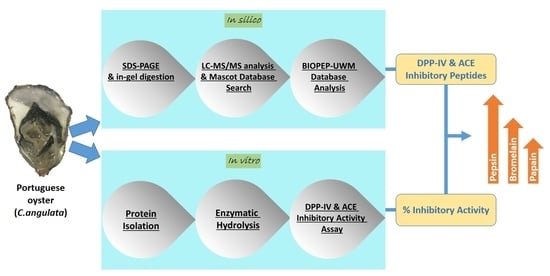
In Silico and In Vitro Assessment of Portuguese Oyster (Crassostrea angulata) Proteins as Precursor of Bioactive Peptides
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identified Proteins From C. angulata
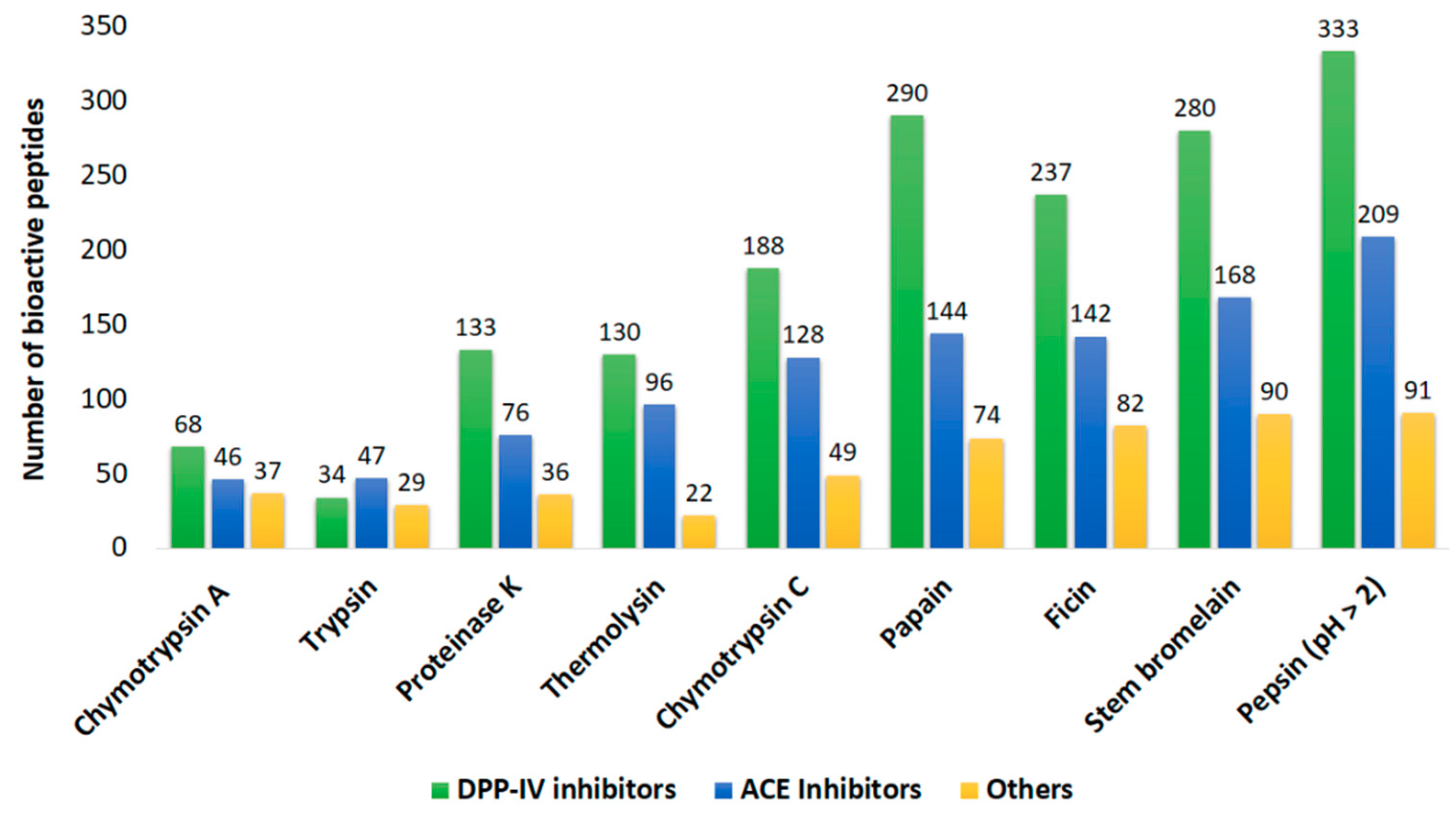
2.2. In Silico Prediction of Potential Bioactivities
2.3. In Vitro Hydrolysis of Oyster Proteins
2.4. Confirmation of Bioactivities Through In Vitro Tests
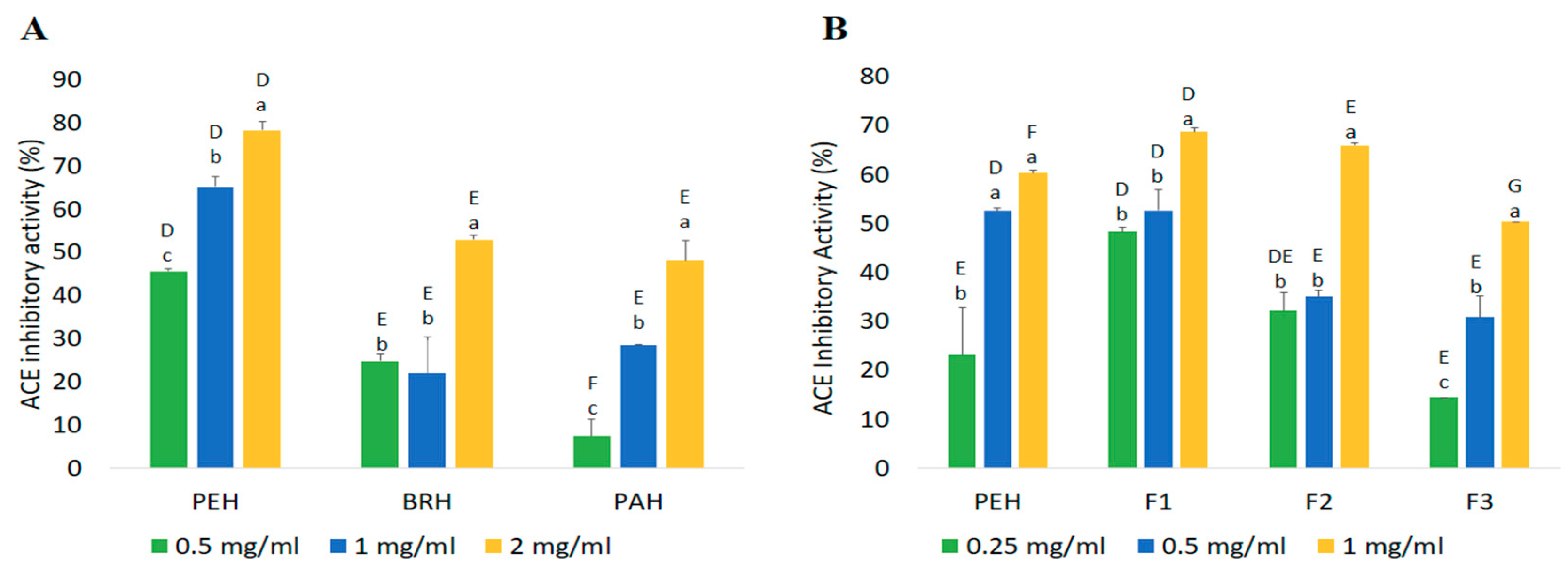
2.4.1. ACE Inhibitory Activity
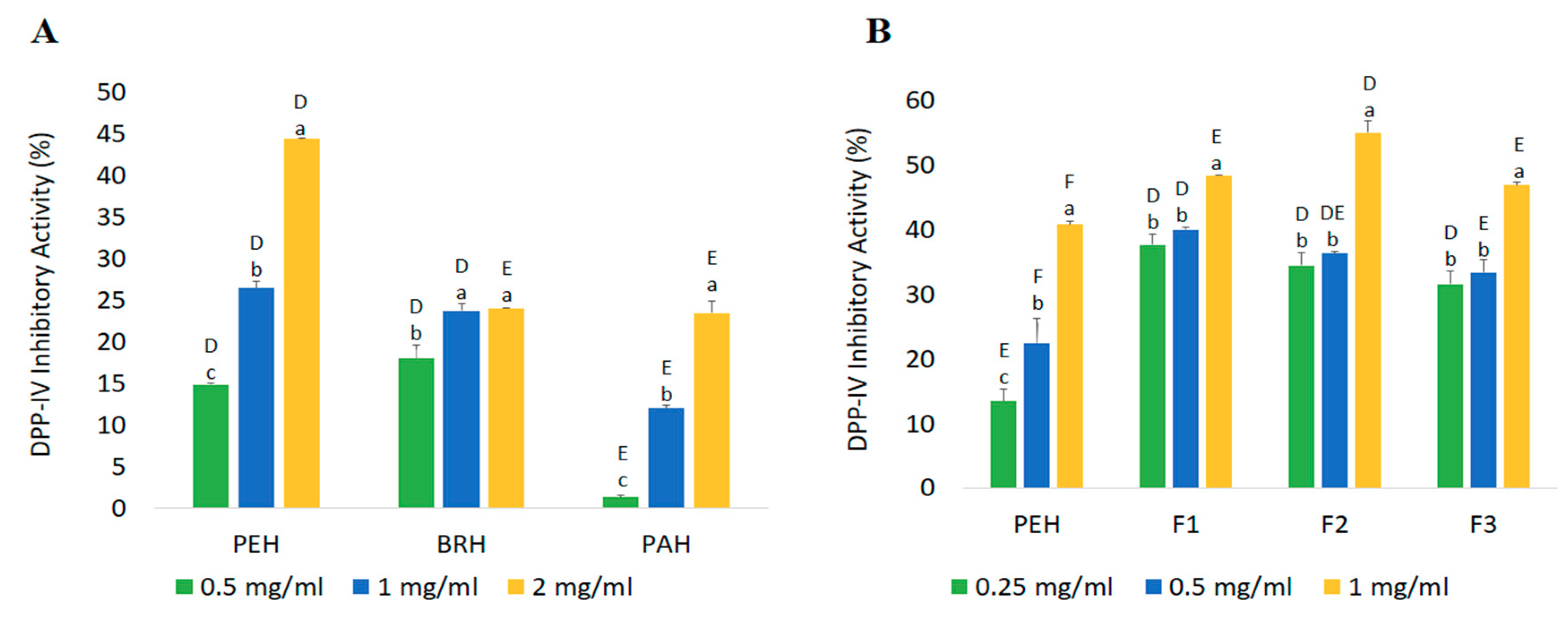
2.4.2. DPP-IV Inhibitory Activity
3. Materials and Methods
3.1. Materials
3.2. Oyster Meat Preparation
3.3. Proteomics Techniques and In Silico Analysis
3.3.1. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-Page) Analysis
3.3.2. Gel Slice and In-Gel Digestion
3.3.3. NanoLC–NanoESI–MS/MS Analysis
3.3.4. Mascot Database Search
3.3.5. BLAST Analysis of Oyster Proteins
3.4. In Vitro Analyses
3.4.1. Protein Isolation
3.4.2. Enzymatic Hydrolysis
3.4.3. Degree of Hydrolysis
3.4.4. Fractionation
3.4.5. Peptide Content Determination
3.4.6. Angiotensin-I Converting Enzyme (ACE) Inhibitory Activity Assay
3.4.7. Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Activity Assay
3.4.8. Calculation of Inhibition Efficiency Ratio
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, C.W.; Liang, C.P.; Huang, F.M.; Hsueh, Y.M. Assessing the human health risks from exposure of inorganic arsenic through oyster (Crassostrea gigas) consumption in Taiwan. Sci. Total Env. 2006, 361, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, J.W.M.; Troost, K.; Fang, J.; Roncarati, A. Global Production of Marine Bivalves. Trends and Challenges. Goods Serv. Mar. Bivalves 2019, 7–26. [Google Scholar] [CrossRef]
- Peralta, E.M.; Tumbokon, B.L.M.; Serrano Jr, A.E. Use of Oyster Processing Byproduct to Replace Fish Meal and Minerals in the Diet of Nile Tilapia Oreochromis niloticus Fry. Isr. J. Aquac. Bamidgeh 2016. [Google Scholar]
