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM)
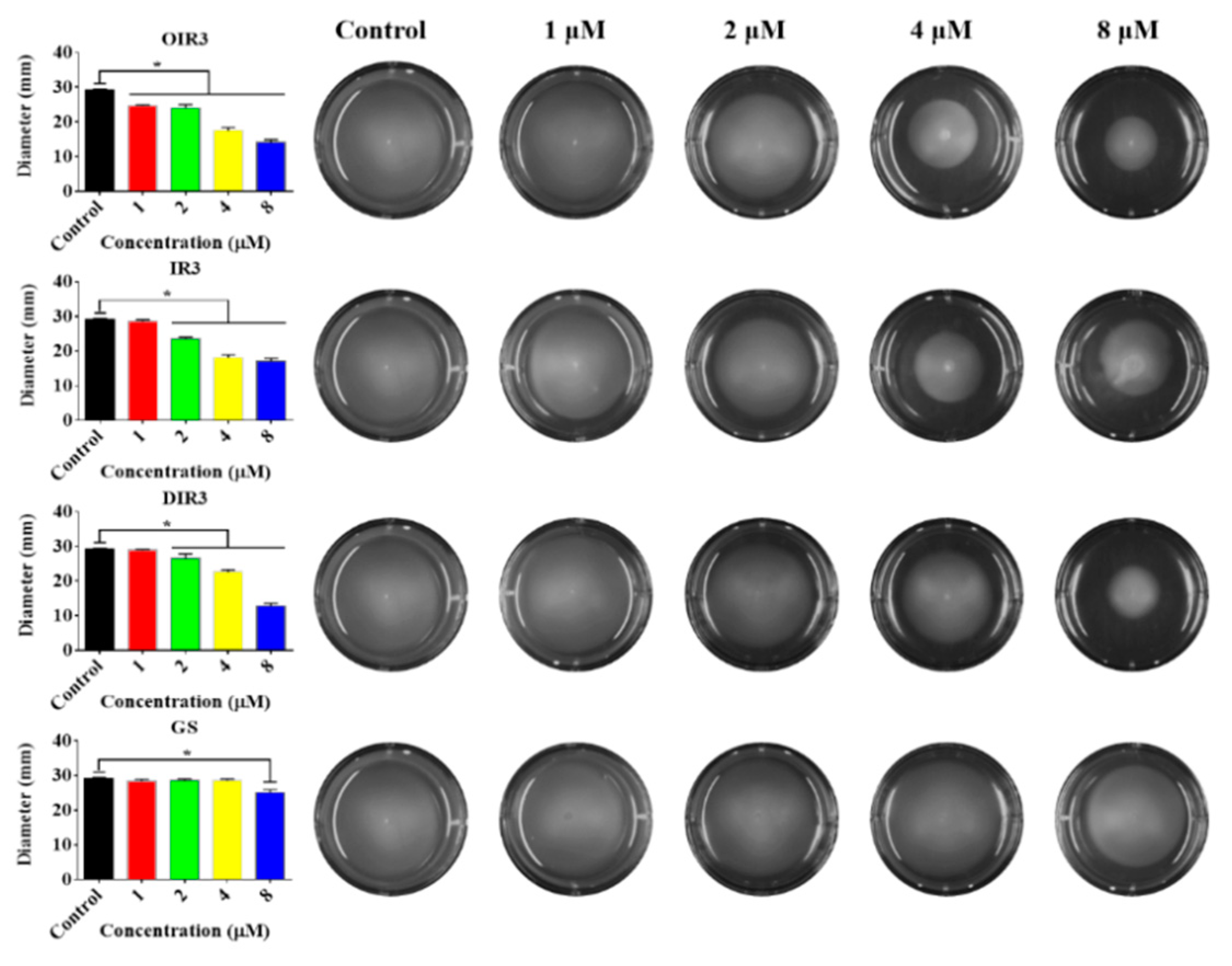
2.12. Swimming Motility Assays
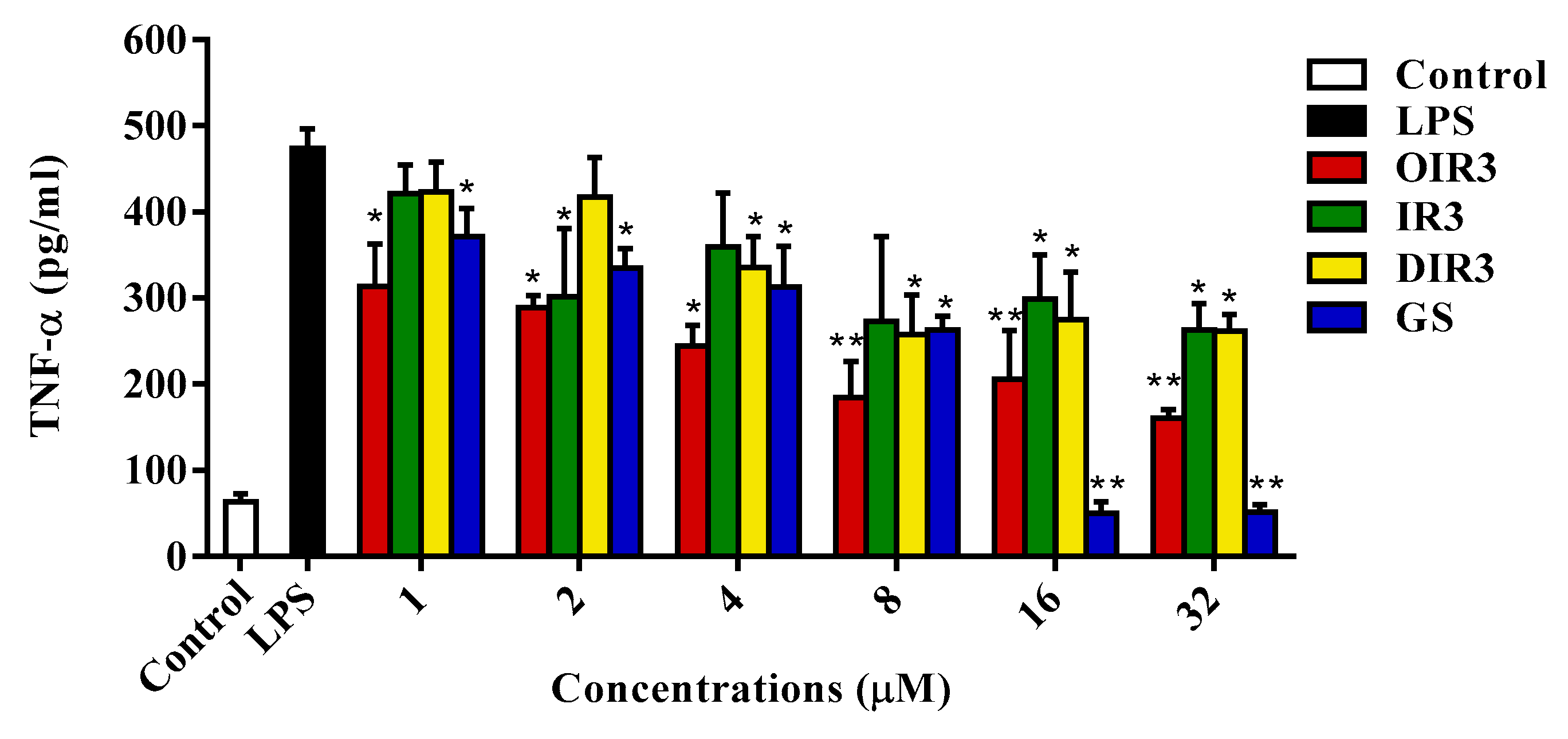
2.13. Anti-Inflammatory Activities
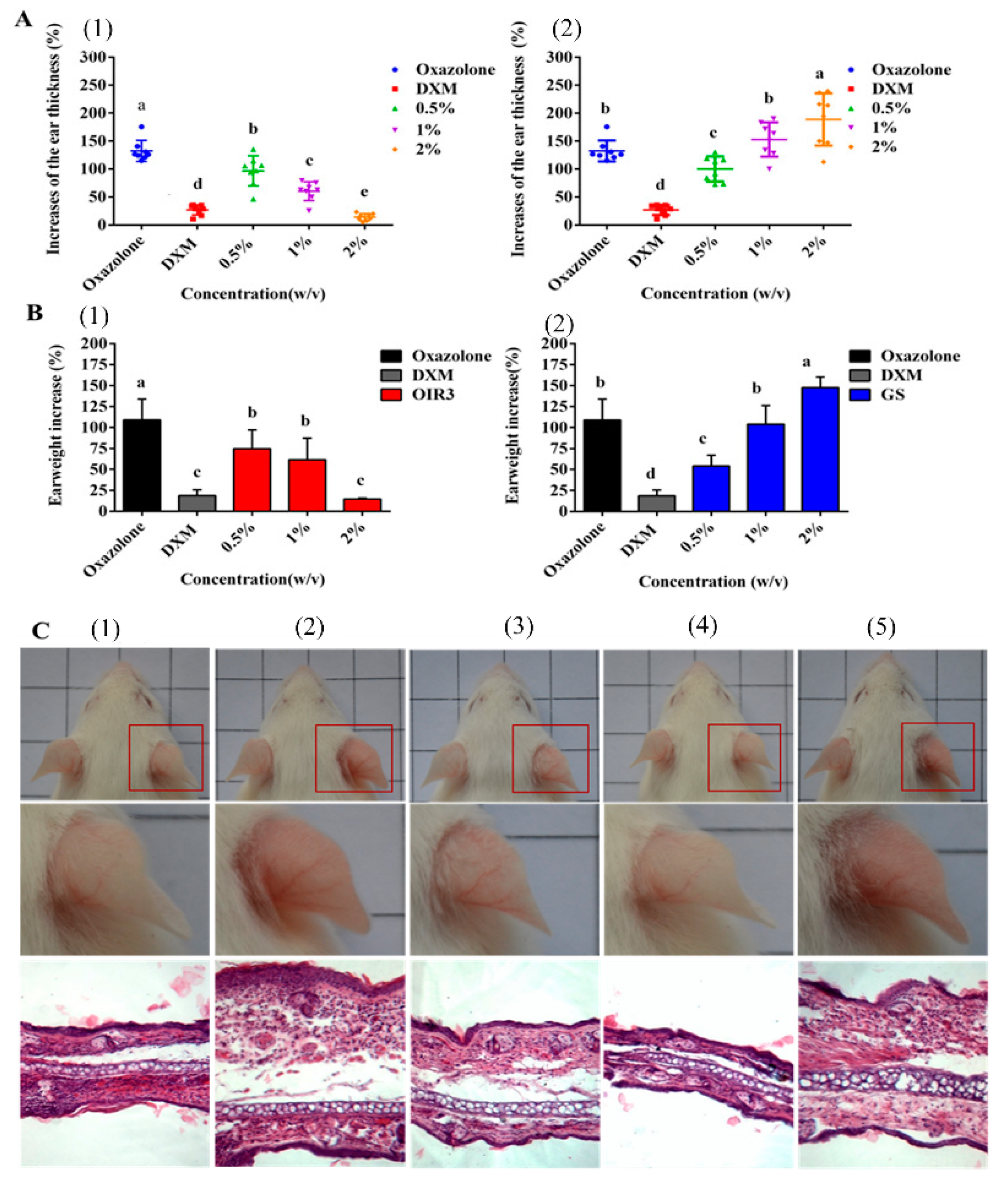
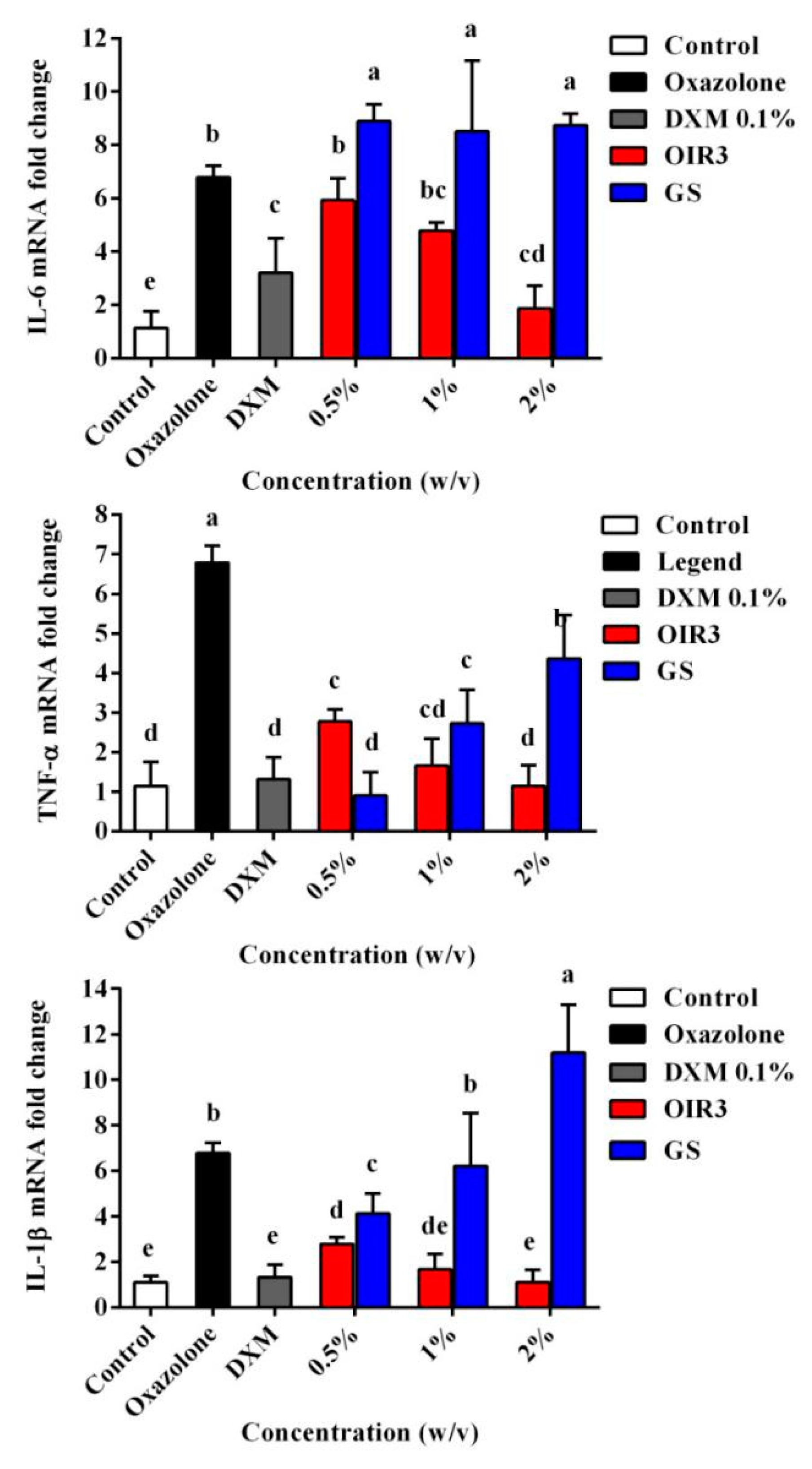
2.14. Effects of Peptides on Oxazolone-Induced Skin Inflammation in Mice
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strains
4.3. Peptide Design and Sequence Analysis
4.4. Synthesis and Characterization of Peptides
4.5. Circular Dichroism (CD) Spectroscopy
4.6. Antimicrobial Assays
4.7. Stability Assays
4.8. Hemolysis Assays
4.9. Cytotoxicity Assay
4.10. Swimming Motility Assays
4.11. Binding Affinities to LPS
4.12. Surface Plasmon Resonance (SPR) Experiments
4.13. Outer Membrane Permeability Assay
4.14. Inner Membrane Permeability Assay
4.15. Cytoplasmic Membrane Electrical Potential Measurement
4.16. Flow Cytometry
4.17. Scanning Electron Microscopy (SEM)
4.18. Transmission Electron Microscopy (TEM)
4.19. Determination of Inflammatory Factors
4.20. Animal Experiments
4.21. Allergic Dermatitis Mode
4.22. Hematoxylin and Eosin (HE) Staining
4.23. Determination of Inflammatory Factors
4.24. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMPs | antimicrobial peptides |
| CD | circular dichroism |
| DMSO | dimethyl sulfoxide |
| EDTA | ethylenediaminetetraacetic acid |
| FACS | fluorescence-assisted cell sorting |
| FBS | fetal bovine serum |
| hRBCs | human red blood cells |
| LPS | lipopolysaccharides |
| MHB | Mueller-Hinton broth |
| MIC | minimum inhibitory concentration |
| MTT | 3-(4,5-dimethylthiozol-2-yl)-2,5-diphenyltetrazolium bromide |
| NF-κB | nuclear factor-κB |
| NO | nitric oxide |
| NPN | N-phenyl-1-naphthylamine |
| OD | optical density |
| PI | propidium iodide |
| qRT-PCR | quantitative real-time PCR |
| SDS | sodium dodecyl sulfate |
| SEM | scanning electronic microscopy |
| TFE | trifluoroethyl alcohol |
| TLR4 | Toll-like receptor-4 |
| DXM | dexamethasone |
References
- Mi, G.; Shi, D.; Herchek, W.; Webster, T.J. Self-assembled arginine-rich peptides as effective antimicrobial agents. J. Biomed. Mater. Res. Part A 2017, 105, 1046–1054. [Google Scholar] [CrossRef]
- Sun, H.; Hong, Y.; Xi, Y.; Zou, Y.; Gao, J.; Du, J. Synthesis, Self-Assembly and Biomedical Applications of Antimicrobial Peptide-Polymer Conjugates. Biomacromolecules 2018, 19, 1701–1720. [Google Scholar] [CrossRef]
- Pérez-Peinado, C.; Dias, S.A.; Domingues, M.M.; Benfield, A.H.; Freire, J.M.; Rádis-Baptista, G.; Gaspar, D.; Marb, C.; Craik, D.J.; Henriques, S.T. Mechanisms of bacterial membrane permeabilization by crotalicidin (Ctn) and its fragment Ctn(15-34), antimicrobial peptides from rattlesnake venom. J. Biol. Chem. 2018, 293, 1536. [Google Scholar] [CrossRef]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef]
