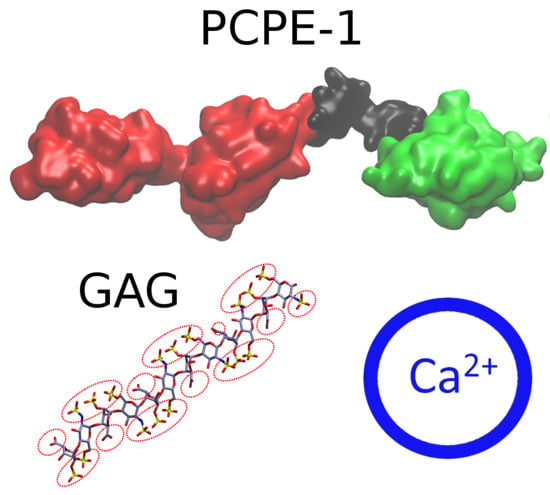
Analysis of Procollagen C-Proteinase Enhancer-1/Glycosaminoglycan Binding Sites and of the Potential Role of Calcium Ions in the Interaction
Abstract
1. Introduction
2. Results and Discussion
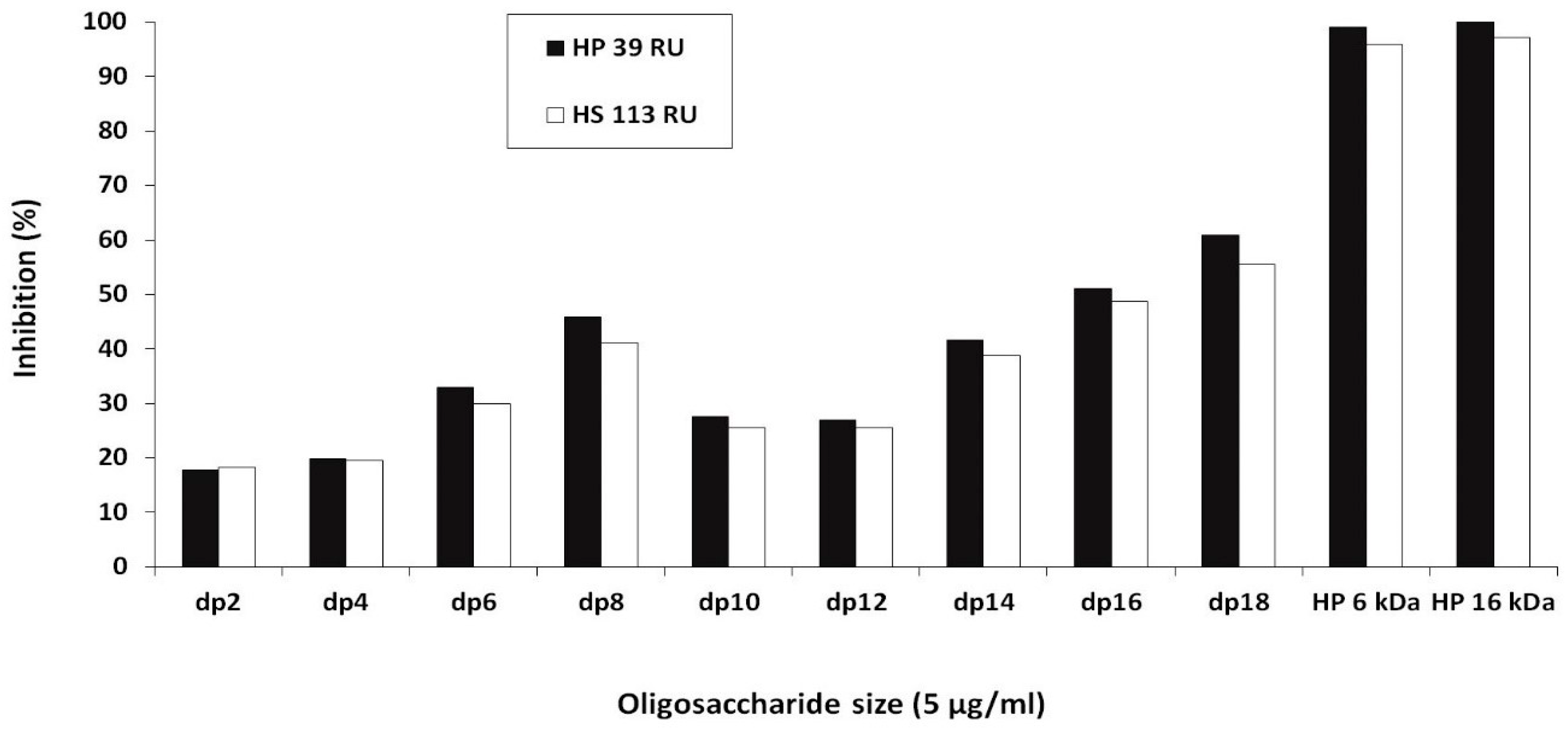
2.1. Experimental Results
2.2. Modeling the Full Structure of PCPE-1
2.3. PCPE-1 Interactions with Glycosaminoglycans
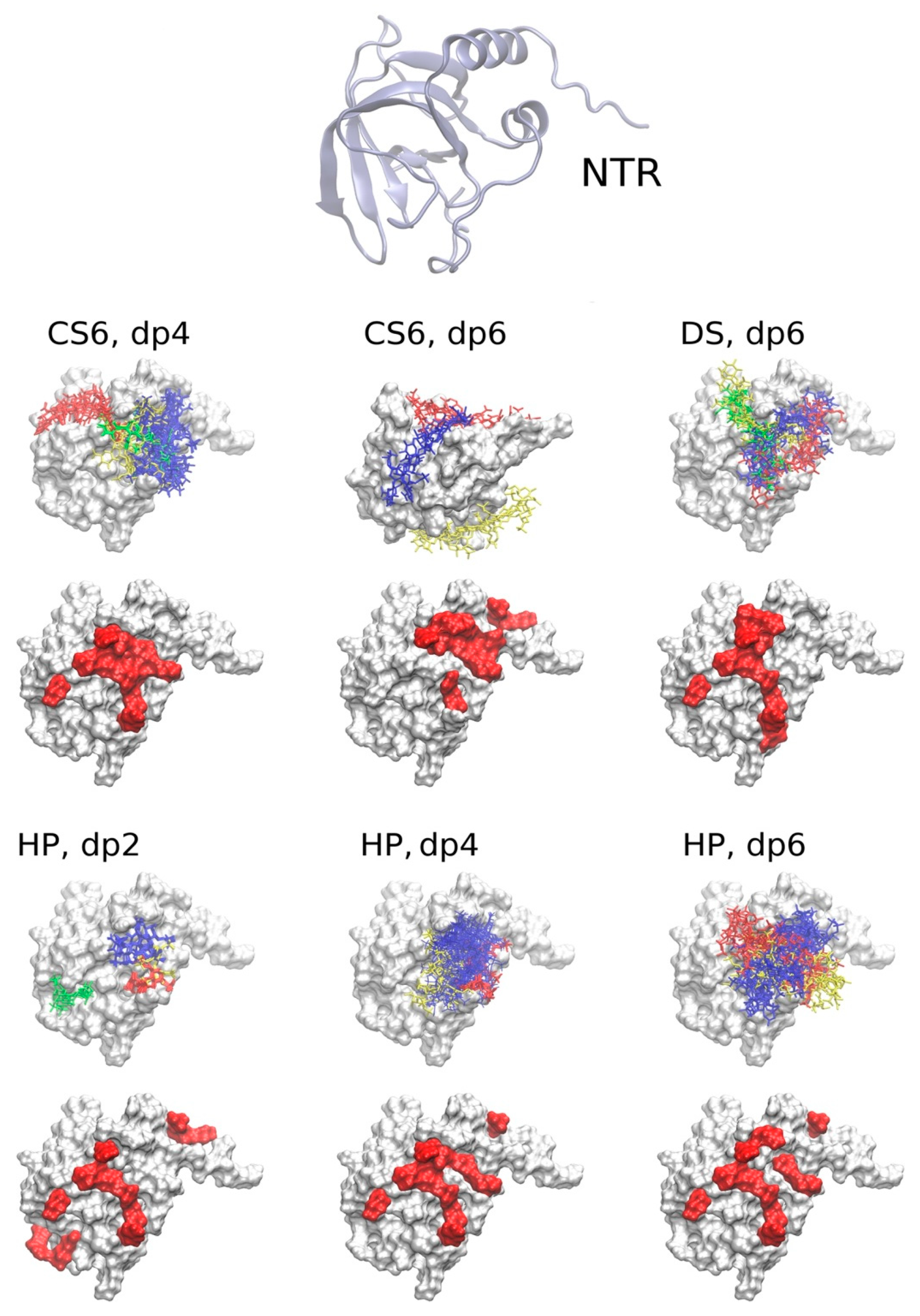
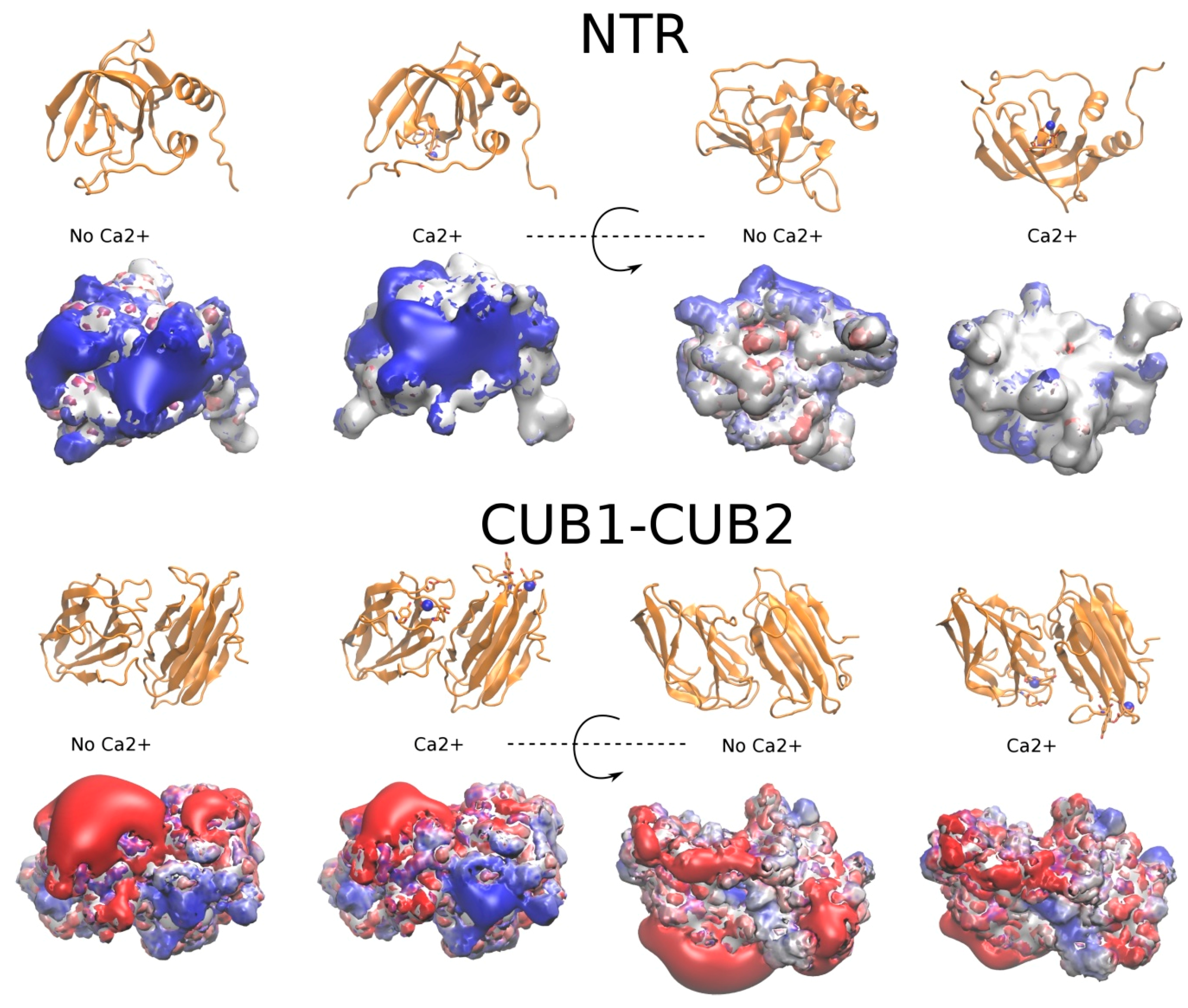
2.3.1. The NTR-Domain
