Obtaining Genome Sequences of Mutualistic Bacteria in Single Microcystis Colonies
Abstract
:1. Introduction
2. Results
2.1. Binning of Single Microcystis Colony Metagenomes
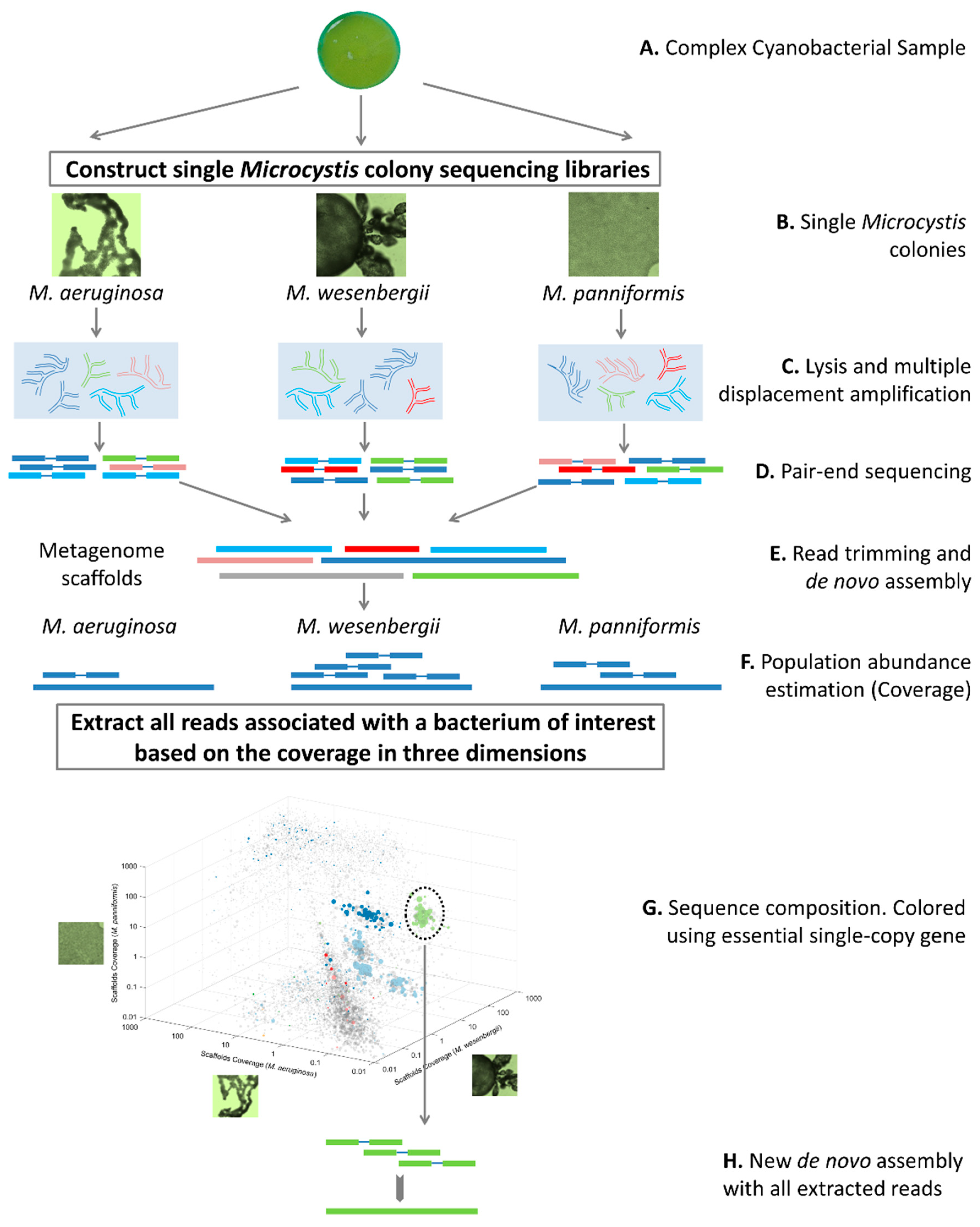
2.2. Pipeline of Obtaining Genomes in Single Microcystis Colonies
2.3. Colony Classification and Sample Preparation
2.4. Mapping Results
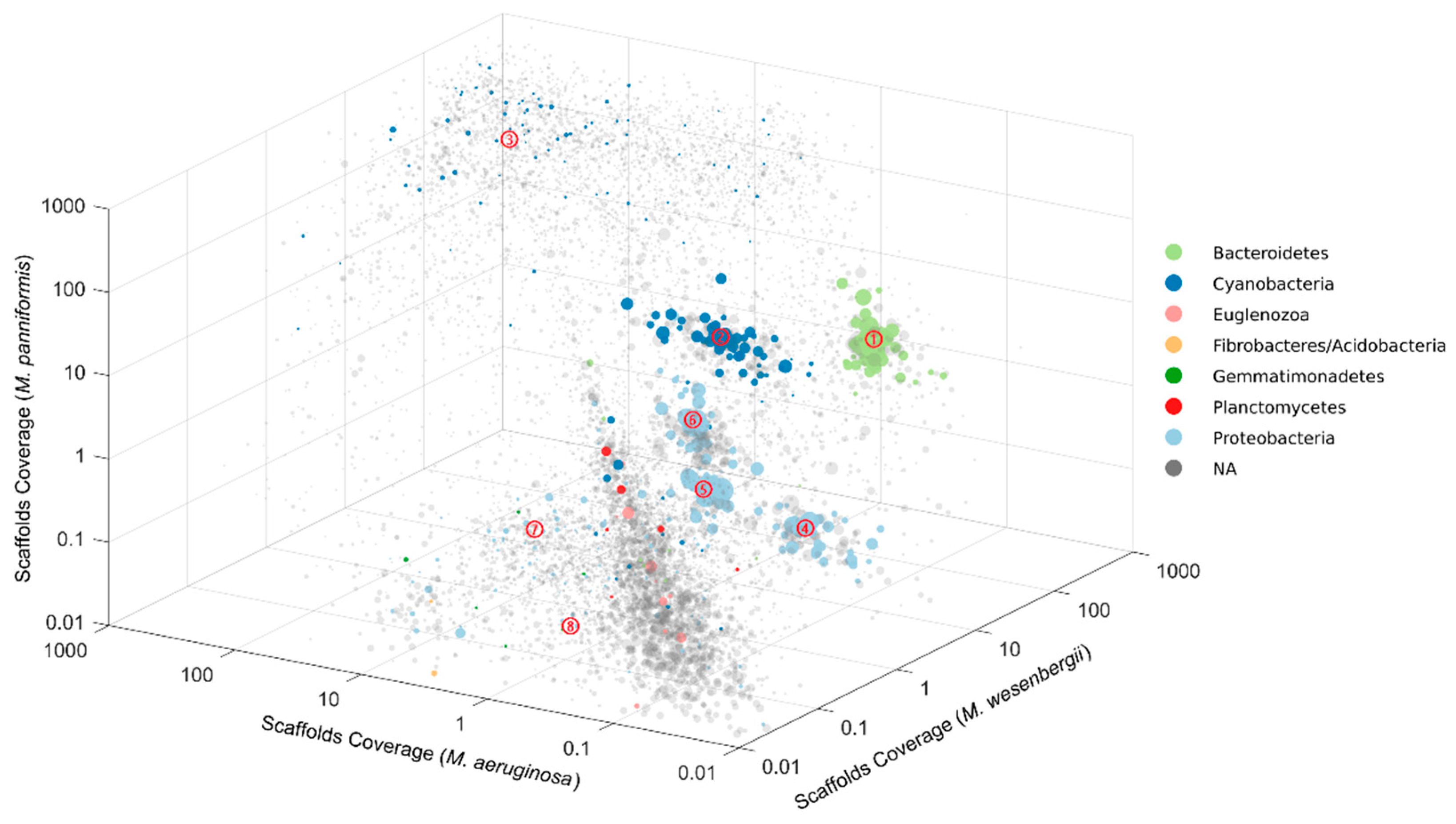
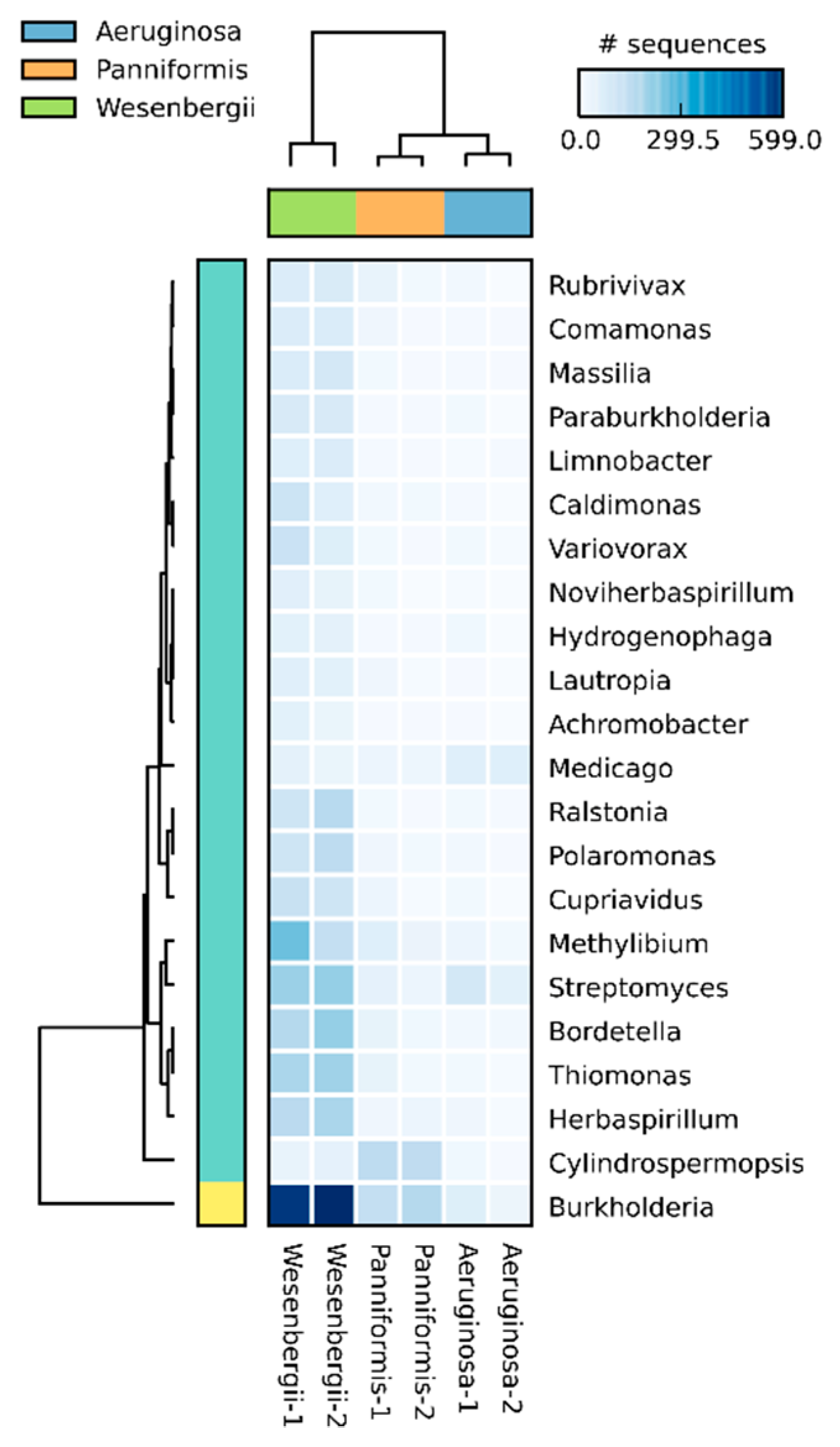
2.5. Clustering Results
2.6. Assembly and Reads Binning
3. Discussion
4. Material and Methods
4.1. Sample Collection, Colony Classification, and Observation
4.2. Colony Lysis and DNA Amplification
4.3. Library Construction and Sequencing
4.4. Read Trimming, Alignment
4.5. Taxonomic Analysis and Clustering
4.6. De Novo Metagenome Assembly and Scaffold Coverage
4.7. Conserved Marker Genes Identification
4.8. Sequence Binning
4.9. Re-Assembly
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDA | multiple displacement amplification |
| 3D | three-dimension |
| SEM | scanning electron microscope |
| µcMDA | micro-channel MDA |
| PBS | phosphate-buffered saline |
| NCBI | National Center for Biotechnology Information |
References
- Blaha, L.; Babica, P.; Marsalek, B. Toxins produced in cyanobacterial water blooms-toxicity and risks. Interdiscip. Toxicol. 2009, 2, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Pope, P.B.; Patel, B.K. Metagenomic analysis of a freshwater toxic cyanobacteria bloom. FEMS Microbiol. Ecol. 2008, 64, 9–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, T.; Nakajima, N.; Okamoto, S.; Suzuki, I.; Tanabe, Y.; Tamaoki, M.; Nakamura, Y.; Kasai, F.; Watanabe, A.; Kawashima, K.; et al. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa nies-843. DNA Res. 2007, 14, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.R. Some aspects of the ecophysiology of cyanobacteria. Ann. Microbiol. 1983, 134, 61–72. [Google Scholar] [CrossRef]
- Frangeul, L.; Quillardet, P.; Castets, A.M.; Humbert, J.F.; Matthijs, H.C.; Cortez, D.; Tolonen, A.; Zhang, C.C.; Gribaldo, S.; Kehr, J.C.; et al. Highly plastic genome of Microcystis aeruginosa pcc 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genom. 2008, 9, 274. [Google Scholar] [CrossRef]