- Tajbakhsh, M.; Karimi, A.; Tohidpour, A.; Abbasi, N.; Fallah, F.; Akhavan, M.M. The antimicrobial potential of a new derivative of cathelicidin from Bungarus fasciatus against methicillin-resistant Staphylococcus aureus. J. Microbiol. 2018, 56, 128–137. [Google Scholar] [CrossRef]
- Yu, K.; Lo, J.C.Y.; Yan, M.; Yang, X.; Brooks, D.E.; Hancock, R.E.W.; Lange, D.; Kizhakkedathu, J.N. Anti-adhesive antimicrobial peptide coating prevents catheter associated infection in a mouse urinary infection model. Biomaterials 2017, 116, 69. [Google Scholar] [CrossRef]
- Ma, Z.; Wei, D.; Yan, P.; Zhu, X.; Shan, A.; Bi, Z. Characterization of cell selectivity, physiological stability and endotoxin neutralization capabilities of α-helix-based peptide amphiphiles. Biomaterials 2015, 52, 517–530. [Google Scholar] [CrossRef]
- Zorzi, A.; Deyle, K.; Heinis, C. Cyclic peptide therapeutics: Past, present and future. Curr. Opin. Chem. Biol. 2017, 38, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.; Nigro, E.; De Biasi, M.G.; Daniele, A.; Morelli, G.; Galdiero, S.; Scudiero, O. Cyclic Peptides as Novel Therapeutic Microbicides: Engineering of Human Defensin Mimetics. Molecules 2017, 22, 1217. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Jiao, Q.; Liu, T.; Tan, S.J.; Jiang, Z.Y. Discovery of a head-to-tail cyclic peptide as the Keap1-Nrf2 protein-protein interaction inhibitor with high cell potency. Eur. J. Med. Chem. 2017, 143, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Ong, Y.S.; Gao, L.; Kalesh, K.A.; Yu, Z.; Wang, J.; Liu, C.; Li, Y.; Sun, H.; Lee, S.S. Recent Advances in Synthesis and Identification of Cyclic Peptides for Bioapplications. Curr. Top. Med. Chem. 2017, 17, 2302–2318. [Google Scholar] [CrossRef]
- Frederick, B.A.; Helfrich, B.A.; Coldren, C.D.; Zheng, D.; Chan, D.; Jr, B.P.; Raben, D. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol. Cancer Ther. 2007, 6, 1683–1691. [Google Scholar] [CrossRef]
- Gause, G.F.; Brazhnikova, M.G. Gramicidin S and its use in the Treatment of Infected Wounds. Nature 1944, 154, 703. [Google Scholar] [CrossRef]
- Marina, R.; Hans André, E.B.; Nicolas Maré, V.; Marietjie, S.; Abré, D.B. Direct surfactin-gramicidin S antagonism supports detoxification in mixed producer cultures of Bacillus subtilis and Aneurinibacillus migulanus. Microbiology 2012, 158(Pt. 12), 3072–3082. [Google Scholar]
- Shao, C.; Tian, H.; Wang, T.; Wang, T.; Chou, S.; Shan, A.; Cheng, B. Central β-turn increases the cell selectivity of imperfectly amphipathic α-helical peptides. Acta Biomater. 2018, 69, 243–255. [Google Scholar] [CrossRef]
