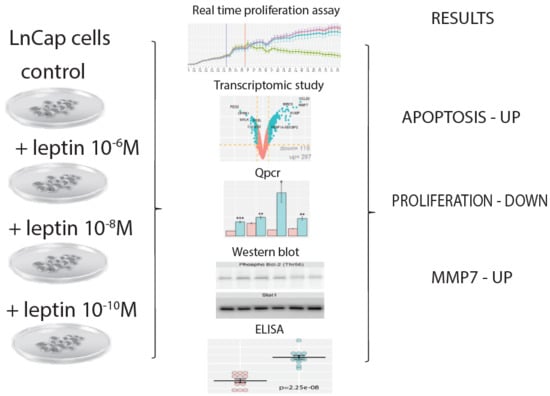
Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations
Abstract
1. Introduction
2. Results
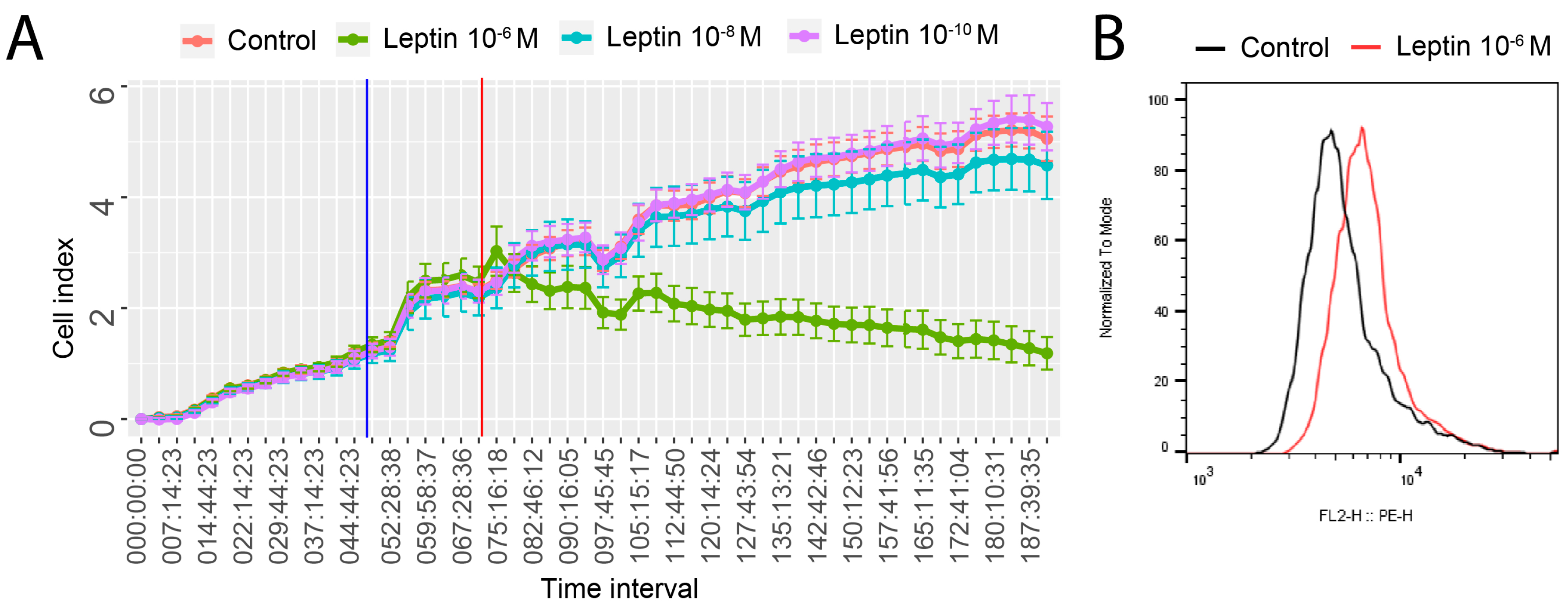
2.1. Leptin at a Dose of 1 × 10−6 M Exerted an Inhibitory Effect on Proliferative Activity of LNCaP Cells and Stimulate Apoptosis
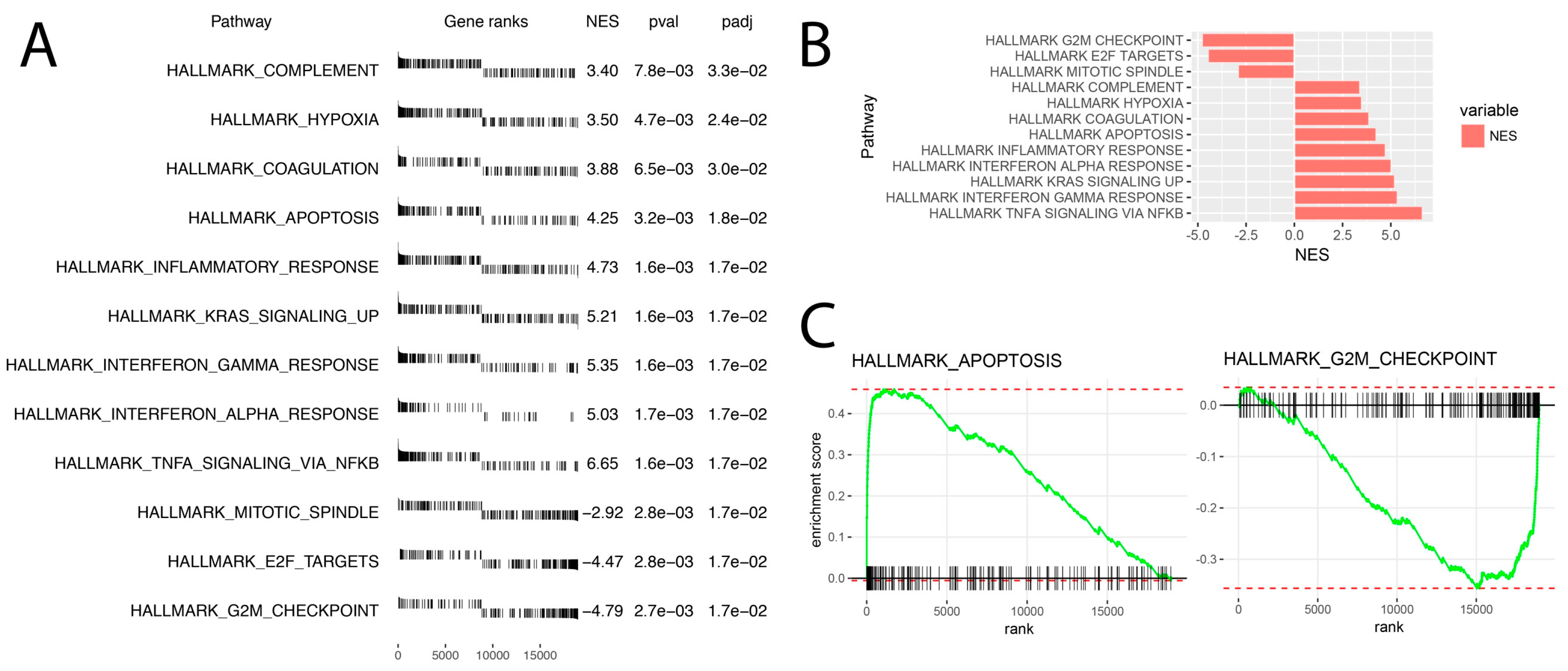
2.2. Leptin at a Dose of 1 × 10−6 M Significantly Modulates the Transcriptomic Profile of LNCaP Cells
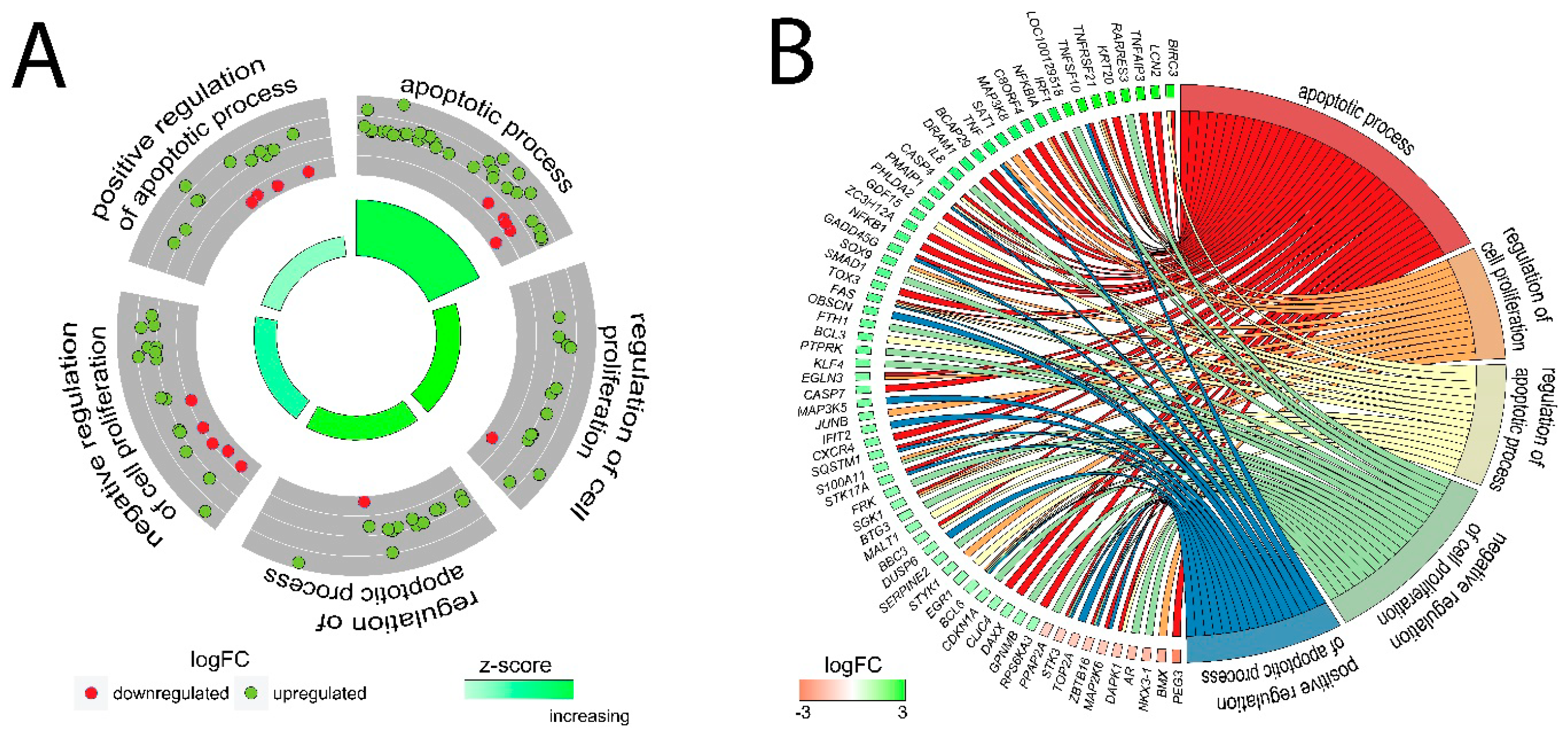
2.3. LEP at 1 × 10−6 M Concentration Exerts a Significant Effect on the Genes Involved in the Regulation of the Following Biological Processes: Apoptosis, Immunological Response and Extracellular Matrix Organisation
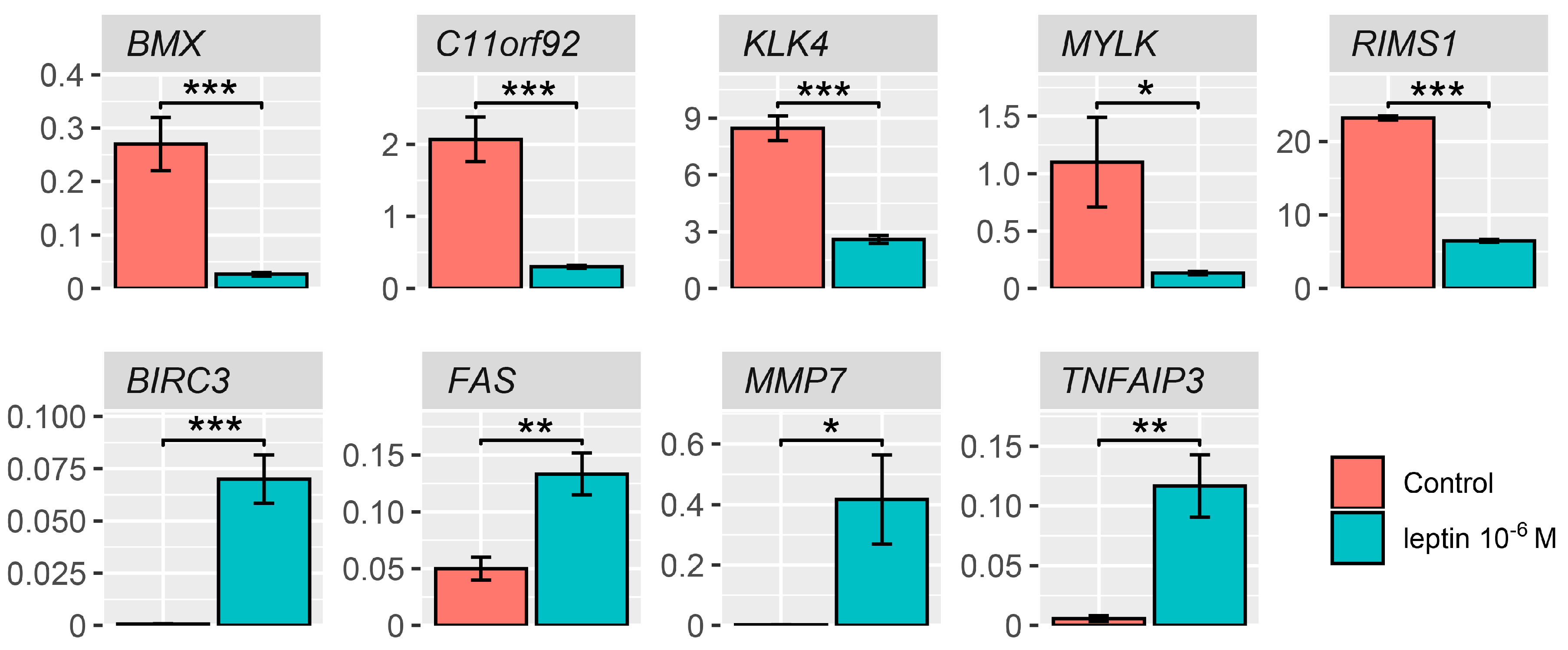
2.4. Detailed Analysis of the LEP-Regulated Gene Expression in LNCaP Cells
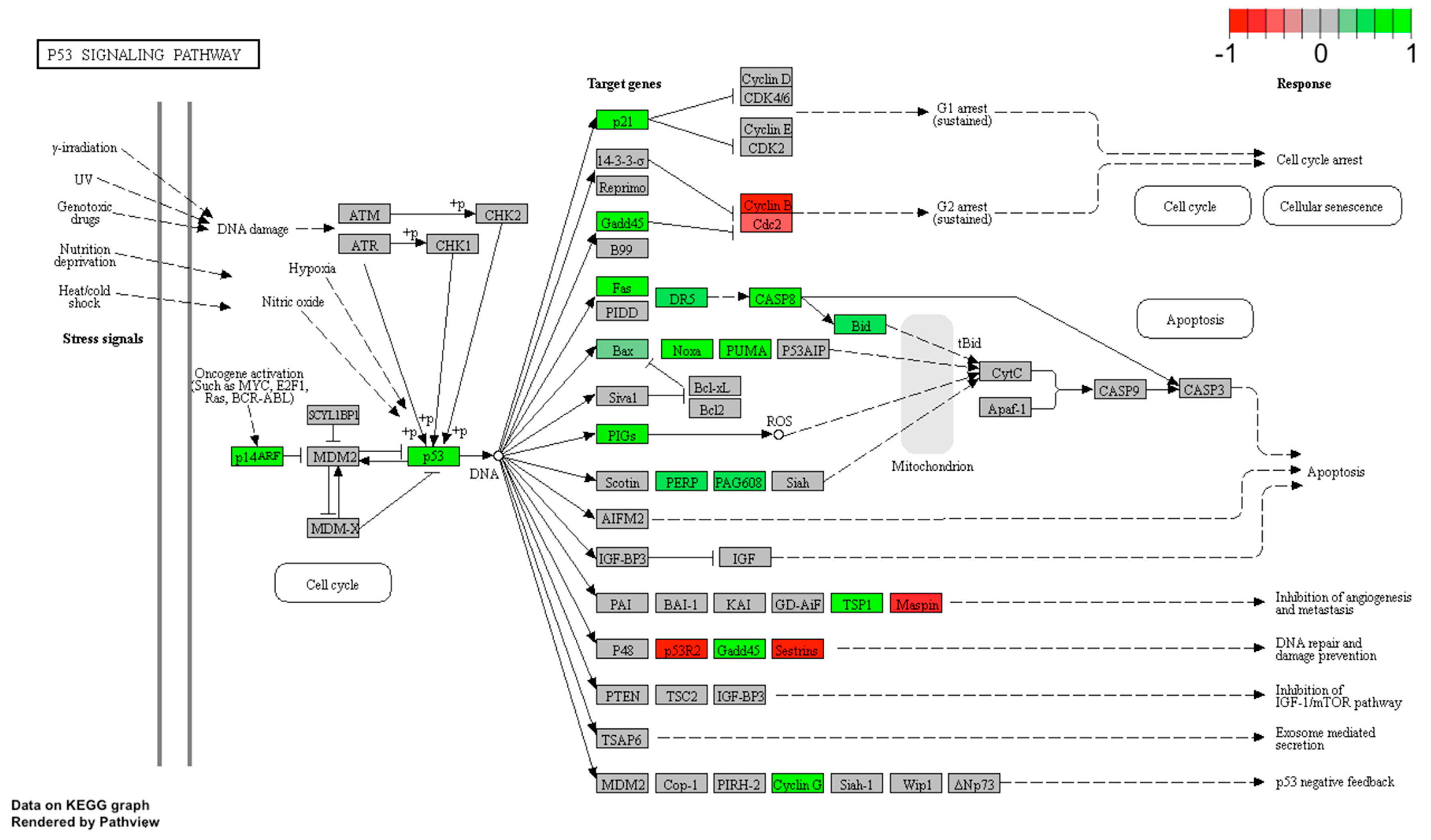
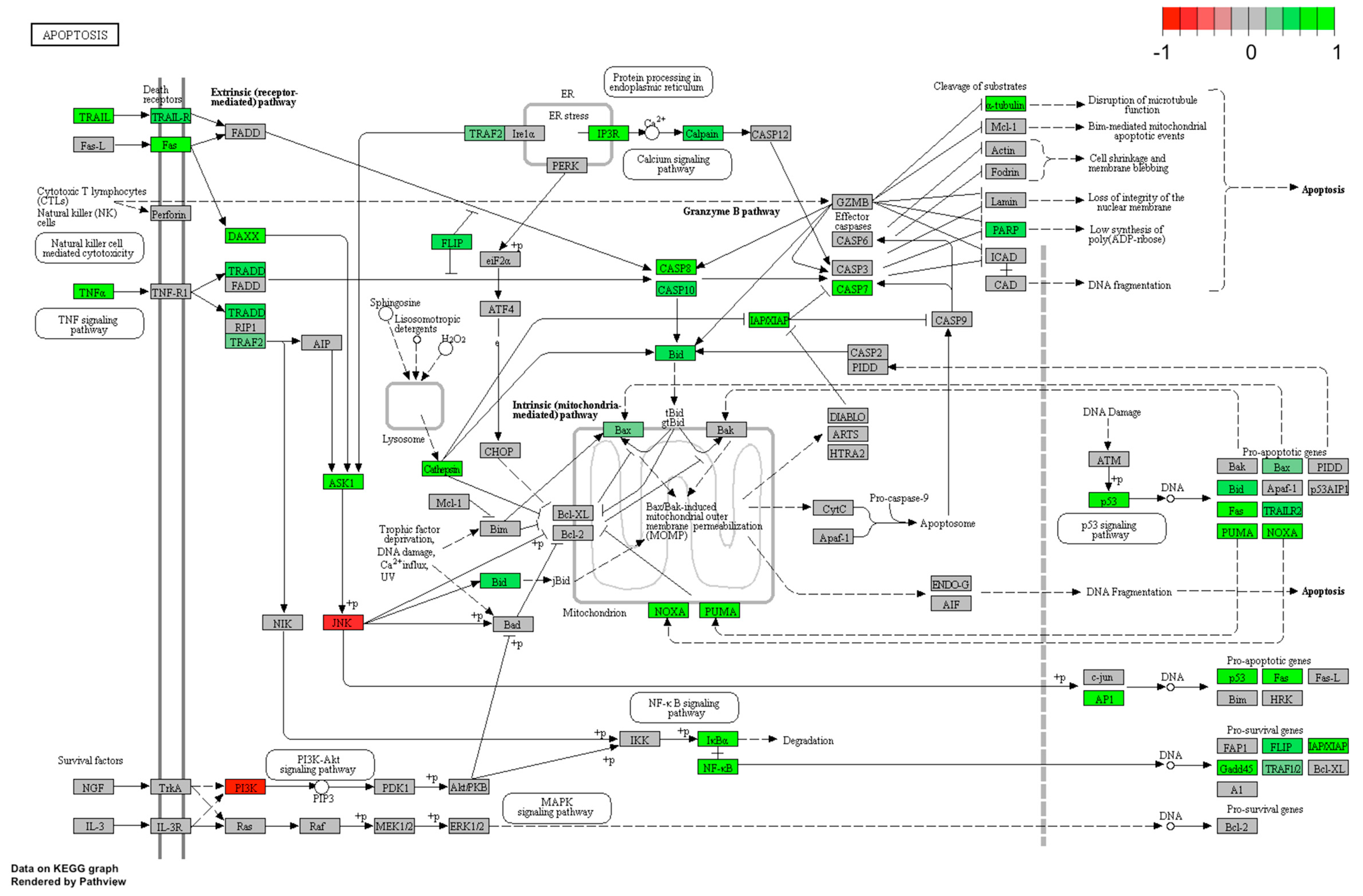
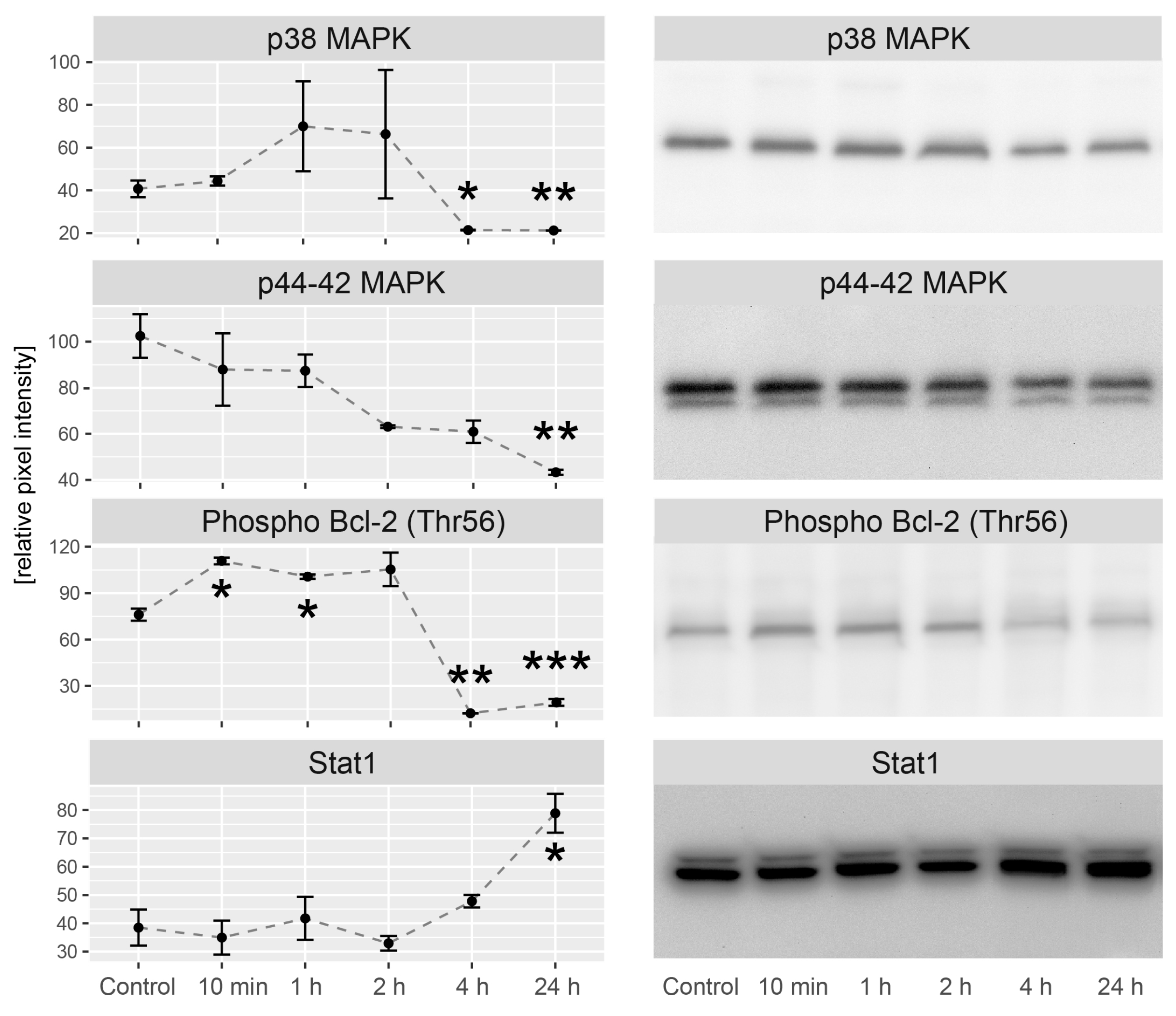
2.5. LEP Regulates Several Key Factors of Signaling Pathways Involved in Apoptosis, Proliferation and Migration
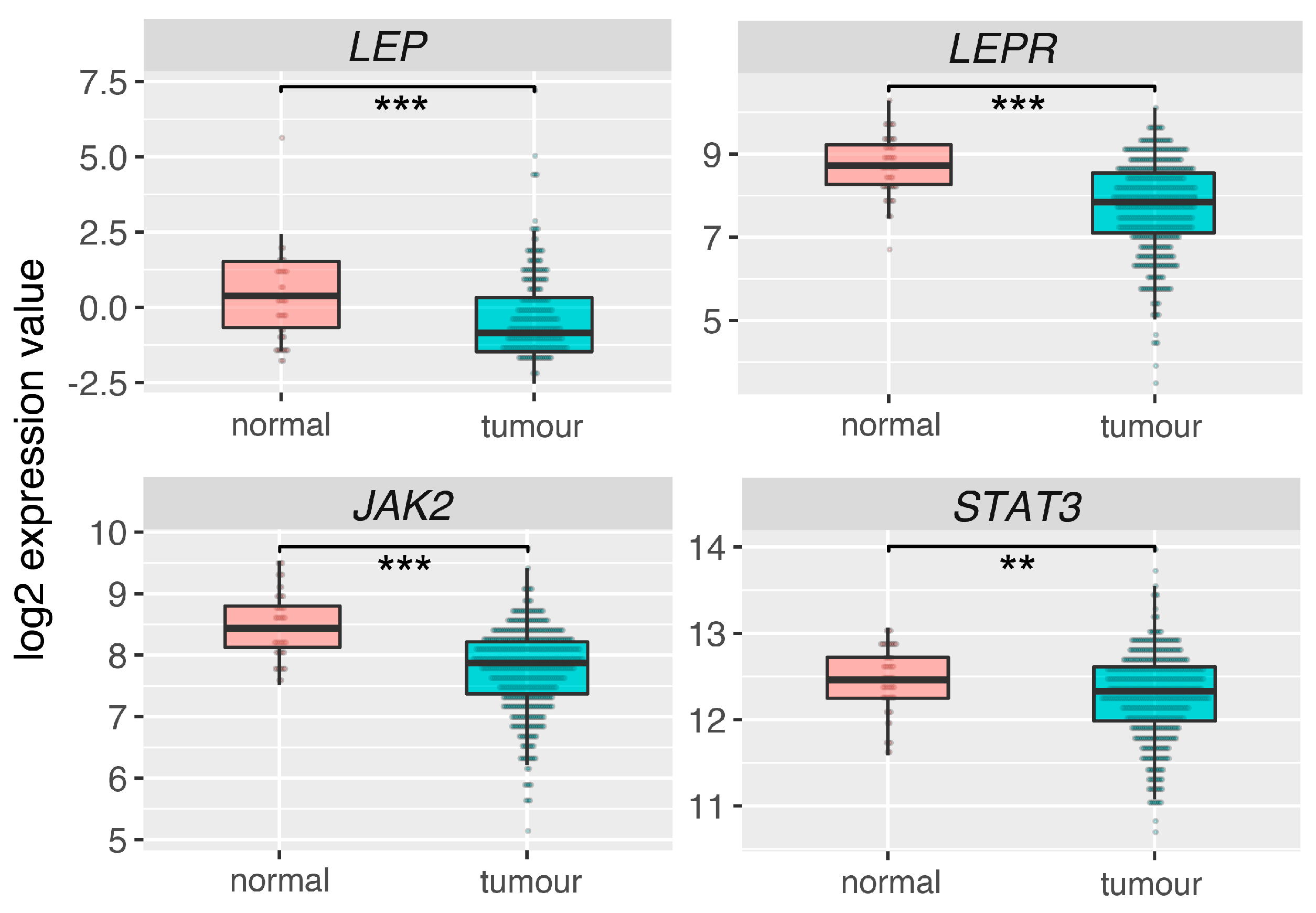
2.6. Leptin (LEP), Leptin Receptor (LEPR) and Its Main Downstream Signaling Genes (JAK2, STAT3) Are Downregulated in Prostate Adrenocarcinoma
3. Discussion
4. Materials and Methods
4.1. Prostate Cancer Cell Line
4.2. Real-Time Proliferation Assay
4.3. Flow Cytometry Analysis of Cleaved PARP-1
4.4. RNA Isolation
4.5. Reverse Transcription
4.6. Q-PCR
4.7. Microarray Expression Study
4.8. Microarray Data Analysis
4.9. Assignment of Differentially Expressed Genes to Relevant Gene Ontology (GO) Terms
4.10. Gene Set Enrichment Analysis (GSEA)
4.11. KEGG Signaling Pathways
4.12. Western Blot Analysis
4.13. ELISA Test—MMP7 Level Detection
4.14. Expression of Leptin System in Normal Prostate and Tumor Tissues—Analysis of The Cancer Genome Atlas (TCGA) Dataset
4.15. Statistics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AR | Androgen receptor |
| BAX | Bcl-2-like protein 4 |
| Bcl-2 | B-cell lymphoma 2 |
| BID | BH3 interacting domain death agonist |
| BIRC3 | Baculoviral IAP repeat containing 3 |
| BMX | BMX non-receptor tyrosine kinase |
| C11orf92 | Chromosome 11 open reading frame 92 |
| CASP3 | Procaspases 3 |
| CASP7 | Procaspases 7 |
| CCL-20 | Chemokine C-Cmotif ligand 20 |
| CNTF | Ciliary neurotrophic factor |
| DAPK1 | Death-associated protein kinase 1 |
| DAVID | Database for annotation, visualization, and integreted discovery |
| DEG | Differentially expressed genes |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial–mesenchymal transition |
| FAS | Fas cell surface death receptor |
| FASL | Fas ligand |
| FBS | Fetal bovine serum |
| FGF | Fibroblast growth factors |
| FSH | Follicle-stimulating hormone |
| GADD45G | Growth arrest and DNA-damage-inducible, gamma |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GO BP | Gene onthlogy biological process |
| GSEA | Gene Set Enrichment Analysis |
| HPRT | Hypoxanthine phosphoribosyltransferase |
| IGF | Insulin-like growth factors |
| IL-11 | Interleukin 11 |
| IL-6 | Interleukin 6 |
| IRS | Insulin receptor substrates |
| JAK 2 | Janus-activated kinase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KLK4 | kallikrein-related peptidase 4 |
| LCN2 | Lipocalin 2 |
| LEP | Leptin |
| LEPR | Leptin receptor |
| LH | Luteinizing hormone |
| LIF | Leukemia inhibitory factor |
| MAPK | Mitogen activated protein kinase |
| MMP7 | Matrix metallopeptidase 7 |
| MYLK | Myosin light chain kinase |