- Zhu, W.; Li, M.; Luo, Y.G.; Dai, X.X.; Guo, L.L.; Xiao, M.; Huang, J.Y.; Tan, X. Vertical distribution of Microcystis colony size in lake taihu: Its role in algal blooms. J. Gt. Lakes Res. 2014, 40, 949–955. [Google Scholar] [CrossRef]
- Shen, H.; Niu, Y.; Xie, P.; Tao, M.; Yang, X. Morphological and physiological changes in Microcystis aeruginosa as a result of interactions with heterotrophic bacteria. Freshw. Biol. 2011, 56, 1065–1080. [Google Scholar] [CrossRef]
- Hugenholtz, P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002, 3, reviews0003.0001-0003.0008. [Google Scholar] [CrossRef]
- Kunin, V.; Copeland, A.; Lapidus, A.; Mavromatis, K.; Hugenholtz, P. A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008, 72, 557–578. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.W.; Jarrod, C.; Philip, H.; Allen, E.E.; Ram, R.J.; Richardson, P.M.; Solovyev, V.V.; Rubin, E.M.; Rokhsar, D.S.; Banfield, J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004, 428, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Héctor, G.M.; Natalia, I.; Victor, K.; Falk, W.; Barry, K.W.; Mchardy, A.C.; Christine, Y.; Shaomei, H.; Salamov, A.A.; Ernest, S. Metagenomic analysis of two enhanced biological phosphorus removal (ebpr) sludge communities. Nat. Biotechnol. 2006, 24, 1263–1269. [Google Scholar]
- Mande, S.S.; Monzoorul Haque, M.; Tarini Shankar, G. Classification of metagenomic sequences: Methods and challenges. Brief Bioinform. 2012, 13, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A.; Heinrich, F.; Bertilsson, S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012, 6, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Steffen, M.M.; Li, Z.; Effler, T.C.; Hauser, L.J.; Boyer, G.L.; Wilhelm, S.W. Comparative metagenomics of toxic freshwater cyanobacteria bloom communities on two continents. PloS ONE 2012, 7, e44002. [Google Scholar] [CrossRef] [PubMed]
- Mou, X.Z.; Jacob, J.S.; Lu, X.X.; Robbins, S.; Sun, S.L.; Ortiz, J.D. Diversity and distribution of free-living and particle-associated bacterioplankton in sandusky bay and adjacent waters of lake erie western basin. J. Gt. Lakes Res. 2013, 39, 352–357. [Google Scholar] [CrossRef]