- Song, Y.M.; Yang, S.T.; Lim, S.S.; Kim, Y.; Hahm, K.S.; Kim, J.I.; Shin, S.Y. Effects of L- or D-Pro incorporation into hydrophobic or hydrophilic helix face of amphipathic alpha-helical model peptide on structure and cell selectivity. Biochem. Biophys. Res. Commun. 2004, 314, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.S.; Lee, B.J. Correlation between the activities of alpha-helical antimicrobial peptides and hydrophobicities represented as RP HPLC retention times. Peptides 2005, 26, 2050–2056. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Kofoed, C.; Li, M.; Gonçalves, J.P.L.; Hansen, J.; Wolfram, M.; Hansen, A.K.; Hansen, C.H.F.; Diness, F.; Schoffelen, S.; et al. Computational Evolution of Threonine-Rich β-Hairpin Peptides Mimicking Specificity and Affinity of Antibodies. ACS Cent. Sci. 2019, 5, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Jobin, V.; Ramakrishnan, N. Antibacterial activity of human neutrophil defensin HNP-1 analogs without cysteines. Antimicrob. Agents Chemother. 2005, 49, 4561. [Google Scholar]
- Gupta, V.K.; Kaushik, A.; Chauhan, D.S.; Ahirwar, R.K.; Sharma, S.; Bisht, D. Anti-mycobacterial activity of some medicinal plants used traditionally by tribes from Madhya Pradesh, India for treating tuberculosis related symptoms. J. Ethnopharmacol. 2018, 227, 113–120. [Google Scholar] [CrossRef]
- Famuyide, I.M.; Aro, A.O.; Fasina, F.O.; Eloff, J.N.; McGaw, L.J. Antibacterial and antibiofilm activity of acetone leaf extracts of nine under-investigated south African Eugenia and Syzygium (Myrtaceae) species and their selectivity indices. BMC Complementary Altern. Med. 2019, 19, 141. [Google Scholar] [CrossRef]
- Ryder, M.P.; Wu, X.; Mckelvey, G.R.; Mcguire, J.; Schilke, K.F. Binding interactions of bacterial lipopolysaccharide and the cationic amphiphilic peptides polymyxin B and WLBU2. Colloids Surf. B Biointerfaces 2014, 120, 81–87. [Google Scholar] [CrossRef]
- Sarmishtha, D.; Hao, Z.; David, D.S.; Croniger, C.M.; Xiaoxia, L.; Stark, G.R. Erlotinib protects against LPS-induced endotoxicity because TLR4 needs EGFR to signal. Proc. Natl. Acad. Sci. USA 2015, 112, 9680. [Google Scholar]
- Yuin, O.Z.; Junchi, C.; Yuan, H.; Kaijin, X.; Zhongkang, J.; Weimin, F.; Yan, Y.Y. Effect of stereochemistry, chain length and sequence pattern on antimicrobial properties of short synthetic β-sheet forming peptide amphiphiles. Biomaterials 2014, 35, 1315–1325. [Google Scholar]
- Wang, J.; Chou, S.; Yang, Z.; Yang, Y.; Wang, Z.; Song, J.; Dou, X.; Shan, A. Combating drug-resistant fungi with novel imperfectly amphipathic palindromic peptides. J. Med. Chem. 2018, 61, 3889–3907. [Google Scholar] [CrossRef]
- Ma, Q.Q.; Lv, Y.F.; Gu, Y.; Dong, N.; Li, D.S.; Shan, A.S. Rational design of cationic antimicrobial peptides by the tandem of leucine-rich repeat. Amino Acids 2013, 44, 1215–1224. [Google Scholar] [CrossRef]
- Dong, N.; Ma, Q.; Shan, A.; Lv, Y.F.; Hu, W.; Gu, Y.; Li, Y. Strand length-dependent antimicrobial activity and membrane-active mechanism of arginine- and valine-rich β-hairpin-like antimicrobial peptides. Antimicrob. Agents Chemother. 2012, 56, 2994. [Google Scholar] [CrossRef]