| NES | Normalized enrichment score |
| NF-κB | Nuclear factor of kappa |
| NOXA | Phorbol-12-myristate-13-acetate-induced protein 1 |
| PARP | Procyclic acidic repetitive proteins |
| PEG3 | Paternally-expressed gene 3 protein |
| PI3K/Akt | Initiating activation of the phosphoinositide 3 kinase |
| PUMA | p53 upregulated modulator of apoptosis |
| RIMS1 | regulating synaptic membrane exocytosis 1 |
| RTCA | Real Time Cell Analyzer |
| Stat | Signal transducer and activator of transcription |
| STK3 | Serine/threonine-protein kinase 3 |
| TCGA | The Cancer Genome Atlas |
| TGF | Transforming growth factor |
| TNF | Tumor necrosis factor |
| TNFAIP3 | Tumor necrosis factor, alpha-induced protein |
| TRADD | Tumor necrosis factor receptor type 1-associated DEATH domain protein |
| TSH | Thyroid-stimulating hormone |
| VEGF | Vascular endothelial growth factor |
| ZBTB16 | Zinc finger and BTB domain-containing protein 16 |
References
- Lee, D.W.; Leinung, M.C.; Rozhavskaya-Arena, M.; Grasso, P. Leptin and the treatment of obesity: Its current status. Eur. J. Pharmacol. 2002, 440, 129–139. [Google Scholar] [CrossRef]
- Bray, G.A. The underlying basis for obesity: Relationship to cancer. J. Nutr. 2002, 132, 3451S–3455S. [Google Scholar] [CrossRef]
- Calle, E.E.; Rodriguez, C.; Walker-Thurmond, K.; Thun, M.J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003, 348, 1625–1638. [Google Scholar] [CrossRef]
- Calle, E.E.; Thun, M.J. Obesity and cancer. Oncogene 2004, 23, 6365–6378. [Google Scholar] [CrossRef] [PubMed]
- Renehan, A.G.; Zwahlen, M.; Egger, M. Adiposity and cancer risk: New mechanistic insights from epidemiology. Nat. Rev. Cancer 2015, 15, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, P.J.; Stambolic, V. Impact of the obesity epidemic on cancer. Annu. Rev. Med. 2015, 66, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, A.; Pisani, P.; Tenet, V.; Wolk, A.; Adami, H.O. Overweight as an avoidable cause of cancer in Europe. Int. J. Cancer 2001, 91, 421–430. [Google Scholar] [CrossRef]
- Tong, X.; Ma, Y.; Zhou, Q.; He, J.; Peng, B.; Liu, S.; Yan, Z.; Yang, X.; Fan, H. Serum and tissue leptin in lung cancer: A meta-analysis. Oncotarget 2017, 8, 19699–19711. [Google Scholar] [CrossRef] [PubMed]
- Oba, J.; Wei, W.; Gershenwald, J.E.; Johnson, M.M.; Wyatt, C.M.; Ellerhorst, J.A.; Grimm, E.A. Elevated Serum Leptin Levels are Associated With an Increased Risk of Sentinel Lymph Node Metastasis in Cutaneous Melanoma. Medicine (Baltimore) 2016, 95, e3073. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Jimenez, F.; Perez-Perez, A.; de la Cruz-Merino, L.; Sanchez-Margalet, V. Obesity and Breast Cancer: Role of Leptin. Front. Oncol. 2019, 9, 596. [Google Scholar] [CrossRef]
- Ribeiro, R.; Monteiro, C.; Catalan, V.; Hu, P.; Cunha, V.; Rodriguez, A.; Gomez-Ambrosi, J.; Fraga, A.; Principe, P.; Lobato, C.; et al. Obesity and prostate cancer: Gene expression signature of human periprostatic adipose tissue. BMC Med. 2012, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Hoda, M.R.; Theil, G.; Mohammed, N.; Fischer, K.; Fornara, P. The adipocyte-derived hormone leptin has proliferative actions on androgen-resistant prostate cancer cells linking obesity to advanced stages of prostate cancer. J. Oncol. 2012, 2012, 280386. [Google Scholar] [CrossRef] [PubMed]
- Hoda, M.R.; Mohammed, N.; Theil, G.; Fischer, K.; Fornara, P. Obesity and prostate cancer. Role of adipocytokines and clinical implications. Urologe A 2012, 51, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Giovannucci, E. Obesity and Prostate Cancer. Recent Results Cancer Res. 2016, 208, 137–153. [Google Scholar] [PubMed]