- Kurmayer, R.; Dittmann, E.; Fastner, J.; Chorus, I. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. In lake wannsee (berlin, germany). Microb Ecol. 2002, 43, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Via-Ordorika, L.; Fastner, J.; Kurmayer, R.; Hisbergues, M.; Dittmann, E.; Komarek, J.; Erhard, M.; Chorus, I. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp. in european freshwater bodies: Detection of microcystins and microcystin genes in individual colonies. Syst. Appl. Microbiol. 2004, 27, 592–602. [Google Scholar] [CrossRef]
- Lara, Y.; Lambion, A.; Menzel, D.; Codd, G.A.; Wilmotte, A. A cultivation-independent approach for the genetic and cyanotoxin characterization of colonial cyanobacteria. Aquat. Microb. Ecol. 2013, 69, 135–143. [Google Scholar] [CrossRef]
- Otsuka, S.; Suda, S.; Li, R.; Matsumoto, S.; Watanabe, M.M. Morphological variability of colonies of Microcystis morphospecies in culture. J. Gen. Appl. Microbiol. 2000, 46, 39–50. [Google Scholar] [CrossRef]
- Doers, M.P.; Parker, D.L. Properties of Microcystis-Aeruginosa and m flos-aquae (cyanophyta) in culture: Taxonomic implications. J. Phycol. 1988, 24, 502–508. [Google Scholar]
- Parker, D.L. Improved procedures for the cloning and purification of Microcystis-cultures (cyanophyta). J. Phycol. 1982, 18, 471–477. [Google Scholar] [CrossRef]
- Parker, D.L.; Mihalick, J.E.; Plude, J.L.; Plude, M.J.; Clark, T.P.; Egan, L.; Flom, J.J.; Rai, L.C.; Kumar, H.D. Sorption of metals by extracellular polymers from the cyanobacterium Microcystis aeruginosa f. Flos-aquae strain c3-40. J. Appl. Phycol. 2000, 12, 219–224. [Google Scholar] [CrossRef]
- Plude, J.L.; Parker, D.L.; Schommer, O.J.; Timmerman, R.J.; Hagstrom, S.A.; Joers, J.M.; Hnasko, R. Chemical characterization of polysaccharide from the slime layer of the cyanobacterium Microcystis-flos-aquae-c3-40. Appl. Environ. Microbiol. 1991, 57, 1696–1700. [Google Scholar] [PubMed]
- Kurmayer, R.; Christiansen, G.; Chorus, I. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis sp. and determines its microcystin net production in lake wannsee. Appl. Environ. Microbiol. 2003, 69, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Rohrlack, T.; Henning, M.; Kohl, J.G. Mechanisms of the inhibitory effect of the cyanobacterium Microcystis aeruginosa on daphnia galeata’s ingestion rate. J. Plankton Res. 1999, 21, 1489–1500. [Google Scholar] [CrossRef]