- Dong, N.; Zhu, X.; Chou, S.; Shan, A.; Li, W.; Jiang, J. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials 2014, 35, 8028–8039. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Hyun, Y.J.; Lee, J.H.; Lim, H.S. Comparison of Cell Permeability of Cyclic Peptoids and Linear Peptoids. ACS Comb. Sci. 2018, 20, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Yang, X.; Zhang, L.; Moon, S.H.; Yousef, A.E. New Paenibacillus strain produces a family of linear and cyclic antimicrobial lipopeptides: Cyclization is not essential for their antimicrobial activity. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Cirac, A.; Moiset, G.; Mika, J.; Koçer, A.; Salvador, P.; Poolman, B.; Marrink, S.; Sengupta, D. The Molecular Basis for Antimicrobial Activity of Pore-Forming CyclicPeptides. Biophys. J. 2011, 100, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- Wessolowski, A.; Bienert, M.; Dathe, M. Antimicrobial activity of arginine- and tryptophan-rich hexapeptides: The effects of aromatic clusters, d-amino acid substitution and cyclization. Chem. Biol. Drug Des. 2010, 64, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Yoneyama, S.; Fujii, N.; Miyajima, K.; Yamada, K.; Kirino, Y.; Anzai, K. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry 1997, 36, 9799. [Google Scholar] [CrossRef] [PubMed]
- Manabe, T.; Kawasaki, K. D-form KLKLLLLLKLK-NH2 peptide exerts higher antimicrobial properties than its L-form counterpart via an association with bacterial cell wall components. Sci. Rep. 2017, 7, 43384. [Google Scholar] [CrossRef] [PubMed]
- Berditsch, M.; Trapp, M.; Afonin, S.; Weber, C.; Misiewicz, J.; Turkson, J.; Ulrich, A.S. Antimicrobial peptide gramicidin S is accumulated in granules of producer cells for storage of bacterial phosphagens. Sci. Rep. 2017, 7, 44324. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef]
- Cornette, J.L.; Cease, K.B.; Margalit, H.; Spouge, J.L.; Berzofsky, J.A.; Delisi, C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J. Mol. Biol. 1987, 195, 659–685. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, J.; Han, J.; Gao, L.; Liu, H.; Lu, Z.; Zhao, H.; Bie, X. Insights into the Antimicrobial Activity and Cytotoxicity of Engineered α-Helical Peptide Amphiphiles. J. Med. Chem. 2016, 59, 10946–10962. [Google Scholar] [CrossRef]
- Cheng, W.; Mingqiang, S.; Neelakshi, G.; Tolbert, W.D.; Fang, C.; Naixin, Z.; Ke, Y.; Aiping, W.; Yongping, S.; Tianmin, C. Design of a potent antibiotic peptide based on the active region of human defensin 5. J. Med. Chem. 2015, 58, 3083–3093. [Google Scholar]
- Zhan, Y.O.; Wiradharma, N.; Yi, Y.Y. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhanced therapeutic potentials. Adv. Drug Deliv Rev. 2014, 78, 28–45. [Google Scholar]
