microRNAs Regulating Human and Mouse Naïve Pluripotency
Abstract
1. Introduction
2. Results
2.1. microRNAs Regulating Human Naïve to Primed ESCs Transition
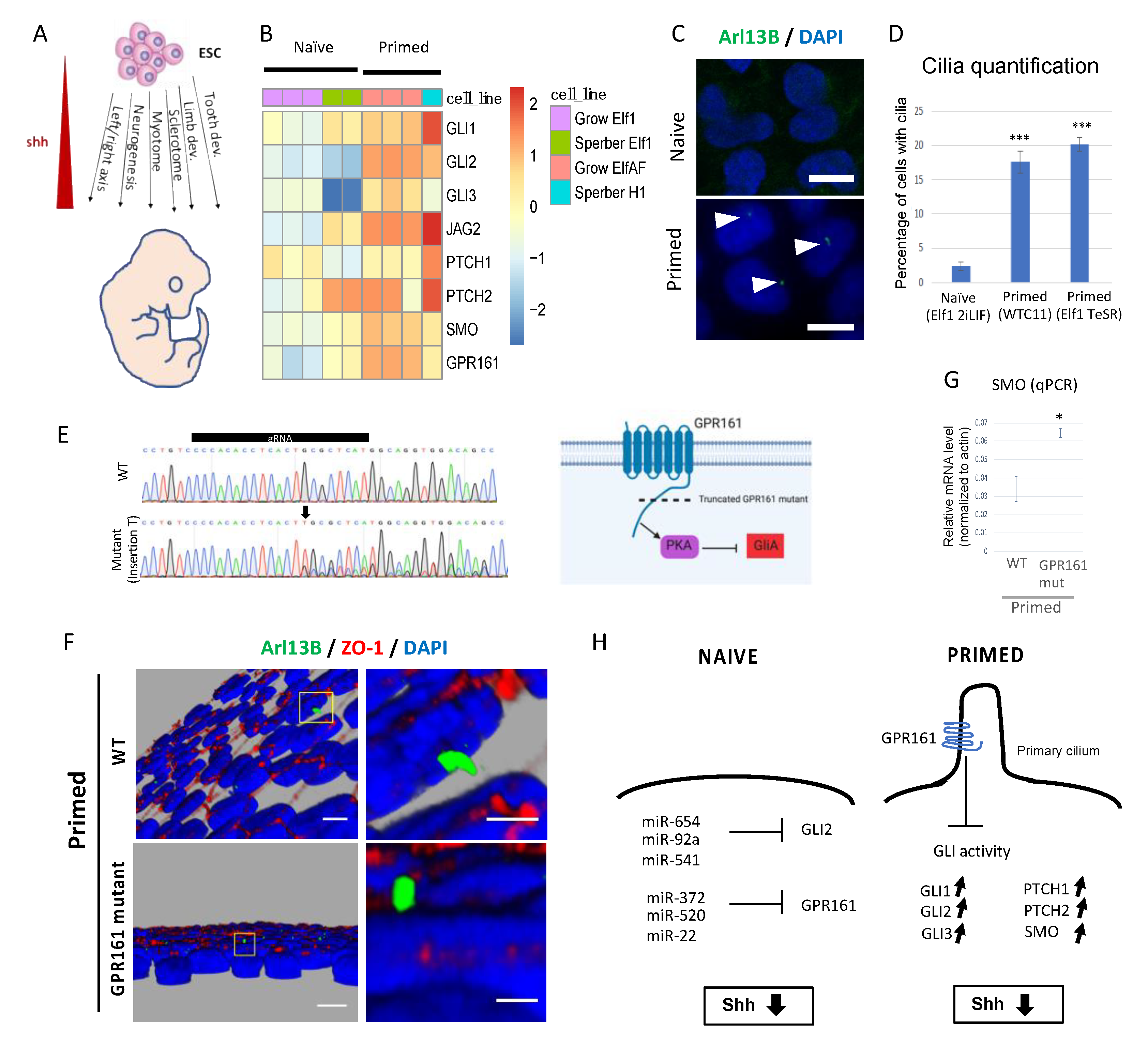
2.2. Hh Pathway in Naïve-to-Primed Transition
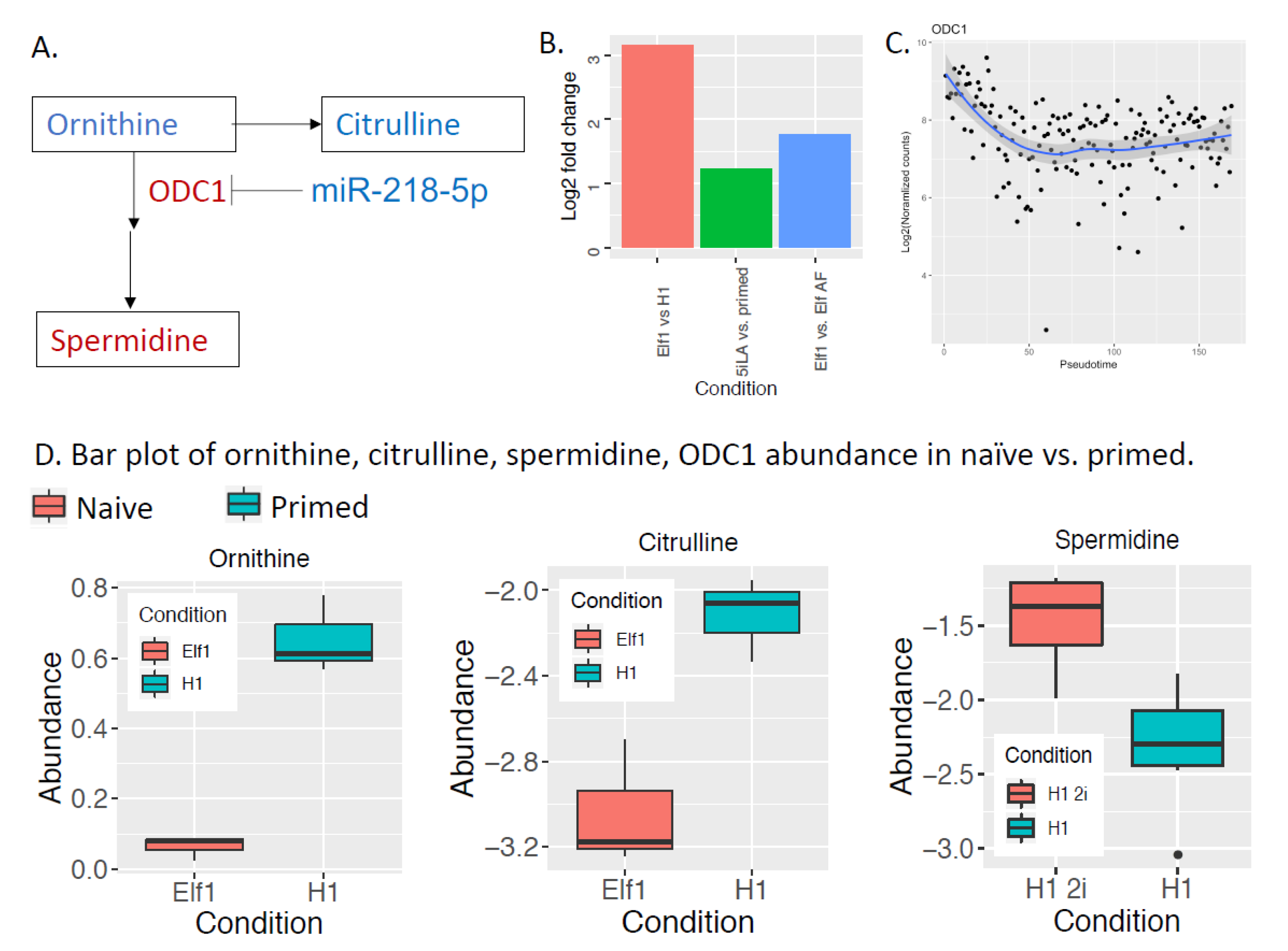
2.3. Metabolism in Naïve-to-Primed Transition
2.4. microRNAs Regulating Mouse Naïve to Primed ESCs Transition
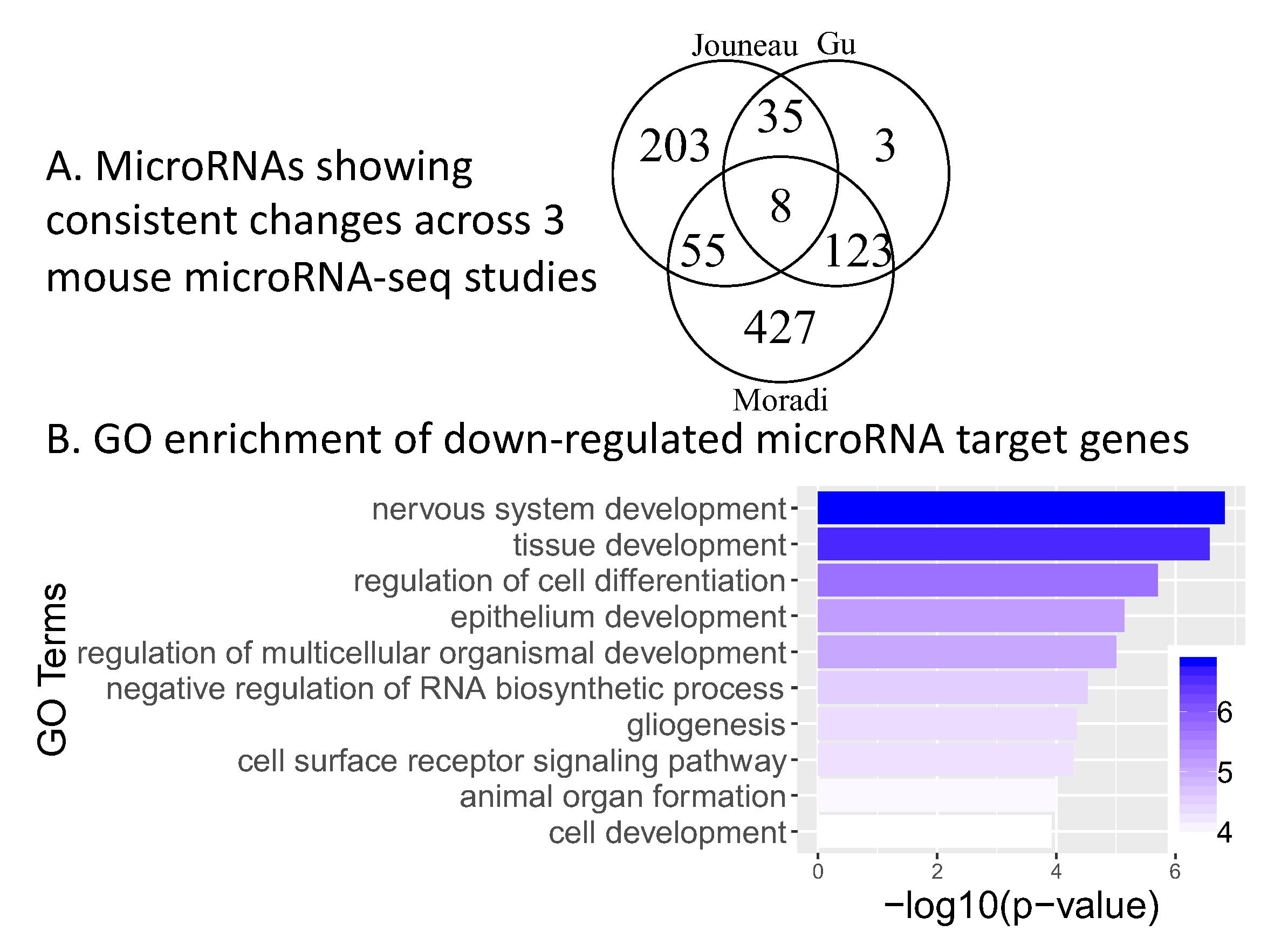
2.5. Consistent microRNA Changes across Human and Mouse Pluripotency
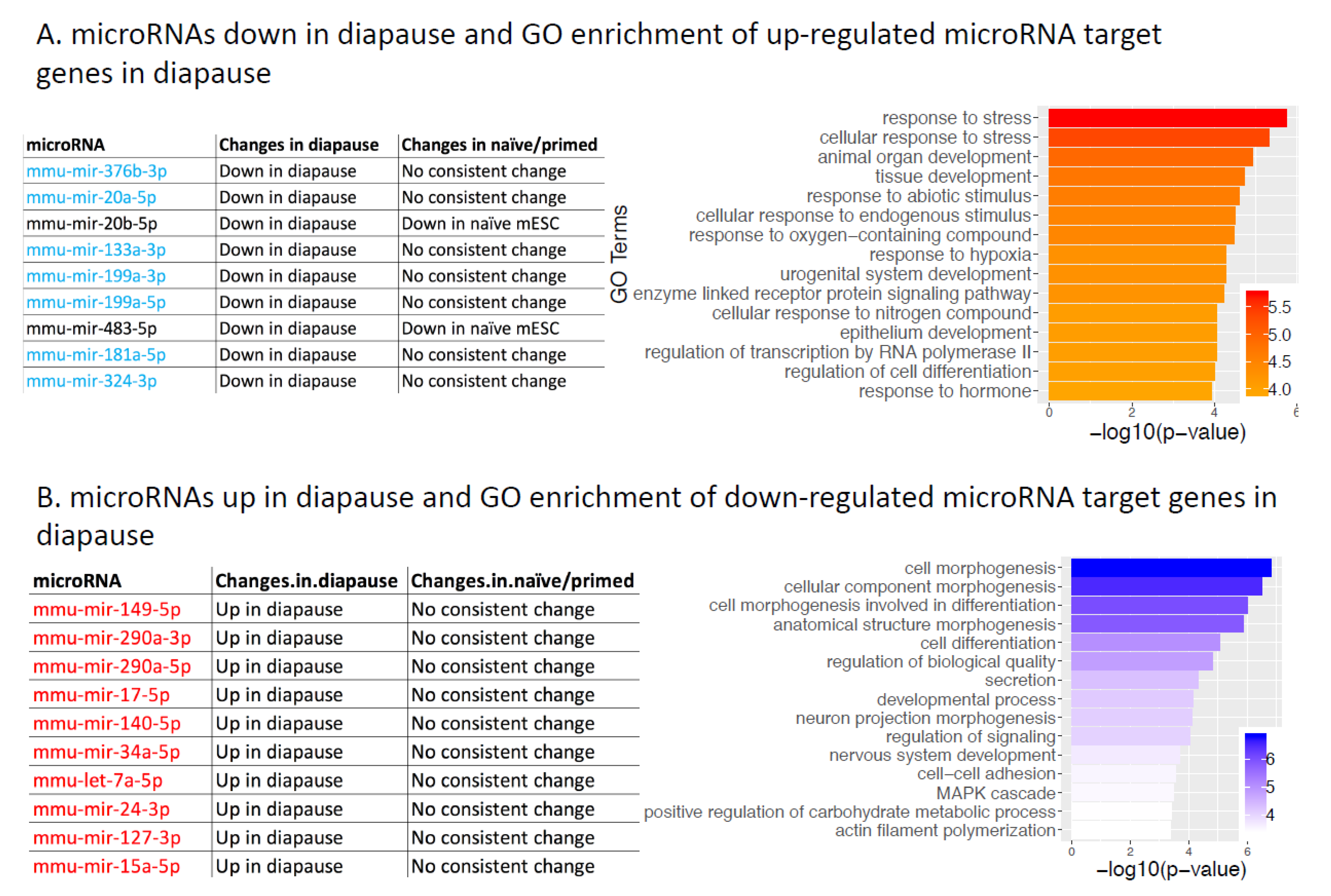
2.6. microRNAs and Their Target Genes Involved in Diapause Regulation
3. Materials and Methods
3.1. Culture of Naïve and Primed Human Pluripotent Stem Cells
3.2. Generation of GPR161 Mutant hESC
3.3. Immunofluorescence Staining and Confocal Imaging
3.4. RT-qPCR Analysis of GPR161 Mutant
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, S.D.; Shcherbata, H.R.; Fischer, K.A.; Nakahara, K.; Carthew, R.W.; Ruohola-Baker, H. Stem cell division is regulated by the microRNA pathway. Nature 2005, 435, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Shcherbata, H.R.; Hatfield, S.; Ward, E.J.; Reynolds, S.; Fischer, K.A.; Ruohola-Baker, H. The MicroRNA pathway plays a regulatory role in stem cell division. Cell Cycle 2006, 5, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Ruohola-Baker, H. Regulation of stem cell populations by microRNAs. Adv. Exp. Med. Biol. 2013, 786, 329–351. [Google Scholar] [PubMed]
- Bar, M.; Wyman, S.K.; Fritz, B.R.; Qi, J.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; Bendoraite, A.; Mitchell, P.S.; Nelson, A.M.; et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 2008, 26, 2496–2505. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, K.T.; Sperber, H.; Ruohola-Baker, H. MicroRNA regulation and role in stem cell maintenance, cardiac differentiation and hypertrophy. Curr. Mol. Med. 2013, 13, 757–764. [Google Scholar] [CrossRef]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar] [CrossRef]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar] [CrossRef]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Baskerville, S.; Shenoy, A.; Babiarz, J.E.; Baehner, L.; Blelloch, R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008, 40, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007, 39, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Yu, J.Y.; Shcherbata, H.R.; Mathieu, J.; Wang, A.J.; Seal, S.; Zhou, W.; Stadler, B.M.; Bourgin, D.; Wang, L.; et al. microRNAs regulate human embryonic stem cell division. Cell Cycle 2009, 8, 3729–3741. [Google Scholar] [CrossRef]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef]
- Kuppusamy, K.T.; Jones, D.C.; Sperber, H.; Madan, A.; Fischer, K.A.; Rodriguez, M.L.; Pabon, L.; Zhu, W.Z.; Tulloch, N.L.; Yang, X.; et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, E2785–E2794. [Google Scholar] [CrossRef]
- Lim, G.B. MicroRNA-directed cardiac repair after myocardial infarction in pigs. Nat. Rev. Cardiol. 2019, 16, 454–455. [Google Scholar] [CrossRef]