- Allott, E.H.; Masko, E.M.; Freedland, S.J. Obesity and prostate cancer: Weighing the evidence. Eur. Urol. 2013, 63, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.; Lopes, C.; Medeiros, R. The link between obesity and prostate cancer: The leptin pathway and therapeutic perspectives. Prostate Cancer Prostatic Dis. 2006, 9, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Malendowicz, W.; Rucinski, M.; Macchi, C.; Spinazzi, R.; Ziolkowska, A.; Nussdorfer, G.G.; Kwias, Z. Leptin and leptin receptors in the prostate and seminal vesicles of the adult rat. Int. J. Mol. Med. 2006, 18, 615–618. [Google Scholar] [CrossRef]
- Malendowicz, W.; Rucinski, M.; Belloni, A.S.; Ziolkowska, A.; Nussdorfer, G.G.; Kwias, Z. Real-time PCR analysis of leptin and leptin receptor expression in the rat prostate, and effects of leptin on prostatic acid phosphatase release. Int. J. Mol. Med. 2006, 18, 1097–1100. [Google Scholar] [CrossRef][Green Version]
- Colli, S.; Silveira Cavalcante, F.; Peixoto Martins, M.; Sampaio, F.J.; da Fonte Ramos, C. Leptin role in the rat prostate ventral lobe. Fertil. Steril. 2011, 95, 1490–1493. [Google Scholar] [CrossRef]
- Habib, C.N.; Al-Abd, A.M.; Tolba, M.F.; Khalifa, A.E.; Khedr, A.; Mosli, H.A.; Abdel-Naim, A.B. Leptin influences estrogen metabolism and accelerates prostate cell proliferation. Life Sci. 2015, 121, 10–15. [Google Scholar] [CrossRef]
- Cioffi, J.A.; Shafer, A.W.; Zupancic, T.J.; Smith-Gbur, J.; Mikhail, A.; Platika, D.; Snodgrass, H.R. Novel B219/OB receptor isoforms: Possible role of leptin in hematopoiesis and reproduction. Nat. Med. 1996, 2, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Malendowicz, W.; Kwias, Z. Leptin receptor isoforms in benign prostatic hyperplasia (Bph). Bph and prostate cancer—No association between plasma concentrations of leptin and prostate specific antigen (pSa). Cent. Eur. J. Urol. 2009, 62, 96–100. [Google Scholar] [CrossRef]
- Leze, E.; Alves-Pereira, J.L.; Colli, S.; Cavalcante, F.S.; Jose Sampaio, F.; da Fonte Ramos, C. Leptin regulates proliferation and apoptosis in human prostate. Sci. World J. 2012, 2012, 842301. [Google Scholar] [CrossRef] [PubMed]
- Szyszka, M.; Tyczewska, M.; Milecka, P.; Jopek, K.; Celichowski, P.; Malendowicz, L.K.; Rucinski, M. Effects of leptin on leptin receptor isoform expression and proliferative activity in human normal prostate and prostate cancer cell lines. Oncol. Rep. 2018, 39, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Onuma, M.; Bub, J.D.; Rummel, T.L.; Iwamoto, Y. Prostate cancer cell-adipocyte interaction: Leptin mediates androgen-independent prostate cancer cell proliferation through c-Jun NH2-terminal kinase. J. Biol. Chem. 2003, 278, 42660–42667. [Google Scholar] [CrossRef] [PubMed]
- Mistry, T.; Digby, J.E.; Desai, K.M.; Randeva, H.S. Leptin and adiponectin interact in the regulation of prostate cancer cell growth via modulation of p53 and bcl-2 expression. BJU Int. 2008, 101, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Deo, D.D.; Rao, A.P.; Bose, S.S.; Ouhtit, A.; Baliga, S.B.; Rao, S.A.; Trock, B.J.; Thouta, R.; Raj, M.H.; Rao, P.N. Differential effects of leptin on the invasive potential of androgen-dependent and -independent prostate carcinoma cells. J. BioMed Biotechnol. 2008, 2008, 163902. [Google Scholar] [CrossRef]