- Taniguchi, M.; Ochiai, A.; Takahashi, K.; Nakamichi, S.I.; Nomoto, T.; Saitoh, E.; Kato, T.; Tanaka, T. Antimicrobial activity and mechanism of action of a novel cationic α-helical octadecapeptide derived from α-amylase of rice. Biopolymers 2015, 104, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Inoue, M. Comprehensive Structure-Activity Relationship Studies of Macrocyclic Natural Products Enabled by Their Total Syntheses. Chem. Rev. 2019, 119, 10002–10031. [Google Scholar] [CrossRef] [PubMed]
- Cutrona, K.J.; Kaufman, B.A.; Figueroa, D.M.; Elmore, D.E. Role of Arginine and Lysine in the Antimicrobial Mechanism of Histone-derived Antimicrobial Peptides. FEBS Lett. 2016, 589, 3915–3920. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef]
- Niv, P.; Yechiel, S. A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 2005, 280, 10378–10387. [Google Scholar]
- Mika, J.T.; Moiset, G.; Cirac, A.D.; Feliu, L.; Bardají, E.; Planas, M.; Sengupta, D.; Marrink, S.J.; Poolman, B. Structural basis for the enhanced activity of cyclic antimicrobial peptides: The case of BPC194. Biochim. Biophys. Acta 2011, 1808, 2197–2205. [Google Scholar] [CrossRef]
- Shao, C.; Li, W.; Tan, P.; Shan, A.; Dou, X.; Ma, D.; Liu, C. Symmetrical Modification of Minimized Dermaseptins to Extend the Spectrum of Antimicrobials with Endotoxin Neutralization Potency. Int. J. Mol. Sci. 2019, 20, 1417. [Google Scholar] [CrossRef]
- Pedersen, R.M.; Grønnemose, R.B.; Stærk, K.; Asferg, C.A.; Andersen, T.B.; Kolmos, H.J.; Møller- Jensen, J.; Andersen, T.E. A Method for Quantification of Epithelium Colonization Capacity by Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 2018, 8, 16. [Google Scholar] [CrossRef]
- Onoue, Y.; Takekawa, N.; Nishikino, T.; Kojima, S.; Homma, M. The role of conserved charged residues in the bidirectional rotation of the bacterial flagellar motor. Microbiologyopen 2018, 7, e00587. [Google Scholar] [CrossRef]
- Moor, K.; Diard, M.; Sellin, M.E.; Felmy, B.; Wotzka, S.Y.; Toska, A.; Bakkeren, E.; Arnoldini, M.; Bansept, F.; Co, A.D.; et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017, 544, 498–502. [Google Scholar] [CrossRef]
- Wang, Q.; Jin, L.; Wang, H.; Tai, S.; Liu, H.; Zhang, D. AWRK6, A Synthetic Cationic Peptide Derived from Antimicrobial Peptide Dybowskin-2CDYa, Inhibits Lipopolysaccharide-Induced Inflammatory Response. Int. J. Mol. Sci. 2018, 19, 600. [Google Scholar] [CrossRef] [PubMed]
- Han, B.H.; Lee, Y.J.; Yoon, J.J.; Choi, E.S.; Namgung, S.; Jin, X.J.; Jeong, D.H.; Kang, D.G.; Lee, H.S. Hwangryunhaedoktang exerts anti-inflammation on LPS-induced NO production by suppressing MAPK and NF-κB activation in RAW264.7 macrophages. J. Integr. Med. 2017, 15, 326–336. [Google Scholar] [CrossRef]