- Gao, F.; Kataoka, M.; Liu, N.; Liang, T.; Huang, Z.P.; Gu, F.; Ding, J.; Liu, J.; Zhang, F.; Ma, Q.; et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat. Commun. 2019, 10, 1802. [Google Scholar] [CrossRef]
- Gabisonia, K.; Prosdocimo, G.; Aquaro, G.D.; Carlucci, L.; Zentilin, L.; Secco, I.; Ali, H.; Braga, L.; Gorgodze, N.; Bernini, F.; et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019, 569, 418–422. [Google Scholar] [CrossRef]
- Miklas, J.W.; Clark, E.; Levy, S.; Detraux, D.; Leonard, A.; Beussman, K.; Showalter, M.R.; Smith, A.T.; Hofsteen, P.; Yang, X.; et al. TFPa/HADHA is required for fatty acid beta-oxidation and cardiolipin re-modeling in human cardiomyocytes. Nat. Commun. 2019, 10, 4671. [Google Scholar] [CrossRef]
- Stadler, B.M.; Ruohola-Baker, H. Small RNAs: Keeping stem cells in line. Cell 2008, 132, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Stadler, B.; Ivanovska, I.; Mehta, K.; Song, S.; Nelson, A.; Tan, Y.; Mathieu, J.; Darby, C.; Blau, C.A.; Ware, C.; et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010, 19, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y.; et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hong, Y.; Xiang, D.; Zhu, P.; Wu, E.; Li, W.; Mosenson, J.; Wu, W.S. MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Rep. 2015, 4, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Ruohola-Baker, H. Metabolic remodeling during the loss and acquisition of pluripotency. Development 2017, 144, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Sperber, H.; Mathieu, J.; Wang, Y.; Ferreccio, A.; Hesson, J.; Xu, Z.; Fischer, K.A.; Devi, A.; Detraux, D.; Gu, H.; et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015, 17, 1523–1535. [Google Scholar] [CrossRef]
- Gu, W.; Gaeta, X.; Sahakyan, A.; Chan, A.B.; Hong, C.S.; Kim, R.; Braas, D.; Plath, K.; Lowry, W.E.; Christofk, H.R. Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 2016, 19, 476–490. [Google Scholar] [CrossRef]
- Cao, Y.; Guo, W.T.; Tian, S.; He, X.; Wang, X.W.; Liu, X.; Gu, K.L.; Ma, X.; Huang, D.; Hu, L.; et al. miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J. 2015, 34, 609–623. [Google Scholar] [CrossRef]
- Manor, Y.S.; Massarwa, R.; Hanna, J.H. Establishing the human naive pluripotent state. Curr. Opin. Genet. Dev. 2015, 34, 35–45. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Ware, C.B. Concise Review: Lessons from Naive Human Pluripotent Cells. Stem Cells 2017, 35, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Smith, A. Pluripotency Deconstructed. Dev. Growth Differ. 2018, 60, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Morgani, S.; Nichols, J.; Hadjantonakis, A.K. The many faces of Pluripotency: In vitro adaptations of a continuum of in vivo states. BMC Dev. Biol. 2017, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Hussein, A.M.; Wang, Y.; Mathieu, J.; Margaretha, L.; Song, C.; Jones, D.C.; Cavanaugh, C.; Miklas, J.W.; Mahen, E.; Showalter, M.R.; et al. Metabolic control over mTOR dependent diapause-like state. Dev. Cell 2019. (accepted). [Google Scholar]