- Pawiro, S. Bivalves: Global production and trade trends. In Safe Management of Shellfish and Harvest Waters; IWA: London, UK, 2010; pp. 11–19. [Google Scholar]
- Sikorski, Z.E.; Pan, B.S. The involvement of proteins and nonprotein nitrogen in postmortem changes in seafoods. Seaf. Proteins 1994, 71–83. [Google Scholar]
- Schutta, M.H. Diabetes and hypertension: Epidemiology of the relationship and pathophysiology of factors associated with these comorbid conditions. J. Cardiometabolic Syndr. 2007, 2, 124–130. [Google Scholar] [CrossRef]
- Meisel, H.; Walsh, D.J.; Murray, B.; FitzGerald, R.J. ACE inhibitory peptides. Nutraceutical proteins Pept. Health Dis. 2005, 273–319. [Google Scholar]
- Ahn, C.B.; Jeon, Y.J.; Kim, Y.T.; Je, J.-Y. Angiotensin I converting enzyme (ACE) inhibitory peptides from salmon byproduct protein hydrolysate by Alcalase hydrolysis. Process. Biochem. 2012, 47, 2240–2245. [Google Scholar] [CrossRef]
- McIntosh, C.H. Incretin-based therapies for type 2 diabetes. Can. J. Diabetes 2008, 32, 131–139. [Google Scholar] [CrossRef]
- Holst, J.J.; Deacon, C.F. Glucagon-like peptide 1 and inhibitors of dipeptidyl peptidase IV in the treatment of type 2 diabetes mellitus. Curr. Opin. Pharmacol. 2004, 4, 589–596. [Google Scholar] [CrossRef]
- Patel, K.; Pate, G., IV. Role of DPP-IV inhibitors in treatment of type II diabetes. IRJP 2010, 1, 19–28. [Google Scholar]
- Hatanaka, T.; Inoue, Y.; Arima, J.; Kumagai, Y.; Usuki, H.; Kawakami, K.; Kimura, M.; Mukaihara, T. Production of dipeptidyl peptidase IV inhibitory peptides from defatted rice bran. Food Chem. 2012, 134, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.H.; Wang, T.Y.; Hung, C.C.; Jao, C.L.; Hsieh, Y.L.; Wu, S.X.; Hsu, K.C. In silico, in vitro and in vivo analyses of dipeptidyl peptidase IV inhibitory activity and the antidiabetic effect of sodium caseinate hydrolysate. Food Funct. 2016, 7, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Deacon, C.F. Dipeptidyl peptidase-4 inhibitors in the treatment of type 2 diabetes: A comparative review. DiabetesObes. Metab. 2011, 13, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Patchett, A.A.; Harris, E.; Tristram, E.W.; Wyvratt, M.J.; Wu, M.T.; Taub, D.; Peterson, E.R.; Ikeler, T.J.; Ten Broeke, J.; Payne, L.G. A new class of angiotensin-converting enzyme inhibitors. Nature 1980, 288, 280. [Google Scholar] [CrossRef]