2.3.2. CUB1-CUB2 Domains
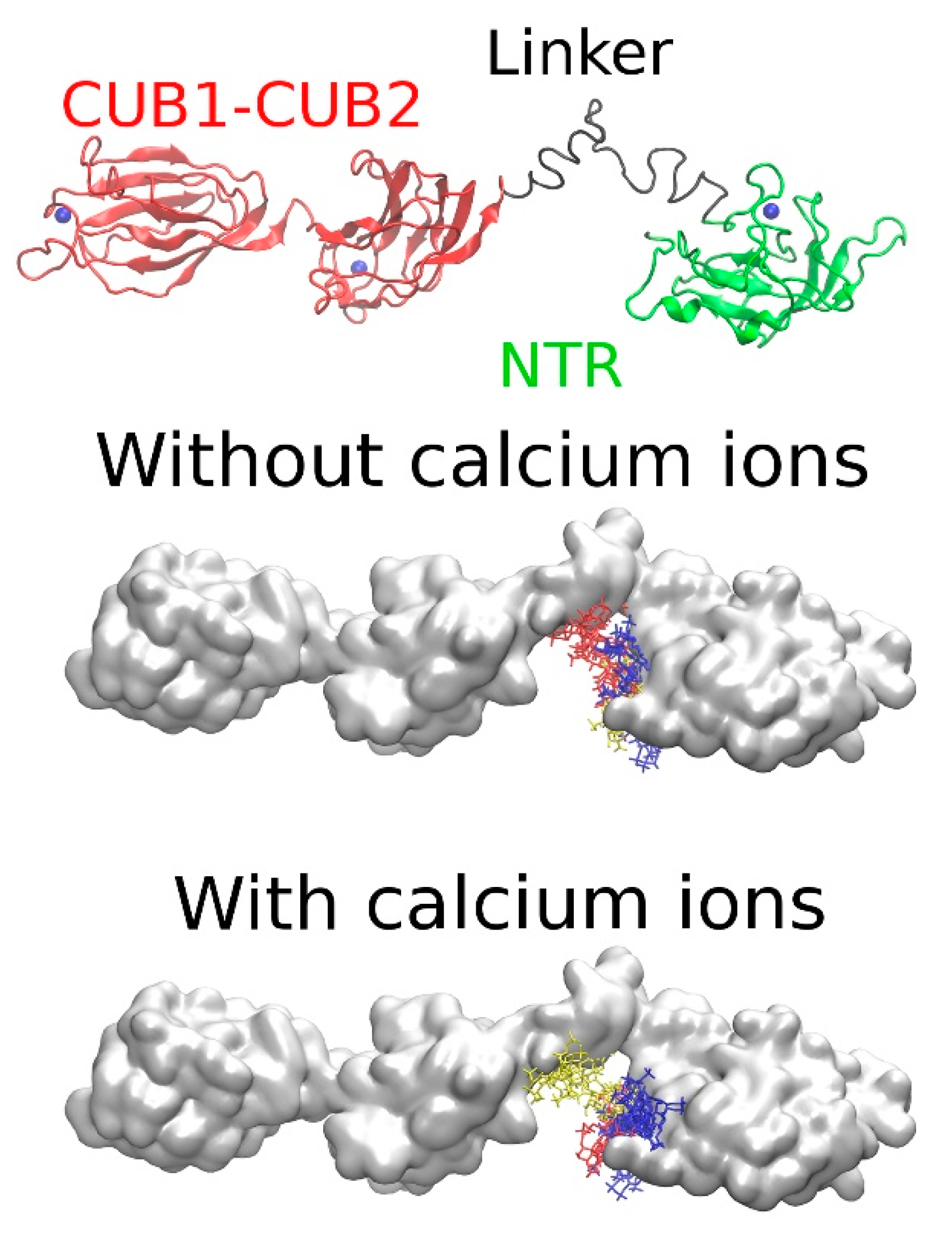
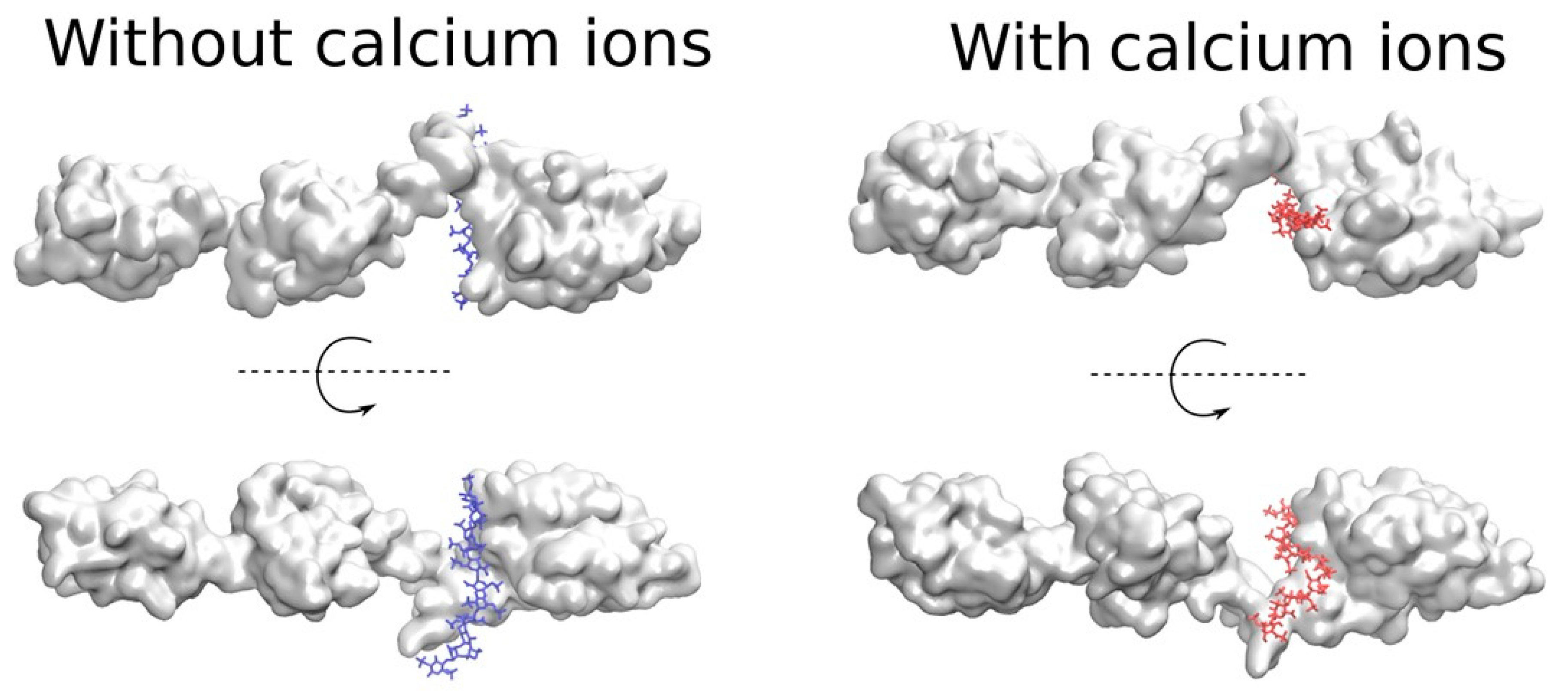
2.3.3. Full PCPE-1
2.4. The Potential Role of Ca2+ in PCPE-1 Interactions with Glycosaminoglycans
2.4.1. Prediction of Ca2+ Binding Sites
2.4.2. PCPE-1 Interactions with Glycosaminoglycans in the Presence of Ca2+ Ions
2.4.3. Predicting Longer GAG Binding Poses Using the Fragment-Based Approach
3. Materials and Methods
3.1. Surface Plasmon Resonance (SPR) Binding Assays
3.2. Structures