- Cobos, C.C.; Bansal, P.S.; Navarro, S.; Wilson, D.; Don, L.; Giacomin, P.; Loukas, A.; Daly, N.L. An engineered cyclic peptide alleviates symptoms of inflammation in a murine model of inflammatory bowel disease. J. Biol. Chem. 2017, 292, 10288–10294. [Google Scholar] [CrossRef] [PubMed]
- Kiatsurayanon, C.; Ogawa, H.; Niyonsaba, F. The role of host defense peptide human β-defensins in the maintenance of skin barriers. Curr. Pharm. Des. 2018, 24, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Y.; Thapa, P.; Espiritu, M.J.; Menon, V.; Bingham, J.P. From nature to creation: Going around in circles, the art of peptide cyclization. Bioorganic Med. Chem. 2017, 26, 1135. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.L.; Macarthur, M.W.; Hutchinson, E.G.; Thornton, J.M. Stereochemical quality of protein structure coordinates. Proteins-Struct. Funct. Bioinform. 2010, 12, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Xia, X.; Xu, L.; Wang, Y.Z. Design of hybrid β-hairpin peptides with enhanced cell specificity and potent anti-inflammatory activity. Biomaterials 2013, 34, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Q.; Dong, N.; Shan, A.S.; Lv, Y.F.; Li, Y.Z.; Chen, Z.H.; Cheng, B.J.; Li, Z.Y. Biochemical property and membrane-peptide interactions of de novo antimicrobial peptides designed by helix-forming units. Amino Acids 2012, 43, 2527–2536. [Google Scholar] [CrossRef]
- Chou, S.; Shao, C.; Wang, J.; Shan, A.; Xu, L.; Dong, N.; Li, Z. Short, multiple-stranded β-hairpin peptides have antimicrobial potency with high selectivity and salt resistance. Acta Biomater. 2016, 30, 78–93. [Google Scholar] [CrossRef]
- Dong, N.; Wang, C.; Zhang, T.; Zhang, L.; Xue, C.; Feng, X.; Bi, C.; Shan, A. Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria. Int. J. Mol. Sci. 2019, 20, 3954. [Google Scholar] [CrossRef]
- Ouberai, M.; Garch, F.E.; Bussiere, A.; Riou, M.; Alsteens, D.; Lins, L.; Baussanne, I.; Dufrêne, Y.F.; Brasseur, R.; Decout, J.L. The Pseudomonas aeruginosa membranes: A target for a new amphiphilic aminoglycoside derivative? Biochim. Biophys. Acta 2011, 1808, 1716–1727. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Wang, X.; Chen, W.; Sun, H.; Wang, J. Lipopolysaccharide induces amyloid formation of antimicrobial peptide HAL-2. Biochim. Biophys. Acta 2014, 1838, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; He, S.; Wang, J.; Yang, Y.; Zhang, L.; Li, Y.; Shan, A. Rational Design of Short Peptide Variants by Using Kunitzin-RE, an Amphibian-Derived Bioactivity Peptide, for Acquired Potent Broad-Spectrum Antimicrobial and Improved Therapeutic Potential of Commensalism Coinfection of Pathogens. J. Med. Chem. 2019, 62, 4586–4605. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Tan, P.; Zhu, Y.; Shao, C.; Shan, A.; Li, L. Highly Stabilized α-Helical Coiled Coils Kill Gram-Negative Bacteria by Multicomplementary Mechanisms under Acidic Condition. ACS Appl. Mater. Interfaces 2019, 11, 22113–22128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, X.; Wang, J. Recent Advances in Antibacterial and Antiendotoxic Peptides or Proteins from Marine Resources. Mar. Drugs 2018, 16, 57. [Google Scholar] [CrossRef] [PubMed]