- Thornberry, N.A.; Gallwitz, B. Mechanism of action of inhibitors of dipeptidyl-peptidase-4 (DPP-4). Best Pract. Res. Clin. Endocrinol. Metab. 2009, 23, 479–486. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Gallagher, E.; Tasdemir, D.; Hayes, M. Heart health peptides from macroalgae and their potential use in functional foods. J. Agric. Food Chem. 2011, 59, 6829–6836. [Google Scholar] [CrossRef]
- Balti, R.; Bougatef, A.; Sila, A.; Guillochon, D.; Dhulster, P.; Nedjar-Arroume, N. Nine novel angiotensin I-converting enzyme (ACE) inhibitory peptides from cuttlefish (Sepia officinalis) muscle protein hydrolysates and antihypertensive effect of the potent active peptide in spontaneously hypertensive rats. Food Chem. 2015, 170, 519–525. [Google Scholar] [CrossRef]
- Dong, X.-P.; Zhu, B.-W.; Zhao, H.-X.; Zhou, D.-Y.; Wu, H.-T.; Yang, J.-F.; Li, D.-M.; Murata, Y. Preparation and in vitro antioxidant activity of enzymatic hydrolysates from oyster (Crassostrea talienwhannensis) meat. Int. J. Food Sci. Technol. 2010, 45, 978–984. [Google Scholar] [CrossRef]
- Lee, H.-J.; Saravana, P.S.; Cho, Y.-N.; Haq, M.; Chun, B.-S. Extraction of bioactive compounds from oyster (Crassostrea gigas) by pressurized hot water extraction. J. Supercrit. Fluids 2018, 141, 120–127. [Google Scholar] [CrossRef]
- Yu, Z.; Wu, S.; Zhao, W.; Ding, L.; Shiuan, D.; Chen, F.; Li, J.; Liu, J. Identification and the molecular mechanism of a novel myosin-derived ACE inhibitory peptide. Food Funct. 2018, 9, 364–370. [Google Scholar] [CrossRef]
- Geirsdóttir, M. Isolation, purification and investigation of peptides from fish proteins with blood pressure decreasing properties. Food Res. Innov. Saf. 2009, 36, 1–28. [Google Scholar]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef] [PubMed]