- Xie, M.; Ren, M.; Yang, C.; Yi, H.; Li, Z.; Li, T.; Zhao, J. Metagenomic analysis reveals symbiotic relationship among bacteria in Microcystis-dominated community. Front. Microbiol. 2016, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Hosono, S.; Fang, L.H.; Wu, X.H.; Faruqi, A.F.; Bray-Ward, P.; Sun, Z.Y.; Zong, Q.L.; Du, Y.F.; Du, J.; et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 2002, 99, 5261–5266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosono, S.; Faruqi, A.F.; Dean, F.B.; Du, Y.F.; Sun, Z.Y.; Wu, X.H.; Du, J.; Kingsmore, S.F.; Egholm, M.; Lasken, R.S. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003, 13, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Guan, R.; Zhang, H.J.; Li, H.; Xiao, P.; Yu, G.L.; Du, L.; Cao, D.M.; Zhu, B.C.; Li, R.H.; et al. Complete genome sequence and genomic characterization of Microcystis panniformis fachb 1757 by third-generation sequencing. Stand. Genom. Sci. 2016, 11, 11. [Google Scholar] [CrossRef]
- Berg, K.A.; Lyra, C.; Sivonen, K.; Paulin, L.; Suomalainen, S.; Tuomi, P.; Rapala, J. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 2009, 3, 314–325. [Google Scholar] [CrossRef]
- Cornet, L.; Meunier, L.; Van Vlierberghe, M.; Leonard, R.R.; Durieu, B.; Lara, Y.; Misztak, A.; Sirjacobs, D.; Javaux, E.J.; Philippe, H.; et al. Consensus assessment of the contamination level of publicly available cyanobacterial genomes. PloS ONE 2018, 13, e0200323. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-J.; Alkawally, M.; Brady, A.L.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Dunfield, P.F. Chryseolinea serpens gen. Nov., sp. Nov., a member of the phylum bacteroidetes isolated from soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 654–660. [Google Scholar]