- Cheong, A.W.; Pang, R.T.; Liu, W.M.; Kottawatta, K.S.; Lee, K.F.; Yeung, W.S. MicroRNA Let-7a and dicer are important in the activation and implantation of delayed implanting mouse embryos. Hum. Reprod. 2014, 29, 750–762. [Google Scholar] [CrossRef]
- Liu, W.M.; Pang, R.T.; Cheong, A.W.; Ng, E.H.; Lao, K.; Lee, K.F.; Yeung, W.S. Involvement of microRNA lethal-7a in the regulation of embryo implantation in mice. PLoS ONE 2012, 7, e37039. [Google Scholar] [CrossRef]
- Jouneau, A.; Ciaudo, C.; Sismeiro, O.; Brochard, V.; Jouneau, L.; Vandormael-Pournin, S.; Coppee, J.Y.; Zhou, Q.; Heard, E.; Antoniewski, C.; et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA 2012, 18, 253–264. [Google Scholar] [CrossRef]
- Moradi, S.; Sharifi-Zarchi, A.; Ahmadi, A.; Mollamohammadi, S.; Stubenvoll, A.; Gunther, S.; Salekdeh, G.H.; Asgari, S.; Braun, T.; Baharvand, H. Small RNA Sequencing Reveals Dlk1-Dio3 Locus-Embedded MicroRNAs as Major Drivers of Ground-State Pluripotency. Stem Cell Rep. 2017, 9, 2081–2096. [Google Scholar] [CrossRef]
- Gu, K.L.; Zhang, Q.; Yan, Y.; Li, T.T.; Duan, F.F.; Hao, J.; Wang, X.W.; Shi, M.; Wu, D.R.; Guo, W.T.; et al. Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 2016, 26, 350–366. [Google Scholar] [CrossRef]
- Ware, C.B.; Wang, L.; Mecham, B.H.; Shen, L.; Nelson, A.M.; Bar, M.; Lamba, D.A.; Dauphin, D.S.; Buckingham, B.; Askari, B.; et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell 2009, 4, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Ware, C.B.; Nelson, A.M.; Mecham, B.; Hesson, J.; Zhou, W.; Jonlin, E.C.; Jimenez-Caliani, A.J.; Deng, X.; Cavanaugh, C.; Cook, S.; et al. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4484–4489. [Google Scholar] [CrossRef] [PubMed]
- Moody, J.D.; Levy, S.; Mathieu, J.; Xing, Y.; Kim, W.; Dong, C.; Tempel, W.; Robitaille, A.M.; Dang, L.T.; Ferreccio, A.; et al. First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor. Proc. Natl. Acad. Sci. USA 2017, 114, 10125–10130. [Google Scholar] [CrossRef] [PubMed]
- Ferreccio, A.; Mathieu, J.; Detraux, D.; Somasundaram, L.; Cavanaugh, C.; Sopher, B.; Fischer, K.; Bello, T.; Hussein, A.M.; Levy, S.; et al. Inducible CRISPR genome editing platform in naive human embryonic stem cells reveals JARID2 function in self-renewal. Cell Cycle 2018, 17, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell 2014, 15, 524–526. [Google Scholar] [CrossRef]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015, 522, 221–225. [Google Scholar] [CrossRef]
- Factor, D.C.; Corradin, O.; Zentner, G.E.; Saiakhova, A.; Song, L.; Chenoweth, J.G.; McKay, R.D.; Crawford, G.E.; Scacheri, P.C.; Tesar, P.J. Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell 2014, 14, 854–863. [Google Scholar] [CrossRef]
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472. [Google Scholar] [CrossRef]