- Umayaparvathi, S.; Arumugam, M.; Meenakshi, S.; Dräger, G.; Kirschning, A.; Balasubramanian, T. Purification and Characterization of Antioxidant Peptides from Oyster (Saccostrea cucullata) Hydrolysate and the Anticancer Activity of Hydrolysate on Human Colon Cancer Cell Lines. Int. J. Pept. Res. Ther. 2013, 20, 231–243. [Google Scholar] [CrossRef]
- Umayaparvathi, S.; Meenakshi, S.; Vimalraj, V.; Arumugam, M.; Sivagami, G.; Balasubramanian, T. Antioxidant activity and anticancer effect of bioactive peptide from enzymatic hydrolysate of oyster (Saccostrea cucullata). Biomed. Prev. Nutr. 2014, 4, 343–353. [Google Scholar] [CrossRef]
- Wang, J.; Hu, J.; Cui, J.; Bai, X.; Du, Y.; Miyaguchi, Y.; Lin, B. Purification and identification of a ACE inhibitory peptide from oyster proteins hydrolysate and the antihypertensive effect of hydrolysate in spontaneously hypertensive rats. Food Chem. 2008, 111, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Tian, X.; Tian, Z. Antimicrobial Capacity and Antioxidant Activity of Enzymatic Hydrolysates of Protein from Rushan Bay Oyster (Crassostrea gigas). J. Food Process. Preserv. 2015, 39, 404–412. [Google Scholar] [CrossRef]
- Cheung, I.W.; Nakayama, S.; Hsu, M.N.; Samaranayaka, A.G.; Li-Chan, E.C. Angiotensin-I converting enzyme inhibitory activity of hydrolysates from oat (Avena sativa) proteins by in silico and in vitro analyses. J. Agric. Food Chem. 2009, 57, 9234–9242. [Google Scholar] [CrossRef]
- Udenigwe, C.C. Bioinformatics approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 137–143. [Google Scholar] [CrossRef]
- Fu, Y.; Therkildsen, M.; Aluko, R.E.; Lametsch, R. Exploration of collagen recovered from animal by-products as a precursor of bioactive peptides: Successes and challenges. Crit. Rev. Food Sci. Nutr. 2019, 59, 2011–2027. [Google Scholar] [CrossRef]
- Coluccio, L.M. Myosins: A Superfamily of Molecular Motors; Springer Science & Business Media: Dordrecht, The Netherlands, 2007; Volume 7. [Google Scholar]