- Shi, L.M.; Huang, Y.X.; Zhang, M.; Shi, X.L.; Cai, Y.F.; Gao, S.L.; Tang, X.M.; Chen, F.Z.; Lu, Y.P.; Kong, F.X. Large buoyant particles dominated by cyanobacterial colonies harbor distinct bacterial communities from small suspended particles and free-living bacteria in the water column. Microbiologyopen 2018, 7, e00608. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Xiao, H.J.; Qiang, W.U.; Wang, S.J.; Pengfu, L.I. Characterization of epiphytic bacteria associated with colonial Microcystis. J. Lake Sci. 2017, 29, 617–624. [Google Scholar]
- Yamamoto, Y.; Niizuma, S.; Kuroda, N.; Sakamoto, M. Occurrence of heterotrophic bacteria causing lysis of cyanobacteria in a eutrophic lake. JPN J. Phycol. 1993, 41, 46–48. [Google Scholar]
- Sedmak, B.; Kosi, G. Microcystins in slovene freshwaters (central europe)—First report. Nat. Toxins 1997, 5, 64–73. [Google Scholar] [CrossRef]
- Vasconcelos, V.; Morais, J.; Vale, M. Microcystins and cyanobacteria trends in a 14 year monitoring of a temperate eutrophic reservoir (aguieira, portugal). J. Environ. Monit. 2011, 13, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Yarmoshenko, L.P.; Kureyshevich, A.V.; Yakushin, V.M. Microcystis botrys and lemmermanniella flexa—New species of cyanoprokaryota for the flora of ukraine in phytoplankton of the kanev reservoir. Hydrobiol. J. 2013, 49, 39–44. [Google Scholar] [CrossRef]
- Agha, R.; del Mar Labrador, M.; de los Ríos, A.; Quesada, A. Selectivity and detrimental effects of epiphytic pseudanabaena on Microcystis colonies. Hydrobiologia 2016, 777, 139–148. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Q.; Simon, P.N.; Liu, J.; Liu, L.; Dai, X.; Zhang, X.; Kuang, J.; Igarashi, Y.; Pan, X.; et al. Distinct network interactions in particle-associated and free-living bacterial communities during a Microcystis aeruginosa bloom in a plateau lake. Front. Microbiol. 2017, 8, 1202. [Google Scholar] [CrossRef]
- Shi, L.M.; Huang, Y.X.; Zhang, M.; Yu, Y.; Lu, Y.P.; Kong, F.X. Bacterial community dynamics and functional variation during the long-term decomposition of cyanobacterial blooms in-vitro. Sci. Total Environ. 2017, 598, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jia, J.; Chen, S.; Huang, T.; Wang, Y.; Zhao, Z.; Feng, J.; Hao, H.; Li, S.; Ma, X. Dynamics of bacterial and fungal communities during the outbreak and decline of an algal bloom in a drinking water reservoir. Int. J. Environ. Res. Public Health 2018, 15, 361. [Google Scholar] [CrossRef] [PubMed]