- Noda, T.; Kikugawa, T.; Tanji, N.; Miura, N.; Asai, S.; Higashiyama, S.; Yokoyama, M. Longterm exposure to leptin enhances the growth of prostate cancer cells. Int. J. Oncol. 2015, 46, 1535–1542. [Google Scholar] [CrossRef]
- Miyazaki, T.; Bub, J.D.; Iwamoto, Y. C-Jun NH(2)-terminal kinase mediates leptin-stimulated androgen-independent prostate cancer cell proliferation via signal transducer and activator of transcription 3 and Akt. Biochim. Biophys. Acta 2008, 1782, 593–604. [Google Scholar] [CrossRef]
- Ghasemi, A.; Saeidi, J.; Azimi-Nejad, M.; Hashemy, S.I. Leptin-induced signaling pathways in cancer cell migration and invasion. Cell. Oncol. 2019, 42, 243–260. [Google Scholar] [CrossRef]
- Dutta, D.; Ghosh, S.; Pandit, K.; Mukhopadhyay, P.; Chowdhury, S. Leptin and cancer: Pathogenesis and modulation. Indian J. Endocrinol. Metab. 2012, 16 (Suppl. 3), S596–S600. [Google Scholar] [PubMed]
- Gorska, E.; Popko, K.; Stelmaszczyk-Emmel, A.; Ciepiela, O.; Kucharska, A.; Wasik, M. Leptin receptors. Eur. J. Med. Res. 2010, 15 (Suppl. 2), 50–54. [Google Scholar]
- Huang, C.Y.; Yu, H.S.; Lai, T.Y.; Yeh, Y.L.; Su, C.C.; Hsu, H.H.; Tsai, F.J.; Tsai, C.H.; Wu, H.C.; Tang, C.H. Leptin increases motility and integrin up-regulation in human prostate cancer cells. J. Cell. Physiol. 2011, 226, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Fruhbeck, G. Intracellular signalling pathways activated by leptin. Biochem. J. 2006, 393 Pt 1, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Samuel-Mendelsohn, S.; Inbar, M.; Weiss-Messer, E.; Niv-Spector, L.; Gertler, A.; Barkey, R.J. Leptin signaling and apoptotic effects in human prostate cancer cell lines. Prostate 2011, 71, 929–945. [Google Scholar] [CrossRef] [PubMed]
- Faggioni, R.; Feingold, K.R.; Grunfeld, C. Leptin regulation of the immune response and the immunodeficiency of malnutrition. FASEB J. 2001, 15, 2565–2571. [Google Scholar] [CrossRef]
- Loffreda, S.; Yang, S.Q.; Lin, H.Z.; Karp, C.L.; Brengman, M.L.; Wang, D.J.; Klein, A.S.; Bulkley, G.B.; Bao, C.; Noble, P.W.; et al. Leptin regulates proinflammatory immune responses. FASEB J. 1998, 12, 57–65. [Google Scholar] [CrossRef]
- Santos-Alvarez, J.; Goberna, R.; Sanchez-Margalet, V. Human leptin stimulates proliferation and activation of human circulating monocytes. Cell. Immunol. 1999, 194, 6–11. [Google Scholar] [CrossRef]
- Malinen, M.; Niskanen, E.A.; Kaikkonen, M.U.; Palvimo, J.J. Crosstalk between androgen and pro-inflammatory signaling remodels androgen receptor and NF-kappaB cistrome to reprogram the prostate cancer cell transcriptome. Nucleic Acids Res. 2017, 45, 619–630. [Google Scholar] [CrossRef]
- Jin, R.J.; Lho, Y.; Connelly, L.; Wang, Y.; Yu, X.; Saint Jean, L.; Case, T.C.; Ellwood-Yen, K.; Sawyers, C.L.; Bhowmick, N.A.; et al. The nuclear factor-kappaB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008, 68, 6762–6769. [Google Scholar] [CrossRef]
- Zeligs, K.P.; Neuman, M.K.; Annunziata, C.M. Molecular Pathways: The Balance between Cancer and the Immune System Challenges the Therapeutic Specificity of Targeting Nuclear Factor-kappaB Signaling for Cancer Treatment. Clin. Cancer Res. 2016, 22, 4302–4308. [Google Scholar] [CrossRef] [PubMed]
- Beider, K.; Abraham, M.; Begin, M.; Wald, H.; Weiss, I.D.; Wald, O.; Pikarsky, E.; Abramovitch, R.; Zeira, E.; Galun, E.; et al. Interaction between CXCR4 and CCL20 pathways regulates tumor growth. PLoS ONE 2009, 4, e5125. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Ge, D.; Liu, S.; Zhang, Q.; Borowsky, A.D.; Melamed, J. Interleukin-17 Induces Expression of Chemokines and Cytokines in Prostatic Epithelial Cells but Does Not Stimulate Cell Growth In Vitro. Int. J. Med. Biol. Front. 2012, 18, 629–644. [Google Scholar] [PubMed]