- Katayama, S.; Shima, J.; Saeki, H. Solubility improvement of shellfish muscle proteins by reaction with glucose and its soluble state in low-ionic-strength medium. J. Agric. Food Chem. 2002, 50, 4327–4332. [Google Scholar] [CrossRef]
- Lacroix, I.M.; Li-Chan, E.C. Evaluation of the potential of dietary proteins as precursors of dipeptidyl peptidase (DPP)-IV inhibitors by an in silico approach. J. Funct. Foods 2012, 4, 403–422. [Google Scholar] [CrossRef]
- Chen, D.W.; Su, J.; Liu, X.L.; Yan, D.M.; Lin, Y.; Jiang, W.M.; Chen, X.H. Amino Acid Profiles of Bivalve Mollusks from Beibu Gulf, China. J. Aquat. Food Prod. Technol. 2012, 21, 369–379. [Google Scholar] [CrossRef]
- Worthington, K. Worthington Enzyme Manual. Worthington Biochemical Corporation. Lakewood NJ 2011. [Google Scholar]
- Menard, R.; Khouri, H.E.; Plouffe, C.; Dupras, R.; Ripoll, D.; Vernet, T.; Tessier, D.C.; Laliberte, F.; Thomas, D.Y.; Storer, A.C. A protein engineering study of the role of aspartate 158 in the catalytic mechanism of papain. Biochemistry 1990, 29, 6706–6713. [Google Scholar] [CrossRef]
- Ramalingam, C.; Srinath, R.; Islam, N.N. Isolation and characterization of bromelain from pineapple (Ananas comosus) and comparing its anti-browning activity on apple juice with commercial anti-browning agents. Elixir. Food Sci. 2012, 45, 7822–7826. [Google Scholar]
- Alarcón, F.; Moyano, F.; Díaz, M. Use of SDS-page in the assessment of protein hydrolysis by fish digestive enzymes. Aquac. Int. 2001, 9, 255–267. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef]
- Asha, K.K.; Remya Kumari, K.R.; Ashok Kumar, K.; Chatterjee, N.S.; Anandan, R.; Mathew, S. Sequence Determination of an Antioxidant Peptide Obtained by Enzymatic Hydrolysis of Oyster Crassostrea madrasensis (Preston). Int. J. Pept. Res. Ther. 2016, 22, 421–433. [Google Scholar] [CrossRef]
- Aluko, R. Bioactive Peptides. Funct. Foods Nutraceuticals 2012, 37–61. [Google Scholar] [CrossRef]
- Tsai, J.-S.; Chen, J.-L.; Pan, B.S. ACE-inhibitory peptides identified from the muscle protein hydrolysate of hard clam (Meretrix lusoria). Process. Biochem. 2008, 43, 743–747. [Google Scholar] [CrossRef]