- Rubinchik, E.; Pasetka, C. Assay Systems for Measurement of Anti-inflammatory Activity. Methods Mol. Biol. 2010, 618, 349. [Google Scholar] [PubMed]
- Sundar, R.; Francis, S.; Hiradhar, P.K.; Rajesh, N.; Devada, S. Effect of Commiphora mukul in chronic oxazolone induced mouse dermatitis model. Int. J. Vet. Sci. 2016. [Google Scholar] [CrossRef]
- Gao, H.; Liu, X.; Sun, W.; Kang, N.; Liu, Y.; Yang, S.; Xu, Q.; Wang, C.; Chen, X. Total tanshinones exhibits anti-inflammatory effects through blocking TLR4 dimerization via the MyD88 pathway. Cell Death Dis. 2017, 8, e3004. [Google Scholar] [CrossRef]

| Peptides | CN a | Sequences | AA | TMW b | MMW c | NC d | RT e (min) |
|---|---|---|---|---|---|---|---|
| OIR1 | 1 | Cyclo-(IRPIRP) | 6 | 732.95 | 732.97 | +2 | 10.32 |
| OIR2 | 2 | Cyclo-(IR)2P(IR)2P | 10 | 1271.62 | 1271.66 | +4 | 10.66 |
| OIR3 | 3 | Cyclo-(IR)3P(IR)3P | 14 | 1810.34 | 1810.35 | +6 | 12.64 |
| IR1 | 4 | IRPIRP | 6 | 750.94 | 750.90 | +2 | 10.28 |
| IR2 | 5 | (IR)2P(IR)2P | 10 | 1289.64 | 1289.60 | +4 | 10.58 |
| IR3 | 6 | (IR)3P(IR)3P | 14 | 1828.31 | 1828.35 | +6 | 12.40 |
| DIR1 | 7 | IRDPIRDP | 6 | 750.94 | 750.95 | +2 | 10.44 |
| DIR2 | 8 | (IR)2DP(IR)2DP | 10 | 1289.64 | 1289.60 | +4 | 10.60 |
| DIR3 | 9 | (IR)3DP(IR)3DP | 14 | 1828.31 | 1828.35 | +6 | 12.82 |
| Strains | MICs a (µM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OIR1 | OIR2 | OIR3 | IR1 | IR2 | IR3 | DIR1 | DIR2 | DIR3 | GS | CH b | CS c | PS d | |
| Gram-negative | |||||||||||||
| E. coli 25922 | >128 | 64 | 2 | >128 | >128 | 16 | >128 | 128 | 4 | 4 | 16 | 0.5 | 128 |
| E. coli UB 1005 | >128 | 64 | 2 | >128 | >128 | 8 | >128 | 128 | 4 | 4 | 4 | 0.5 | 64 |
| P. aeruginosa 27853 | >128 | >128 | 8 | >128 | >128 | 8 | >128 | >128 | 8 | 4 | >128 | 0.5 | 128 |
| S. typhimurium 7731 | >128 | >128 | 4 | >128 | >128 | 16 | >128 | >128 | 4 | 8 | 8 | 0.5 | 0.5 |
| S. typhimurium 14028 | >128 | >128 | 4 | >128 | >128 | 16 | >128 | >128 | 16 | 16 | 16 | 4 | 16 |
| Gram-positive | |||||||||||||
| S. aureus 29213 | >128 | >128 | 4 | >128 | >128 | 64 | >128 | >128 | 8 | 2 | 8 | 64 | 64 |
| S. aureus 25923 | >128 | >128 | 4 | >128 | >128 | 64 | >128 | >128 | 64 | 2 | 32 | 64 | 32 |
| MRSA43300 e | >128 | >128 | 4 | >128 | >128 | 64 | >128 | >128 | 64 | 2 | 32 | 64 | 32 |
| S. epidermidis 12228 | >128 | >128 | 4 | >128 | >128 | >128 | >128 | >128 | 128 | 2 | 32 | 128 | 0.5 |
| nGM (–) f1 | 256 | 147.0 | 3.5 | 256 | 256 | 12.1 | 256 | 194.0 | 6.1 | 6.1 | 18.4 | 0.8 | 24.3 |
| pGM (+) f2 | 256 | 256 | 4.0 | 256 | 256 | 90.5 | 256 | 256 | 45.3 | 2 | 22.6 | 76.1 | 13.5 |