- Zhang, Q.; Liu, S.; Parajuli, K.R.; Zhang, W.; Zhang, K.; Mo, Z.; Liu, J.; Chen, Z.; Yang, S.; Wang, A.R.; et al. Interleukin-17 promotes prostate cancer via MMP7-induced epithelial-to-mesenchymal transition. Oncogene 2017, 36, 687–699. [Google Scholar] [CrossRef]
- Pajouh, M.S.; Nagle, R.B.; Breathnach, R.; Finch, J.S.; Brawer, M.K.; Bowden, G.T. Expression of metalloproteinase genes in human prostate cancer. J. Cancer Res. Clin. Oncol. 1991, 117, 144–150. [Google Scholar] [CrossRef]
- Knox, J.D.; Wolf, C.; McDaniel, K.; Clark, V.; Loriot, M.; Bowden, G.T.; Nagle, R.B. Matrilysin expression in human prostate carcinoma. Mol. Carcinog. 1996, 15, 57–63. [Google Scholar] [CrossRef]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef]
- Powell, W.C.; Fingleton, B.; Wilson, C.L.; Boothby, M.; Matrisian, L.M. The metalloproteinase matrilysin proteolytically generates active soluble Fas ligand and potentiates epithelial cell apoptosis. Curr. Biol. 1999, 9, 1441–1447. [Google Scholar] [CrossRef]
- Mitsiades, N.; Yu, W.H.; Poulaki, V.; Tsokos, M.; Stamenkovic, I. Matrix metalloproteinase-7-mediated cleavage of Fas ligand protects tumor cells from chemotherapeutic drug cytotoxicity. Cancer Res. 2001, 61, 577–581. [Google Scholar]
- Tung, M.C.; Hsieh, S.C.; Yang, S.F.; Cheng, C.W.; Tsai, R.T.; Wang, S.C.; Huang, M.H.; Hsieh, Y.H. Knockdown of lipocalin-2 suppresses the growth and invasion of prostate cancer cells. Prostate 2013, 73, 1281–1290. [Google Scholar] [CrossRef]
- Mahadevan, N.R.; Rodvold, J.; Almanza, G.; Perez, A.F.; Wheeler, M.C.; Zanetti, M. ER stress drives Lipocalin 2 upregulation in prostate cancer cells in an NF-kappaB-dependent manner. BMC Cancer 2011, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Somasundar, P.; Yu, A.K.; Vona-Davis, L.; McFadden, D.W. Differential effects of leptin on cancer in vitro. J. Surg. Res. 2003, 113, 50–55. [Google Scholar] [CrossRef]
- Somasundar, P.; Frankenberry, K.A.; Skinner, H.; Vedula, G.; McFadden, D.W.; Riggs, D.; Jackson, B.; Vangilder, R.; Hileman, S.M.; Vona-Davis, L.C. Prostate cancer cell proliferation is influenced by leptin. J. Surg. Res. 2004, 118, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Hoda, M.R.; Popken, G. Mitogenic and anti-apoptotic actions of adipocyte-derived hormone leptin in prostate cancer cells. BJU Int. 2008, 102, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Umesaki, N. Leptin regulates the proliferation and apoptosis of human endometrial epithelial cells. Int. J. Mol. Med. 2008, 22, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Azain, M.J.; Compton, M.M.; Hartzell, D.L.; Hausman, G.J.; Baile, C.A. Brain administration of leptin causes deletion of adipocytes by apoptosis. Endocrinology 1998, 139, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.S.; Hong, J.S.; Kim, S.W.; Koh, J.M.; An, C.S.; Choi, J.Y.; Cheng, S.L. Leptin induces apoptosis via ERK/cPLA2/cytochrome c pathway in human bone marrow stromal cells. J. Biol. Chem. 2003, 278, 21920–21929. [Google Scholar] [CrossRef]
- Zhu, M.L.; Kyprianou, N. Androgen receptor and growth factor signaling cross-talk in prostate cancer cells. Endocr. Relat. Cancer 2008, 15, 841–849. [Google Scholar] [CrossRef]
- Russell, P.J.; Bennett, S.; Stricker, P. Growth factor involvement in progression of prostate cancer. Clin. Chem. 1998, 44, 705–723. [Google Scholar]