- Byun, H.-G.; Kim, S.-K. Structure and activity of angiotensin I converting enzyme inhibitory peptides derived from Alaskan pollack skin. BMB Rep. 2002, 35, 239–243. [Google Scholar] [CrossRef]
- Lee, S.-H.; Qian, Z.-J.; Kim, S.-K. A novel angiotensin I converting enzyme inhibitory peptide from tuna frame protein hydrolysate and its antihypertensive effect in spontaneously hypertensive rats. Food Chem. 2010, 118, 96–102. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Food-derived dipeptidyl-peptidase IV inhibitors as a potential approach for glycemic regulation–current knowledge and future research considerations. Trends Food Sci. Technol. 2016, 54, 1–16. [Google Scholar] [CrossRef]
- Ketnawa, S.; Suwal, S.; Huang, J.Y.; Liceaga, A.M. Selective separation and characterisation of dual ACE and DPP-IV inhibitory peptides from rainbow trout (Oncorhynchus mykiss) protein hydrolysates. Int. J. Food Sci. Technol. 2018, 54, 1062–1073. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Huang, B.-B.; Lin, H.-C.; Chang, Y.-W. Analysis of proteins and potential bioactive peptides from tilapia (Oreochromis spp.) processing co-products using proteomic techniques coupled with BIOPEP database. J. Funct. Foods 2015, 19, 629–640. [Google Scholar] [CrossRef]
- Jun, S.Y.; Park, P.J.; Jung, W.K.; Kim, S.K. Purification and characterization of an antioxidative peptide from enzymatic hydrolysate of yellowfin sole (Limanda aspera) frame protein. Eur. Food Res. Technol. 2004, 219, 20–26. [Google Scholar]
- Charoenphun, N.; Cheirsilp, B.; Sirinupong, N.; Youravong, W. Calcium-binding peptides derived from tilapia (Oreochromis niloticus) protein hydrolysate. Eur. Food Res. Technol. 2013, 236, 57–63. [Google Scholar] [CrossRef]
- Raghavan, S.; Kristinsson, H.G. ACE-inhibitory activity of tilapia protein hydrolysates. Food Chem. 2009, 117, 582–588. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Lin, Y.-S.; Hou, W.-C.; Aluko, R.E. Kinetics of the inhibition of renin and angiotensin I-converting enzyme by flaxseed protein hydrolysate fractions. J. Funct. Foods 2009, 1, 199–207. [Google Scholar] [CrossRef]