- Coman, C.; Bica, A.; Druga, B.; Barbu-Tudoran, L.; Dragos, N. Methodological constraints in the molecular biodiversity study of a thermomineral spring cyanobacterial mat: A case study. Antonie Van Leeuwenhoek 2011, 99, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Cai, Y.; Kong, F.; Yu, Y. Specific association between bacteria and buoyant Microcystis colonies compared with other bulk bacterial communities in the eutrophic lake taihu, china. Environ. Microbiol. Rep. 2012, 4, 669–678. [Google Scholar]
- Spits, C.; Le Caignec, C.; de Rycke, M.; Van Haute, L.; van Steirteghem, A.; Liebaers, I.; Sermon, K. Optimization and evaluation of single-cell whole, genome multiple displacement amplification. Hum. Mutat. 2006, 27, 496–503. [Google Scholar] [CrossRef]
- Pinard, R.; de Winter, A.; Sarkis, G.J.; Gerstein, M.B.; Tartaro, K.R.; Plant, R.N.; Egholm, M.; Rothberg, J.M.; Leamon, J.H. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genom. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Shi, X.L.; Qiao, Y.; Chen, L.; Duan, M.Q.; Hou, Y.; Ge, Q.Y.; Tao, Y.H.; Tu, J.; Lu, Z.H. 1d-reactor decentralized mda for uniform and accurate whole genome amplification. Anal. Chem. 2017, 89, 10147–10152. [Google Scholar] [CrossRef]
- Li, J.J.; Lu, N.; Tao, Y.H.; Duan, M.Q.; Qiao, Y.; Xu, Y.M.; Ge, Q.Y.; Bi, C.W.; Fu, J.Y.; Tu, J.; et al. Accurate and sensitive single-cell-level detection of copy number variations by micro-channel multiple displacement amplification (mu cmda). Nanoscale 2018, 10, 17933–17941. [Google Scholar] [CrossRef]
- Sidore, A.M.; Lan, F.; Lim, S.W.; Abate, A.R. Enhanced sequencing coverage with digital droplet multiple displacement amplification. Nucleic Acids Res. 2016, 44, 66. [Google Scholar] [CrossRef]
- Rhee, M.; Light, Y.K.; Meagher, R.J.; Singh, A.K. Digital droplet multiple displacement amplification (ddmda) for whole genome sequencing of limited DNA samples. PloS ONE 2016, 11, e0153699. [Google Scholar] [CrossRef]