- Wu, Y.; Zhao, W.; Zhao, J.; Pan, J.; Wu, Q.; Zhang, Y.; Bauman, W.A.; Cardozo, C.P. Identification of androgen response elements in the insulin-like growth factor I upstream promoter. Endocrinology 2007, 148, 2984–2993. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen receptor in prostate cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zhang, S.; Burguera, B.G.; Couce, M.E.; Osamura, R.Y.; Kulig, E.; Lloyd, R.V. Leptin and leptin receptor expression in rat and mouse pituitary cells. Endocrinology 2000, 141, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Kosior-Korzecka, U.; Bobowiec, R. Leptin effect on nitric oxide and GnRH-induced FSH secretion from ovine pituitary cells in vitro. J. Physiol. Pharmacol. 2006, 57, 637–647. [Google Scholar] [PubMed]
- Weil, C.; Le Bail, P.Y.; Sabin, N.; Le Gac, F. In vitro action of leptin on FSH and LH production in rainbow trout (Onchorynchus mykiss) at different stages of the sexual cycle. Gen. Comp. Endocrinol. 2003, 130, 2–12. [Google Scholar] [CrossRef]
- Chen, C.; Roh, S.G.; Nie, G.Y.; Loneragan, K.; Xu, R.W.; Ruan, M.; Clarke, L.J.; Goding, J.W.; Gertler, A. The in vitro effect of leptin on growth hormone secretion from primary cultured ovine somatotrophs. Endocrine 2001, 14, 73–78. [Google Scholar] [CrossRef]
- Melen-Mucha, G.; Lawnicka, H. Leptin promotes the growth of Colon 38 cancer cells and interferes with the cytotoxic effect of fluorouracil in vitro. Endokrynol. Pol. 2007, 58, 2–6. [Google Scholar] [PubMed]
- Szyszka, M.; Paschke, L.; Tyczewska, M.; Rucinski, M.; Grabowska, P.; Malendowicz, L.K. Lack of expression of preproorexin and orexin receptors genes in human normal and prostate cancer cell lines. Folia Histochem. Cytobiol. 2015, 53, 333–341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paschke, L.; Zemleduch, T.; Rucinski, M.; Ziolkowska, A.; Szyszka, M.; Malendowicz, L.K. Adiponectin and adiponectin receptor system in the rat adrenal gland: Ontogenetic and physiologic regulation, and its involvement in regulating adrenocortical growth and steroidogenesis. Peptides 2010, 31, 1715–1724. [Google Scholar] [CrossRef]
- Jopek, K.; Tyczewska, M.; Celichowski, P.; Malendowicz, L.K.; Rucinski, M. Transcriptome Profile in Unilateral Adrenalectomy-Induced Compensatory Adrenal Growth in the Rat. Int. J. Mol. Sci. 2018, 19, 1111. [Google Scholar] [CrossRef]
- Jopek, K.; Celichowski, P.; Szyszka, M.; Tyczewska, M.; Milecka, P.; Malendowicz, L.K.; Rucinski, M. Transcriptome Profile of Rat Adrenal Evoked by Gonadectomy and Testosterone or Estradiol Replacement. Front. Endocrinol. (Lausanne) 2017, 8, 26. [Google Scholar] [CrossRef]
- Trejter, M.; Hochol, A.; Tyczewska, M.; Ziolkowska, A.; Jopek, K.; Szyszka, M.; Malendowicz, L.K.; Rucinski, M. Visinin-like peptide 1 in adrenal gland of the rat. Gene expression and its hormonal control. Peptides 2015, 63, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska-Kempisty, H.; Rucinski, M.; Borys, S.; Kucinska, M.; Kaczmarek, M.; Zawierucha, P.; Wierzchowski, M.; Lazewski, D.; Murias, M.; Jodynis-Liebert, J. 3′-hydroxy-3,4,5,4′-tetramethoxystilbene, the metabolite of resveratrol analogue DMU-212, inhibits ovarian cancer cell growth in vitro and in a mice xenograft model. Sci. Rep. 2016, 6, 32627. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Fresno, C.; Fernandez, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef]
- Benjamini, Y.; Cohen, R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 2017, 18, 91–104. [Google Scholar] [CrossRef]