- Wu, S.M.; Choo, A.B.; Yap, M.G.; Chan, K.K. Role of Sonic hedgehog signaling and the expression of its components in human embryonic stem cells. Stem Cell Res. 2010, 4, 38–49. [Google Scholar] [CrossRef]
- Li, Q.; Lex, R.K.; Chung, H.; Giovanetti, S.M.; Ji, Z.; Ji, H.; Person, M.D.; Kim, J.; Vokes, S.A. The Pluripotency Factor NANOG Binds to GLI Proteins and Represses Hedgehog-mediated Transcription. J. Biol. Chem. 2016, 291, 7171–7182. [Google Scholar] [CrossRef] [PubMed]
- Kiprilov, E.N.; Awan, A.; Desprat, R.; Velho, M.; Clement, C.A.; Byskov, A.G.; Andersen, C.Y.; Satir, P.; Bouhassira, E.E.; Christensen, S.T.; et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J. Cell Biol. 2008, 180, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Bangs, F.K.; Schrode, N.; Hadjantonakis, A.K.; Anderson, K.V. Lineage specificity of primary cilia in the mouse embryo. Nat. Cell Biol. 2015, 17, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Freedman, B.S.; Lam, A.Q.; Sundsbak, J.L.; Iatrino, R.; Su, X.; Koon, S.J.; Wu, M.; Daheron, L.; Harris, P.C.; Zhou, J.; et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J. Am. Soc. Nephrol. 2013, 24, 1571–1586. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Detraux, D.; Kuppers, D.; Wang, Y.; Cavanaugh, C.; Sidhu, S.; Levy, S.; Robitaille, A.M.; Ferreccio, A.; Bottorff, T.; et al. Folliculin regulates mTORC1/2 and WNT pathways in early human pluripotency. Nat. Commun. 2019, 10, 632. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Wen, X.; Ratti, N.; Loktev, A.; Rangell, L.; Scales, S.J.; Jackson, P.K. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 2013, 152, 210–223. [Google Scholar] [CrossRef]
- Matteson, P.G.; Desai, J.; Korstanje, R.; Lazar, G.; Borsuk, T.E.; Rollins, J.; Kadambi, S.; Joseph, J.; Rahman, T.; Wink, J.; et al. The orphan G protein-coupled receptor, Gpr161, encodes the vacuolated lens locus and controls neurulation and lens development. Proc. Natl. Acad. Sci. USA 2008, 105, 2088–2093. [Google Scholar] [CrossRef]
- Bachmann, V.A.; Mayrhofer, J.E.; Ilouz, R.; Tschaikner, P.; Raffeiner, P.; Rock, R.; Courcelles, M.; Apelt, F.; Lu, T.W.; Baillie, G.S.; et al. Gpr161 anchoring of PKA consolidates GPCR and cAMP signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 7786–7791. [Google Scholar] [CrossRef]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef]
- Indrieri, A.; van Rahden, V.A.; Tiranti, V.; Morleo, M.; Iaconis, D.; Tammaro, R.; D’Amato, I.; Conte, I.; Maystadt, I.; Demuth, S.; et al. Mutations in COX7B cause microphthalmia with linear skin lesions, an unconventional mitochondrial disease. Am. J. Hum. Genet. 2012, 91, 942–949. [Google Scholar] [CrossRef]