- Lacroix, I.M.; Li-Chan, E.C. Inhibition of dipeptidyl peptidase (DPP)-IV and alpha-glucosidase activities by pepsin-treated whey proteins. J. Agric. Food Chem. 2013, 61, 7500–7506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, R.; Chen, X.; Zeng, Z.; Ma, H.; Chen, S. Dipeptidyl Peptidase IV-Inhibitory Peptides Derived from Silver Carp (Hypophthalmichthys molitrix Val.) Proteins. J. Agric. Food Chem. 2016, 64, 831–839. [Google Scholar] [CrossRef]
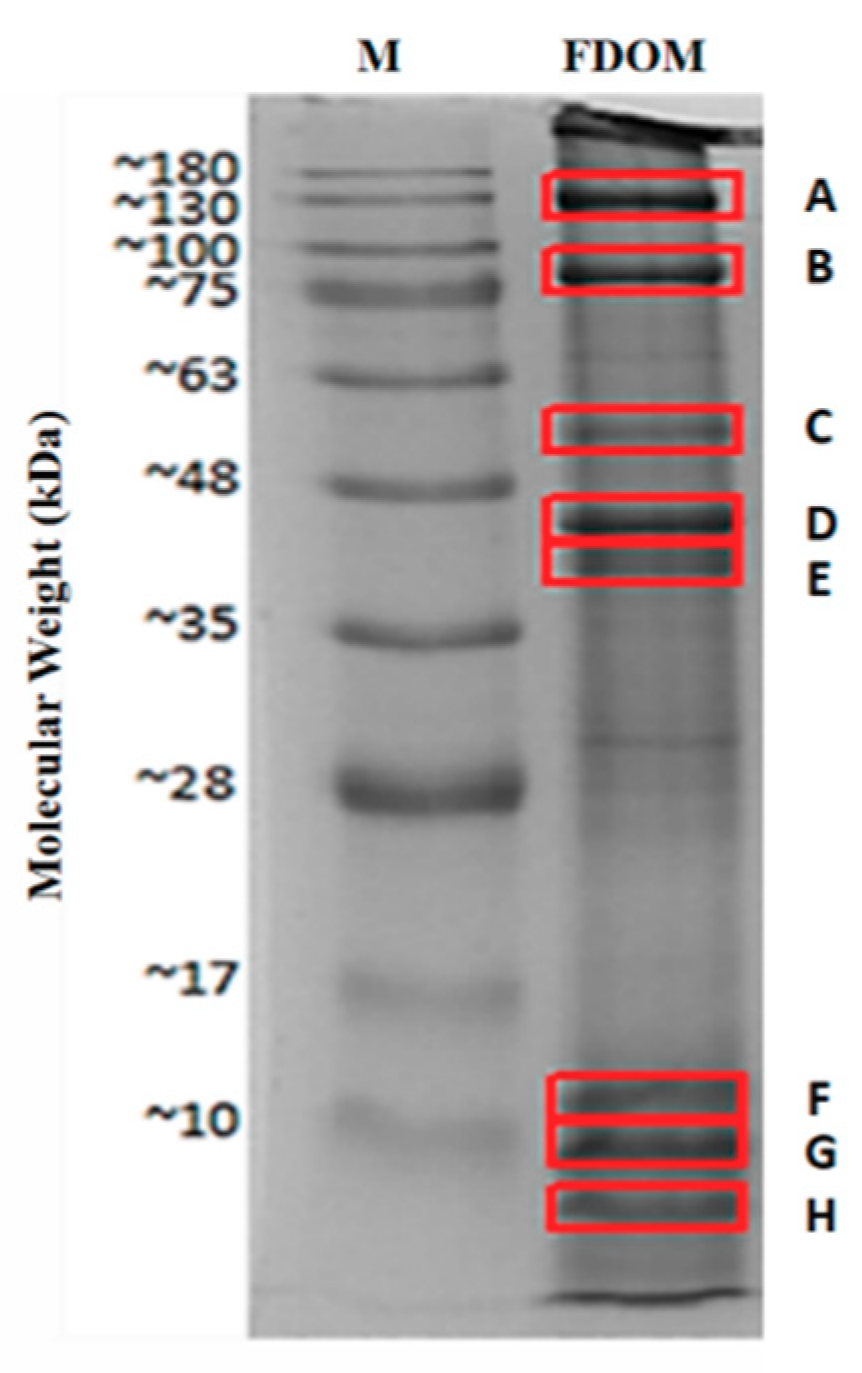
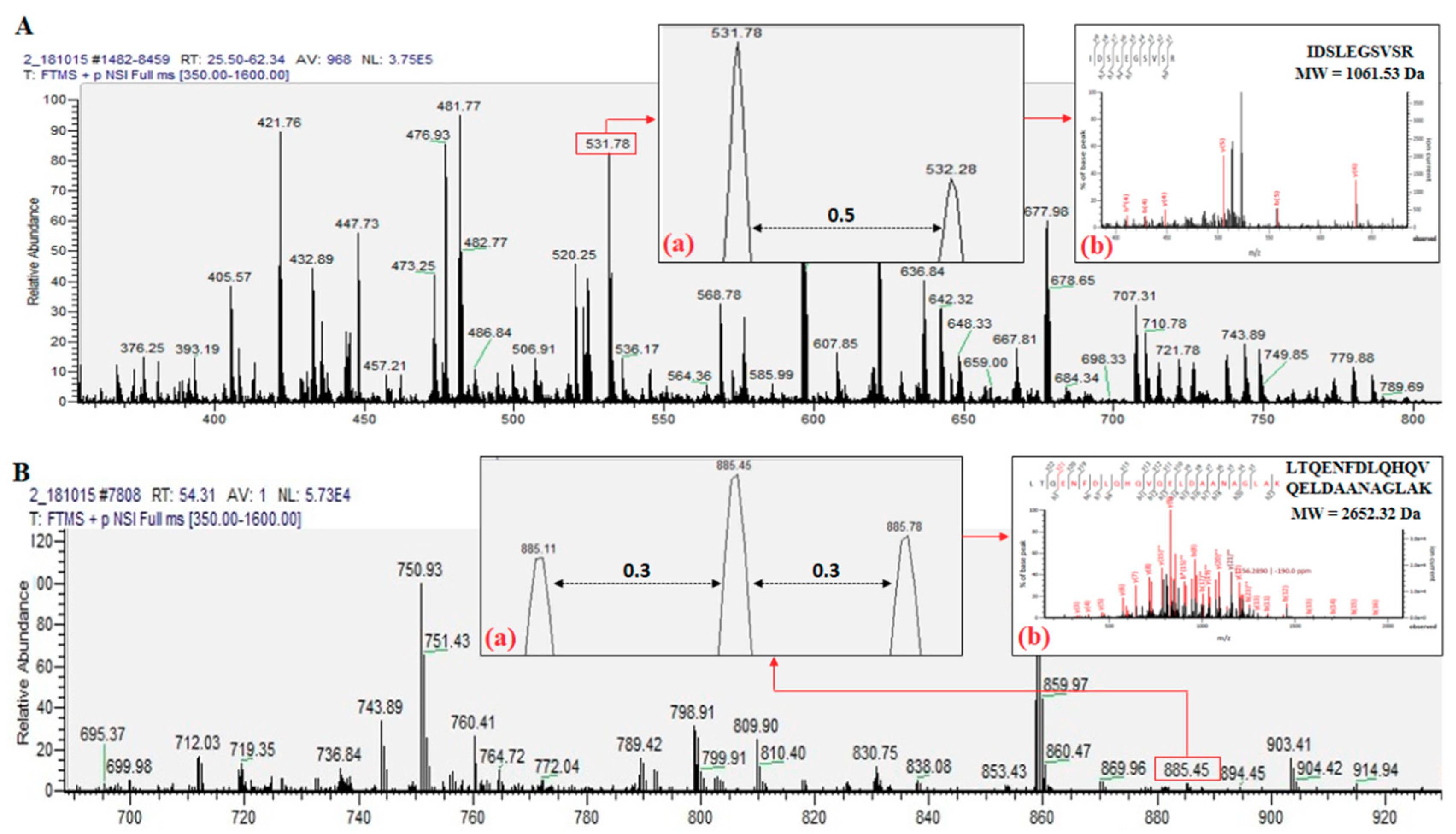
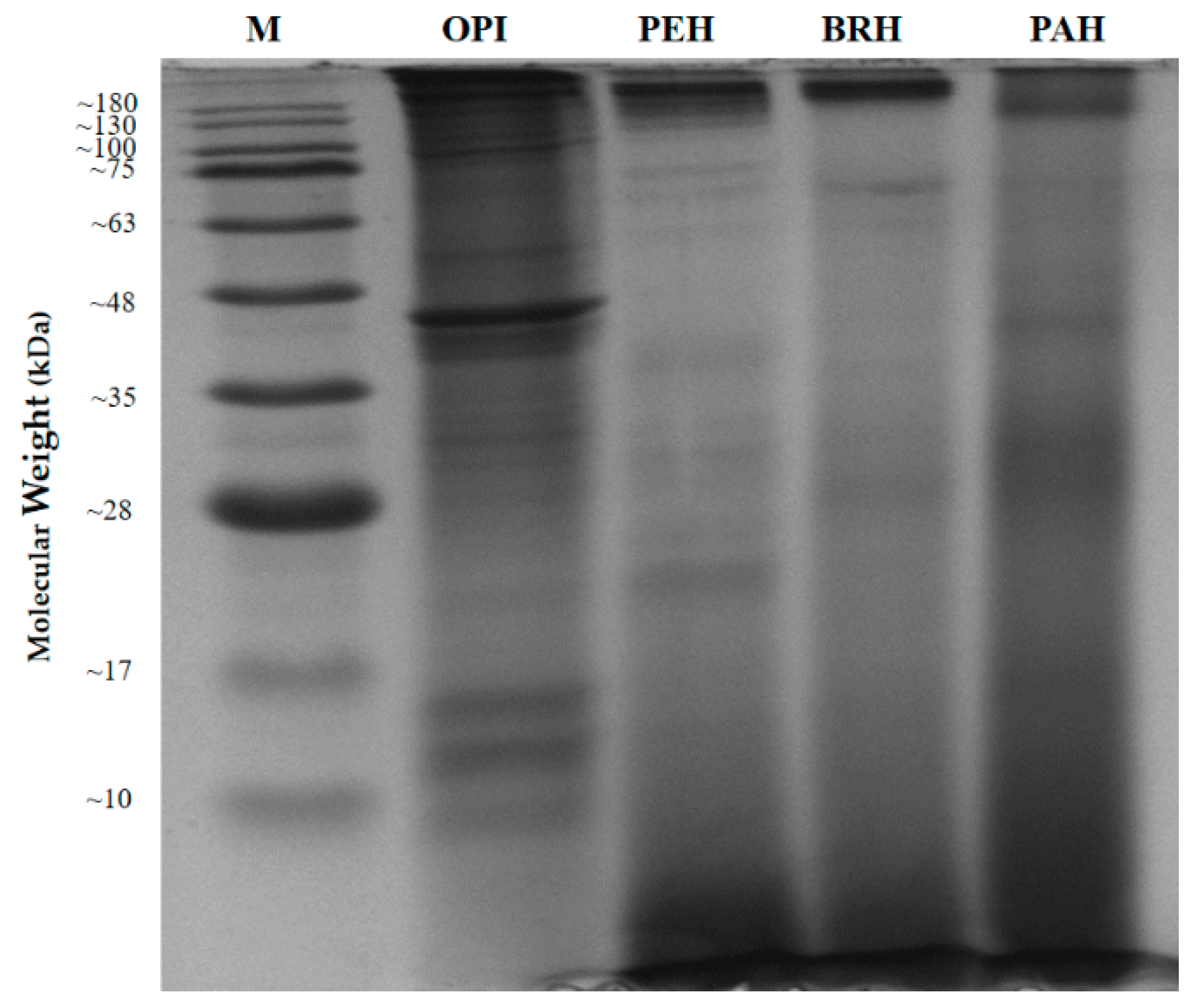
| Protein Name | Accession Number (NCBI) | Protein Score | Sequence Coverage (%) | Amino Acid Length | Molecular Weights from NCBI Database (kDa) | Molecular Weights Estimated from SDS-PAGE (kDa) |
|---|---|---|---|---|---|---|
| Myosin heavy chain, striated muscle isoform X1 * | XP_011442515.1 | 4670 | 48% | 1901 | 222.66 | 130.00 (A) |
| 82.50 (B) | ||||||
| Paramyosin isoform X2 * | XP_011429256.1 | 2736 | 65% | 851 | 102.21 | 82.50 (B) |
| Myosin heavy chain, striated muscle | EKC37566.1 | 789 | 20% | 2001 | 229.67 | 55.14 (C) |
| 44.55 (D) | ||||||
| 40.75 (E) | ||||||
| 11.66 (F) | ||||||
| 9.15 (G) | ||||||
| 5.76 (H) | ||||||
| Actin * | EKC30049.1 | 1272 | 58% | 351 | 41.80 | 44.55 (D) |
| Tropomyosin isoform X1 * | XP_019925727.1 | 579 | 70% | 251 | 33.02 | 40.75 (E) |
| Hypothetical protein CGI_10010027 | EKC40031.1 | 220 | 27% | 151 | 20.58 | 11.66 (F) |
| Myosin regulatory light chain B, smooth adductor muscle isoform X2 * | XP_011417566.1 | 112 | 50% | 151 | 18.63 | 9.15 (G) |
| 11.66 (F) | ||||||
| 5.76 (H) |
| Protein Name | Number of Potential Bioactive Peptides | ||
|---|---|---|---|
| ACE Inhibitor | DPP-IV Inhibitor | Other Activities * | |
| myosin heavy chain, striated muscle isoform X1 | 721 | 1147 | 395 |
| paramyosin isoform X2 | 294 | 517 | 147 |
| actin | 196 | 246 | 69 |
| tropomyosin isoform X1 | 97 | 165 | 56 |