- Brinig, M.M.; Lepp, P.W.; Ouverney, C.C.; Armitage, G.C.; Relman, D.A. Prevalence of bacteria of division tm7 in human subgingival plaque and their association with disease. Appl. Environ. Microbiol. 2003, 69, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Marcy, Y.; Ouverney, C.; Bik, E.M.; Losekann, T.; Ivanova, N.; Martin, H.G.; Szeto, E.; Platt, D.; Hugenholtz, P.; Relman, D.A.; et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated tm7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 2007, 104, 11889–11894. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, M.; Hugenholtz, P.; Skarshewski, A.; Nielsen, K.L.; Tyson, G.W.; Nielsen, P.H. Genome sequences of rare, uncultured bacteria obtained by differential-coverage binning of multiple metagenomes. Nat. Biotechnol. 2013, 31, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Sharon, I.; Morowitz, M.J.; Thomas, B.C.; Costello, E.K.; Relman, D.A.; Banfield, J.F. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013, 23, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Song, L.; Li, R. Taxonomic notes on water bloom forming Microcystis species (cyanophyta) from china—An example from samples of the dianchi lake. Acta Phytotaxon. Sin. 2007, 45, 727–741. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. Ncbi reference sequences (refseq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, 61–65. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using diamond. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Gorska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. Megan community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. Stamp: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; LoCascio, P.F.; Hauser, L.J.; Uberbacher, E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 2012, 28, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.L.; Rusch, D.B.; Yooseph, S.; Lombardo, M.J.; Richter, R.A.; Valas, R.; Novotny, M.; Yee-Greenbaum, J.; Selengut, J.D.; Haft, D.H.; et al. Genomic insights to sar86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012, 6, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. Checkm: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]

| Aeruginosa-1 | Aeruginosa-2 | Wesenbergii-1 | Wesenbergii-2 | Panniformis-1 | Panniformis-2 | |
|---|---|---|---|---|---|---|
| Total reads (million) | 94.68 | 77.94 | 45.23 | 165.66 | 106.81 | 91.77 |