- Izreig, S.; Samborska, B.; Johnson, R.M.; Sergushichev, A.; Ma, E.H.; Lussier, C.; Loginicheva, E.; Donayo, A.O.; Poffenberger, M.C.; Sagan, S.M.; et al. The miR-17 approximately 92 microRNA Cluster Is a Global Regulator of Tumor Metabolism. Cell Rep. 2016, 16, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Choi, M.; Margineantu, D.; Margaretha, L.; Hesson, J.; Cavanaugh, C.; Blau, C.A.; Horwitz, M.S.; Hockenbery, D.; Ware, C.; et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012, 31, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ratanasirintrawoot, S.; Chandrasekaran, S.; Wu, Z.; Ficarro, S.B.; Yu, C.; Ross, C.A.; Cacchiarelli, D.; Xia, Q.; Seligson, M.; et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 2016, 19, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhu, M.; Lv, P.; Cheng, L.; Wang, Q.; Tian, P.; Yan, Z.; Wen, B. The Long Noncoding RNA Lncenc1 Maintains Naive States of Mouse ESCs by Promoting the Glycolysis Pathway. Stem Cell Rep. 2018, 11, 741–755. [Google Scholar] [CrossRef]
- Bahat, A.; Goldman, A.; Zaltsman, Y.; Khan, D.H.; Halperin, C.; Amzallag, E.; Krupalnik, V.; Mullokandov, M.; Silberman, A.; Erez, A.; et al. MTCH2-mediated mitochondrial fusion drives exit from naive pluripotency in embryonic stem cells. Nat. Commun. 2018, 9, 5132. [Google Scholar] [CrossRef]
- Cornacchia, D.; Zhang, C.; Zimmer, B.; Chung, S.Y.; Fan, Y.; Soliman, M.A.; Tchieu, J.; Chambers, S.M.; Shah, H.; Paull, D.; et al. Lipid Deprivation Induces a Stable, Naive-to-Primed Intermediate State of Pluripotency in Human PSCs. Cell Stem Cell 2019, 25, 120–136 e10. [Google Scholar] [CrossRef]
- Zhang, D.; Zhao, T.; Ang, H.S.; Chong, P.; Saiki, R.; Igarashi, K.; Yang, H.; Vardy, L.A. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 2012, 26, 461–473. [Google Scholar] [CrossRef]
- Zhao, T.; Goh, K.J.; Ng, H.H.; Vardy, L.A. A role for polyamine regulators in ESC self-renewal. Cell Cycle 2012, 11, 4517–4523. [Google Scholar] [CrossRef]
- James, C.; Zhao, T.Y.; Rahim, A.; Saxena, P.; Muthalif, N.A.; Uemura, T.; Tsuneyoshi, N.; Ong, S.; Igarashi, K.; Lim, C.Y.; et al. MINDY1 Is a Downstream Target of the Polyamines and Promotes Embryonic Stem Cell Self-Renewal. Stem Cells 2018, 36, 1170–1178. [Google Scholar] [CrossRef]
- Zhu, K.; Ding, H.; Wang, W.; Liao, Z.; Fu, Z.; Hong, Y.; Zhou, Y.; Zhang, C.Y.; Chen, X. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget 2016, 7, 28075–28085. [Google Scholar] [CrossRef]
- Bjornsen, L.P.; Hadera, M.G.; Zhou, Y.; Danbolt, N.C.; Sonnewald, U. The GLT-1 (EAAT2; slc1a2) glutamate transporter is essential for glutamate homeostasis in the neocortex of the mouse. J. Neurochem. 2014, 128, 641–649. [Google Scholar] [CrossRef] [PubMed]
- TeSlaa, T.; Chaikovsky, A.C.; Lipchina, I.; Escobar, S.L.; Hochedlinger, K.; Huang, J.; Graeber, T.G.; Braas, D.; Teitell, M.A. alpha-Ketoglutarate Accelerates the Initial Differentiation of Primed Human Pluripotent Stem Cells. Cell Metab. 2016, 24, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Contreras, L.; Ramirez, L.; Du, J.; Hurley, J.B.; Satrustegui, J.; de la Villa, P. Deficient glucose and glutamine metabolism in Aralar/AGC1/Slc25a12 knockout mice contributes to altered visual function. Mol. Vis. 2016, 22, 1198–1212. [Google Scholar] [PubMed]