| tGM f3 | 256 | 188.1 | 3.7 | 256 | 256 | 22.6 | 256 | 219.5 | 14.8 | 3.7 | 20.2 | 5.9 | 18.7 |
| MHC5 g | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 8 | >128 | >128 | >128 |
| nSI (–) h1 | 1.0 | 1.7 | 73.1 | 1.0 | 1.0 | 21.2 | 1.0 | 1.3 | 42.0 | 1.3 | 13.9 | 320.0 | 10.5 |
| pSI (+) h2 | 1.0 | 1.0 | 64.0 | 1.0 | 1.0 | 2.8 | 1.0 | 1.0 | 5.7 | 4.0 | 11.3 | 3.4 | 19.0 |
| tSI h3 [20,21] | 1.0 | 1.4 | 69.2 | 1.0 | 1.0 | 11.3 | 1.0 | 1.2 | 17.3 | 2.2 | 12.7 | 43.4 | 13.7 |
| Peptides | Control b | NaCl b | KCl b | NH4Cl b | MgCl2 b | CaCl2 b | ZnCl2 b | FeCl3 b | Serum c | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gram-negative strain E. coli ATCC 25922 | 25% | 50% | ||||||||
| OIR3 | 2 | 4 | 2 | 2 | 2 | 4 | 2 | 2 | 4 | 8 |
| IR3 | 16 | 32 | 16 | 16 | 16 | 32 | 16 | 16 | 32 | 64 |
| DIR3 | 4 | 16 | 16 | 8 | 8 | 16 | 8 | 8 | 8 | 16 |
| GS | 4 | 16 | 4 | 4 | 8 | 16 | 4 | 4 | 32 | 64 |
| Gram-positive strain S. aureus ATCC 29213 | ||||||||||
| OIR3 | 4 | 8 | 4 | 4 | 4 | 8 | 4 | 4 | 8 | 16 |
| IR3 | 64 | 32 | 16 | 16 | 16 | 32 | 16 | 16 | 32 | 64 |
| DIR3 | 8 | 16 | 8 | 8 | 8 | 16 | 8 | 8 | 8 | 16 |
| GS | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 32 | >64 |
| Target Gene | Sequence (5′–3′) |
|---|---|
| TNF-α | Forward: CCTATGTCTCAGCCTCTTCTCAT |
| Reverse: CACTTGGTGGTTTGCTACGA | |
| IL-6 | Forward: GGAGAGGAGACTTCACAGAGGA |
| Reverse: ATTTCCACGATTTCCCAGAGA | |
| IL-1β | Forward: TGAAATGCCACCTTTTGACAG |
| Reverse: CCACAGCCACAATGAGTGATAC | |
| mGAPDH | Forward: TGTTCCTACCCCCAATGTGT |
| Reverse: TGTGAGGGAGATGCTCAGTG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, N.; Wang, C.; Li, X.; Guo, Y.; Li, X. Simplified Head-to-Tail Cyclic Polypeptides as Biomaterial-Associated Antimicrobials with Endotoxin Neutralizing and Anti-Inflammatory Capabilities. Int. J. Mol. Sci. 2019, 20, 5904. https://doi.org/10.3390/ijms20235904
Dong N, Wang C, Li X, Guo Y, Li X. Simplified Head-to-Tail Cyclic Polypeptides as Biomaterial-Associated Antimicrobials with Endotoxin Neutralizing and Anti-Inflammatory Capabilities. International Journal of Molecular Sciences. 2019; 20(23):5904. https://doi.org/10.3390/ijms20235904
Chicago/Turabian StyleDong, Na, Chensi Wang, Xinran Li, Yuming Guo, and Xiaoli Li. 2019. "Simplified Head-to-Tail Cyclic Polypeptides as Biomaterial-Associated Antimicrobials with Endotoxin Neutralizing and Anti-Inflammatory Capabilities" International Journal of Molecular Sciences 20, no. 23: 5904. https://doi.org/10.3390/ijms20235904
APA StyleDong, N., Wang, C., Li, X., Guo, Y., & Li, X. (2019). Simplified Head-to-Tail Cyclic Polypeptides as Biomaterial-Associated Antimicrobials with Endotoxin Neutralizing and Anti-Inflammatory Capabilities. International Journal of Molecular Sciences, 20(23), 5904. https://doi.org/10.3390/ijms20235904