| Coverage Ratio of NIES-843* | 67.87% | 69.71% | 83.83% | 78.41% | 80.56% | 75.70% |
| Coverage Ratio of FACHB-1757 * | 68.20% | 70.35% | 87.09% | 94.30% | 99.89% | 95.32% |
| Figure ID | No. Contigs | Total Length (bp) | GC (%) | No. Essential Genes | No. Duplicated Essential Genes | Standardized Relative Abundance* (%) | Genus | Phylogenetic Affiliation | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aeruginosa | wesenbergii | panniformis | Overall | ||||||||
| 1 | 149 | 3,487,294 | 50.16 | 101/105 | 1 | 0.004 | 2.250 | 0.116 | 1.004 | Chryseolinea | Bacteroidetes |
| 2 | 309 | 4,301,792 | 42.03 | 102/106 | 5 | 0.190 | 0.528 | 0.095 | 0.443 | Pseudanabaena | Cyanobacteriaidetes |
| 3 | 3434 | 10,835,247 | 43.20 | 81/106 | 40 | 9.356 | 23.222 | 8.004 | 15.344 | Microcystis | Cyanobacteroidetes |
| 4 | 160 | 2,863,377 | 67.60 | 98/106 | 5 | 0.003 | 0.567 | 0.064 | 0.376 | Brevundimonas | Proteobacteria |
| 5 | 39 | 1,954,923 | 54.00 | 102/106 | 1 | 0.007 | 0.435 | 0.004 | 0.126 | Cupriavidus | Proteobacteria |
| 6 | 219 | 3,678,702 | 68.52 | 103/106 | 2 | 0.040 | 0.774 | 0.094 | 0.842 | Burkholderia | Proteobacteria |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, J.; Chen, L.; Gao, S.; Zhang, J.; Bi, C.; Tao, Y.; Lu, N.; Lu, Z. Obtaining Genome Sequences of Mutualistic Bacteria in Single Microcystis Colonies. Int. J. Mol. Sci. 2019, 20, 5047. https://doi.org/10.3390/ijms20205047
Tu J, Chen L, Gao S, Zhang J, Bi C, Tao Y, Lu N, Lu Z. Obtaining Genome Sequences of Mutualistic Bacteria in Single Microcystis Colonies. International Journal of Molecular Sciences. 2019; 20(20):5047. https://doi.org/10.3390/ijms20205047
Chicago/Turabian StyleTu, Jing, Liang Chen, Shen Gao, Junyi Zhang, Changwei Bi, Yuhan Tao, Na Lu, and Zuhong Lu. 2019. "Obtaining Genome Sequences of Mutualistic Bacteria in Single Microcystis Colonies" International Journal of Molecular Sciences 20, no. 20: 5047. https://doi.org/10.3390/ijms20205047
APA StyleTu, J., Chen, L., Gao, S., Zhang, J., Bi, C., Tao, Y., Lu, N., & Lu, Z. (2019). Obtaining Genome Sequences of Mutualistic Bacteria in Single Microcystis Colonies. International Journal of Molecular Sciences, 20(20), 5047. https://doi.org/10.3390/ijms20205047