- Jalil, M.A.; Begum, L.; Contreras, L.; Pardo, B.; Iijima, M.; Li, M.X.; Ramos, M.; Marmol, P.; Horiuchi, M.; Shimotsu, K.; et al. Reduced N-acetylaspartate levels in mice lacking aralar, a brain- and muscle-type mitochondrial aspartate-glutamate carrier. J. Biol. Chem. 2005, 280, 31333–31339. [Google Scholar] [CrossRef]
- Hore, T.A.; von Meyenn, F.; Ravichandran, M.; Bachman, M.; Ficz, G.; Oxley, D.; Santos, F.; Balasubramanian, S.; Jurkowski, T.P.; Reik, W. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naive pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. USA 2016, 113, 12202–12207. [Google Scholar] [CrossRef]
- Gao, Y.; Han, Z.; Li, Q.; Wu, Y.; Shi, X.; Ai, Z.; Du, J.; Li, W.; Guo, Z.; Zhang, Y. Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating microRNA expression. FEBS J. 2015, 282, 685–699. [Google Scholar] [CrossRef]
- Gennarino, V.A.; D’Angelo, G.; Dharmalingam, G.; Fernandez, S.; Russolillo, G.; Sanges, R.; Mutarelli, M.; Belcastro, V.; Ballabio, A.; Verde, P.; et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome. Res. 2012, 22, 1163–1172. [Google Scholar] [CrossRef]
- Chowdhury, F.; Li, Y.; Poh, Y.C.; Yokohama-Tamaki, T.; Wang, N.; Tanaka, T.S. Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS ONE 2010, 5, e15655. [Google Scholar] [CrossRef]
- Gusdon, A.M.; Zhu, J.; Van Houten, B.; Chu, C.T. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol. Dis. 2012, 45, 962–972. [Google Scholar] [CrossRef]
- Forristal, C.E.; Wright, K.L.; Hanley, N.A.; Oreffo, R.O.; Houghton, F.D. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction 2010, 139, 85–97. [Google Scholar] [CrossRef]
- Tillander, V.; Arvidsson Nordstrom, E.; Reilly, J.; Strozyk, M.; Van Veldhoven, P.P.; Hunt, M.C.; Alexson, S.E. Acyl-CoA thioesterase 9 (ACOT9) in mouse may provide a novel link between fatty acid and amino acid metabolism in mitochondria. Cell Mol. Life Sci. 2014, 71, 933–948. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.P.; Jain, N.; Cao, X. Stress-induced immediate-early gene, egr-1, involves activation of p38/JNK1. Oncogene 1998, 16, 2915–2926. [Google Scholar] [CrossRef] [PubMed]
- Kitamuro, T.; Takahashi, K.; Ogawa, K.; Udono-Fujimori, R.; Takeda, K.; Furuyama, K.; Nakayama, M.; Sun, J.; Fujita, H.; Hida, W.; et al. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J. Biol. Chem. 2003, 278, 9125–9133. [Google Scholar] [CrossRef] [PubMed]
- Molloy, M.E.; Lewinska, M.; Williamson, A.K.; Nguyen, T.T.; Kuser-Abali, G.; Gong, L.; Yan, J.; Little, J.B.; Pandolfi, P.P.; Yuan, Z.M. ZBTB7A governs estrogen receptor alpha expression in breast cancer. J. Mol. Cell Biol. 2018, 10, 273–284. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef]
- Nakamura, T.; Okamoto, I.; Sasaki, K.; Yabuta, Y.; Iwatani, C.; Tsuchiya, H.; Seita, Y.; Nakamura, S.; Yamamoto, T.; Saitou, M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 2016, 537, 57–62. [Google Scholar] [CrossRef]
- Miyaoka, Y.; Chan, A.H.; Judge, L.M.; Yoo, J.; Huang, M.; Nguyen, T.D.; Lizarraga, P.P.; So, P.L.; Conklin, B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 2014, 11, 291–293. [Google Scholar] [CrossRef]