- Walter, W.; Sanchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Damian, D.; Gorfine, M. Statistical concerns about the GSEA procedure. Nat. Genet. 2004, 36, 663. [Google Scholar] [CrossRef]
- Sergushichev, A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv 2016. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Paschke, L.; Jopek, K.; Szyszka, M.; Tyczewska, M.; Ziolkowska, A.; Rucinski, M.; Malendowicz, L.K. ZFP91: A Noncanonical NF-kappaB Signaling Pathway Regulator with Oncogenic Properties Is Overexpressed in Prostate Cancer. BioMed. Res. Int. 2016, 2016, 6963582. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Deng, M.; Bragelmann, J.; Kryukov, I.; Saraiva-Agostinho, N.; Perner, S. FirebrowseR: An R client to the Broad Institute’s Firehose Pipeline. Database (Oxford) 2017, 2017, baw160. [Google Scholar] [CrossRef]



| Cell Type | Method of Detection | Dose of LEP and Time | Effect | Reference |
|---|---|---|---|---|
| LNCaP-FGC, DU145, PC-3 | [3H]thymidine incorporation MTT assay | LEP (0.1–12.5 µg/mL) for 20 h LEP (12.5 µg/mL) for 5 days | stimulation of proliferation in DU145 and PC-3 cells but not in LNCaP-FGC cell | Onuma et al. (2003) [25] |
| DU145 and PC-3 | MTT assay | LEP (0.4 ng/mL and 4.0 ng/mL) for 24 and 48 h | in both cell lines LEP inhibited growth | Somasundar et al. (2003) [52] |
| DU145 and PC-3 | MTT assay ELISA | LEP (4 or 40 ng/mL) for 24 and 72 h LEP (4 or 40 ng/mL) for 24 h | stimulation of proliferation in both cell lines increased apoptosis in both cell lines | Somasundar et al. (2004) [53] |
| DU145 | [3H]thymidine incorporation | LEP (12.5 μg/mL) for 20 h | stimulation of proliferation | Miyazaki et al. (2008) [29] |
| LNCaP, DU-145 and PC-3 | [3H]thymidine incorporation | LEP (20–200 ng/mL) for 144h | stimulation of proliferation of LNCaP cells, no effects on PC-3 and Du-145 cells | Deo et al. (2008) [27] |
| DU145 and PC-3 | XTT colorimetric assay (tetrazolium based assay) Cell death detection ELISA plus® assay | LEP (5–100 ng/mL) for up to 48 h LEP (100 ng/mL) for 24 h | stimulation of proliferation in both cell lines anti-apoptotic effects | Hoda & Popken (2008) [54] |
| LNCaP and PC3 | CellTiter 96® AQueous One Solution Cell Proliferation Assay (tetrazolium based assay) | LEP (0.01–100 nM) for 48 h | proliferation in LNCaP cells unaffected, in PC3 cells significantly increased | Mistry et al. (2008) [26] |
| LNCaP-FGC, DU-145, PC-3, and PC-3 cells stably expressing AR (androgen receptor)—PC-3/AR | apoptosis effector protein caspase 3 levels, cleavage of the DNA repair enzyme, and the numbers of apoptotic cells visualized by Hoechst 33342 | LEP (1 ng/mL) for 24 h | In all studied cell lines statistically significant pro-apoptotic effects of LEP | Samuel-Mendelsohn et al. (2011) [35] |
| samples of human hyperplastic prostate tissue | cell proliferation evaluated by immunohistochemistry for PCNA, RT-PCR (expression of apoptosis related genes) | incubation with LEP (50 ng/mL) for 3 h | stimulation of cell proliferation (ca 3-folds) and BAX expression, lowered expression levels of BCL-2 and BCL-X | Leze et al. (2012) [23] |
| LNCaP, DU145 and PC-3 | WST-8 assays and a Cell Counting kit-8 | LEP (up to 1000 ng/mL) for 48 h | in all cell lines no effects on cell proliferation, LEP (100 ng/mL) - cell number notably increased between days 7–42 of culture | Noda et al. (2015) [28] |