| myosin regulatory light chain B, smooth adductor muscle isoform X2 | 83 | 104 | 39 |
| Total | 1391 | 2179 | 706 |
| Hydrolysate | Hydrolysis Conditions | Maximum DH (%) | Yield * (%) | Peptide Content (mg/mL) | |||
|---|---|---|---|---|---|---|---|
| E/S Ratio | Time (h) | pH | Temp. (°C) | ||||
| PEH | 1:100 | 4 | 2 | 37 | 22.20 ± 0.97 a | 84.69 | 2.42 ± 0.06 a |
| BRH | 1:100 | 4 | 7 | 50 | 17.86 ± 0.08 b | 35.14 | 1.53 ± 0.03 b |
| PAH | 1:100 | 4 | 7 | 65 | 18.57 ± 0.61 b | 27.79 | 2.28 ± 0.03 a |
| Fractions | Peptide Content (mg/mL) | Yield * (%) | Inhibition Efficiency Ratio a (%/mg/mL) | |
|---|---|---|---|---|
| DPP-IV Inhibitory Peptides | ACE Inhibitory Peptides | |||
| PEH | 2.42 ± 0.06 a | 84.69 | 16.86 | 24.92 |
| F1 | 0.32 ± 0.06 c | 10.95 | 153.00 | 217.05 |
| F2 | 0.45 ± 0.02 c | 2.05 | 123.26 | 147.60 |
| F3 | 1.73 ± 0.00 b | 54.74 | 27.11 | 29.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez, H.L.R.; Peralta, J.P.; Tejano, L.A.; Chang, Y.-W. In Silico and In Vitro Assessment of Portuguese Oyster (Crassostrea angulata) Proteins as Precursor of Bioactive Peptides. Int. J. Mol. Sci. 2019, 20, 5191. https://doi.org/10.3390/ijms20205191
Gomez HLR, Peralta JP, Tejano LA, Chang Y-W. In Silico and In Vitro Assessment of Portuguese Oyster (Crassostrea angulata) Proteins as Precursor of Bioactive Peptides. International Journal of Molecular Sciences. 2019; 20(20):5191. https://doi.org/10.3390/ijms20205191
Chicago/Turabian StyleGomez, Honey Lyn R., Jose P. Peralta, Lhumen A. Tejano, and Yu-Wei Chang. 2019. "In Silico and In Vitro Assessment of Portuguese Oyster (Crassostrea angulata) Proteins as Precursor of Bioactive Peptides" International Journal of Molecular Sciences 20, no. 20: 5191. https://doi.org/10.3390/ijms20205191
APA StyleGomez, H. L. R., Peralta, J. P., Tejano, L. A., & Chang, Y.-W. (2019). In Silico and In Vitro Assessment of Portuguese Oyster (Crassostrea angulata) Proteins as Precursor of Bioactive Peptides. International Journal of Molecular Sciences, 20(20), 5191. https://doi.org/10.3390/ijms20205191