- He, B.; Zhang, H.; Wang, J.; Liu, M.; Sun, Y.; Guo, C.; Lu, J.; Wang, H.; Kong, S. Blastocyst activation engenders transcriptome reprogram affecting X-chromosome reactivation and inflammatory trigger of implantation. Proc. Natl. Acad. Sci. USA 2019, 116, 16621–16630. [Google Scholar] [CrossRef]
- Chen, M.; Reed, R.R.; Lane, A.P. Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense. Cell Stem Cell 2019, 25, 501–513 e5. [Google Scholar] [CrossRef]

| Study | Type | Condition | Growth Media | Accession Number |
|---|---|---|---|---|
| Sperber H. et al. [27] | microRNA-seq | Naïve hESC | 2iL-I-F | GSE60995 |
| Sperber H. et al. [27] | mRNA-seq | Naïve and primed hESC | 2iL-I-F/KSR + FGF | GSE60995 |
| Jouneau A. et al. [38] | microRNA-seq | Mouse ESC and EpiSC | 15% FBS-LIF/AF | Supplemental data from original paper |
| Gu et al. [40] | microRNA-seq | Mouse ESC differentiation time course | N2B27-2iLIF/KSR + FGF | Supplemental data from original paper |
| Moradi S. et al. [39] | microRNA-seq | Mouse ground ESC and naïve ESC | N2B27 2iLIF or 3iLIF/15% FBS-LIF | GSE87174 |
| ENCODE project | microRNA-seq | primed ESC | TeSR | https://www.encodeproject.org |
| Roadmap Epigenome project | microRNA-seq | primed ESC | TeSR | http://www.roadmapepigenomics.org |
| Theunissen et al. [45] | mRNA-seq | Naïve/primed hESC | 5iLA/serum | GSE59435 |
| Grow et al. [47] | mRNA-seq | Naïve/primed hESC | 2iLIF/KSR + FGF | GSE63570 |
| Factor et al. [48] | mRNA-seq | Mouse ESC and EpiSC | KSR LIF/KSR + FGF | GSE57409 |
| Hussein et al. [35] | mRNA-seq | Mouse pre-implantation, diapause, post-implantation | From embryos | |
| Liu et al. [37] | microRNA microarray | Mouse diapause and re-activated embryos | From embryos | Supplemental data from original paper |
| Naive | Primed | Assay | References |
|---|---|---|---|
| Glycolysis ↑ | Glycolysis ↑ | RNAseq, Metabolomics, Seahorse Flux Analyzer, TMRE | Zhou et al. [62] Takashima et al. [59] Sperber et al. [27] Gu et al. [28] Zhang et al. [63] Sun et al. [64] Baha et al. [65] Cornacchia et al. [66] |
| OXPHOS ↑ | OXPHOS ↓ | ||
| Fatty acid synthesis ↓ | Fatty acid synthesis ↑ | RNAseq, Metabolomics, Seahorse Flux Analyzer, Oil RedO and Bodipy staining | Sperber et al. [27] Cornacchia et al. [66] |
| Fatty acid oxidation ↑ | Fatty acid oxidation ↓ |
| Naive | Primed | Assay | References |
|---|---|---|---|
| FA synthesis, activation, elongation, desaturation (FASN, ACSL1, ELOVL7, SCD)  miR-10a | FA transporter (rate limiting FAO) CPT1A  miR-106b-5p, miR-20a-5p, miR-302c-3p, miR-17-5p, miR-20b-5p, miR-106a-5p, miR-302a-3p, miR-150-5p, miR-16-5p, miR-93-5p, miR-4517 | RNAseq, microRNAseq, RT-qPCR | Sperber et al. [27] |
| ID | log2FC.Ref_Epigenome | log2FC.ENCODE | padj.Ref_Epigenome | padj.ENCODE | Jouneau | Gu | Moradi | How Many Consistent |
|---|---|---|---|---|---|---|---|---|
| hsa-mir-143, hsa-miR-143-3p | 4.10 | 7.50 | 2.932 × 10−3 | 1.163 × 10−7 | 2.16 | 1.70 | 0.79 | 4 |
| hsa-mir-200c, hsa-miR-200c-3p | −6.60 | −8.97 | 8.956 × 10−96 | 5.294 × 10−93 | 6.44 | −1.03 | −0.88 | 3 |