| human normal prostate (PrEC, PrSC, PrSMC) and prostate cancer (DU145, LNCaP, PC3) cell lines | proliferative activity was determined by RTCA (real-time cell analyzer) | LEP (1 × 10−6, 1 × 10−8 and 1 × 10−10 M) for at least 70 h | lowered proliferation rate of LNCaP cells - at 1 × 10−6 M LEP concentration, increased proliferation rate of DU145 cells at the same concentration, increased proliferation rate of PrSC cells at 1 × 10−8 and 1 × 10−10 M LEP concentrations. In all remaining tests LEP did not influence the proliferation rate of the studied cells | Szyszka et al. (2018) [24] |
| Gene Symbol | Genbank Accession Number | Primer | Primer Sequence (5′–3′) | Position | PCR Product Size (bp) |
|---|---|---|---|---|---|
| BMX | NM_203281.3 | S | CATCGGACACCATCTACCAG | 2053–2072 | 278 |
| BMX non-receptor tyrosine kinase | A | CTTTTGTTTCCTGCCTTGTTC | 2310–2330 | ||
| C11orf92 | NM_001302644.1 | S | AGCAAGAATATCACCGTGAAGCA | 129–151 | 184 |
| chromosome 11 open reading frame 92 | A | ACCACGATGTCGGGTAACTC | 293–312 | ||
| KLK4 | NM_004917.4 | S | CTCTATGACCCGCTGTACCAC | 541–561 | 123 |
| kallikrein-related peptidase 4 | A | CACAAGGCCCTGCAAGTACC | 644–663 | ||
| MYLK | NM_053025.4 | S | AAGTGCTGCTAGATTTGACT | 5781–5800 | 136 |
| myosin light chain kinase | A | AATTAAAGAGCAGTTCCCGTC | 5896–5916 | ||
| RIMS1 | NM_014989.5 | S | AATATTTCCTGGAGTGCGACTGG | 4830–4852 | 179 |
| regulating synaptic membrane exocytosis 1 | A | GGCTTCGTGCTCTAATGACTT | 4988–5008 | ||
| MMP7 | NM_002423.4 | S | ATGATATTAAAGGCATTCAGA | 799–819 | 281 |
| matrix metallopeptidase 7 | A | TTTATTGACATCTACCCACT | 1060–1079 | ||
| TNF IP3 | NM_001270508.1 | S | CCATCATTTTGTACCCTTG | 1132–1150 | 280 |
| TNF alpha induced protein 3 transcript variant 1 | A | TTCAAGTAATCATCTACCAG | 1391–1411 | ||
| FAS | NM_000043.5 | S | CAAAAGTGTTAATGCCCAA | 378–396 | 299 |
| Fas cell surface death receptor transcript variant 1 | A | TGCAGTTTATTTCCACTTC | 658–676 | ||
| BIRC3 | NM_001165.4 | S | TAGTAAAAGGAAATATTGCAG | 4294–4314 | 100 |
| baculoviral IAP repeat containing 3 transcript variant 1 | A | TATTTTATGTCCTGTTGCAC | 4374–4393 | ||
| HPRT | NM_000194.2 | S | GCCATCACATTGTAGCCCTC | 343–362 | 172 |
| hypoxanthine phosphoribosyltransferase 1 | A | ACTTTTATGTCCCCTGTTGACT | 493–514 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szyszka, M.; Paschke, L.; Tyczewska, M.; Jopek, K.; Celichowski, P.; Milecka, P.; Sultanova, G.; Stelcer, E.; Malinska, A.; Malendowicz, L.K.; et al. Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations. Int. J. Mol. Sci. 2019, 20, 5412. https://doi.org/10.3390/ijms20215412
Szyszka M, Paschke L, Tyczewska M, Jopek K, Celichowski P, Milecka P, Sultanova G, Stelcer E, Malinska A, Malendowicz LK, et al. Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations. International Journal of Molecular Sciences. 2019; 20(21):5412. https://doi.org/10.3390/ijms20215412
Chicago/Turabian StyleSzyszka, Marta, Lukasz Paschke, Marianna Tyczewska, Karol Jopek, Piotr Celichowski, Paulina Milecka, Gulnara Sultanova, Ewelina Stelcer, Agnieszka Malinska, Ludwik K. Malendowicz, and et al. 2019. "Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations" International Journal of Molecular Sciences 20, no. 21: 5412. https://doi.org/10.3390/ijms20215412
APA StyleSzyszka, M., Paschke, L., Tyczewska, M., Jopek, K., Celichowski, P., Milecka, P., Sultanova, G., Stelcer, E., Malinska, A., Malendowicz, L. K., & Rucinski, M. (2019). Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations. International Journal of Molecular Sciences, 20(21), 5412. https://doi.org/10.3390/ijms20215412