| hsa-mir-205, hsa-miR-205-5p | 3.23 | 9.28 | 4.715 × 10−12 | 1.392 × 10−88 | 2.23 | 0.93 | −1.28 | 3 |
| hsa-mir-302c, hsa-miR-302c-3p | −1.68 | −6.40 | 5.552 × 10−13 | 1.086 × 10−66 | −5.47 | −1.12 | 2.63 | 3 |
| hsa-mir-20b, hsa-miR-20b-5p | −1.93 | −5.37 | 1.923 × 10−16 | 2.778 × 10−51 | 0.00 | −1.60 | −0.75 | 3 |
| hsa-mir-335,hsa-miR-335-5p | −1.76 | −5.07 | 1.075 × 10−13 | 6.196 × 10−47 | 0.00 | −1.00 | −0.74 | 3 |
| hsa-mir-302a, hsa-miR-302a-3p | −1.50 | −4.46 | 1.090 × 10−10 | 8.146 × 10−39 | −5.15 | −2.08 | 1.67 | 3 |
| hsa-mir-412, hsa-miR-412-5p | 7.17 | 15.02 | 3.547 × 10−64 | 1.038 × 10−28 | −2.60 | 0.92 | 3.34 | 3 |
| hsa-mir-433, hsa-miR-433-3p | 5.38 | 15.02 | 6.536 × 10−39 | 7.545 × 10−21 | 0.00 | 0.62 | 4.29 | 3 |
| hsa-mir-495, hsa-miR-495-3p | 4.68 | 7.47 | 2.492 × 10−34 | 1.098 × 10−19 | 0.00 | 0.60 | 4.27 | 3 |
| hsa-mir-582, hsa-miR-582-3p | 2.72 | 5.91 | 7.063 × 10−12 | 4.664 × 10−14 | 1.40 | 0.96 | 0.48 | 3 |
| hsa-mir-196a-1, hsa-miR-196a-5p | 8.96 | 15.02 | 1.038 × 10−15 | 1.389 × 10−13 | 0.00 | 2.46 | 0.78 | 3 |
| hsa-mir-196a-2, hsa-miR-196a-5p | 8.69 | 15.02 | 2.845 × 10−12 | 2.660 × 10−12 | 0.00 | 2.46 | 0.78 | 3 |
| hsa-mir-200a, hsa-miR-200a-5p | −3.37 | −4.71 | 1.511 × 10−6 | 5.775 × 10−9 | 0.00 | −0.89 | −1.59 | 3 |
| hsa-mir-1247, hsa-miR-1247-3p | 3.40 | 15.02 | 1.677 × 10−8 | 4.527 × 10−6 | 2.85 | 0.00 | 2.17 | 3 |
| hsa-mir-708, hsa-miR-708-3p | 1.13 | 1.45 | 5.997 × 10−5 | 3.027 × 10−4 | 1.87 | 0.72 | 0.08 | 3 |
| hsa-mir-199b, hsa-miR-199b-3p | 3.34 | 4.27 | 4.641 × 10−2 | 6.808 × 10−3 | 2.54 | 0.30 | 1.01 | 3 |
| hsa-let-7f-2, hsa-let-7f-5p | 4.79 | 4.43 | 1.363 × 10−2 | 8.710 × 10−3 | 0.00 | 1.97 | 0.78 | 3 |
| hsa-let-7f-1, hsa-let-7f-5p | 4.72 | 4.31 | 1.455 × 10−2 | 9.959 × 10−3 | 0.00 | 1.97 | 0.78 | 3 |
| hsa-mir-182, hsa-miR-182-3p | 3.19 | 4.10 | 7.430 × 10−4 | 2.571 × 10−2 | 0.00 | 0.64 | 0.88 | 3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Hussein, A.M.; Somasundaram, L.; Sankar, R.; Detraux, D.; Mathieu, J.; Ruohola-Baker, H. microRNAs Regulating Human and Mouse Naïve Pluripotency. Int. J. Mol. Sci. 2019, 20, 5864. https://doi.org/10.3390/ijms20235864
Wang Y, Hussein AM, Somasundaram L, Sankar R, Detraux D, Mathieu J, Ruohola-Baker H. microRNAs Regulating Human and Mouse Naïve Pluripotency. International Journal of Molecular Sciences. 2019; 20(23):5864. https://doi.org/10.3390/ijms20235864
Chicago/Turabian StyleWang, Yuliang, Abdiasis M. Hussein, Logeshwaran Somasundaram, Rithika Sankar, Damien Detraux, Julie Mathieu, and Hannele Ruohola-Baker. 2019. "microRNAs Regulating Human and Mouse Naïve Pluripotency" International Journal of Molecular Sciences 20, no. 23: 5864. https://doi.org/10.3390/ijms20235864
APA StyleWang, Y., Hussein, A. M., Somasundaram, L., Sankar, R., Detraux, D., Mathieu, J., & Ruohola-Baker, H. (2019). microRNAs Regulating Human and Mouse Naïve Pluripotency. International Journal of Molecular Sciences, 20(23), 5864. https://doi.org/10.3390/ijms20235864