3.2.1. Protein Structures
3.2.2. Glycosaminoglycan Structures
3.3. Electrostatic Potential Calculations
3.4. Coarse-Grained MD Simulations
3.5. Molecular Docking
3.5.1. Autodock 3
3.5.2. Fragment-Based Approach
3.6. All-Atom MD Simulations and MM-GBSA Free Energy Calculations
3.7. Ca2+ Ion Position Prediction
3.7.1. Molecular Dynamics Approach
3.7.2. FoldX and IonCom
3.8. Visualization and Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD3 | Autodock 3 |
| BMP-1 | Bone morphogenetic protein-1 |
| CG | Coarse-grained |
| CS | Chondroitin sulfate |
| CUB | Complement, sea urchin protein Uegf, BMP-1 |
| DS | Dermatan sulfate |
| GAG | Glycosaminoglycan |
| HP | Heparin |
| HS | Heparan sulfate |
| MD | Molecular dynamics |
| MM-GBSA | Molecular mechanics-generalized born surface area |
| mTLD | Mammalian tolloid |
| NTR | Netrin-like domain |
| PBSA | Poisson‒Boltzmann surface area |
| PCPE-1 | Procollagen C-proteinase enhancer-1 |
| SAXS | Small angle X-ray scattering |
| UNRES | United residue force field |
References
- Esko, J.D.; Kimata, K.; Lindahl, U. Proteoglycans and Sulfated Glycosaminoglycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; pp. 1–784. [Google Scholar]
- Pomin, V.H.; Mulloy, B. Glycosaminoglycans and Proteoglycans. Pharmaceuticals 2018, 11, 27. [Google Scholar] [CrossRef]
- Proudfoot, A.E. Chemokines and Glycosaminoglycans. Front. Immunol. 2015, 6, 246. [Google Scholar] [CrossRef]
- Shute, J. Glycosaminoglycan and chemokine/growth factor interactions. Handb. Exp. Pharmacol. 2012, 207, 307–324. [Google Scholar]
- Iozzo, R.V.; Zoellerm, J.J.; Nyströmm, A. Basement membrane proteoglycans: Modulators Par Excellence of cancer growth and angiogenesis. Mol. Cells 2009, 27, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Almond, A. Multiscale modeling of glycosaminoglycan structure and dynamics: Current methods and challenges. Curr. Opin. Struct. Biol. 2018, 50, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Pichert, A.; Samsonov, S.A.; Theisgen, S.; Thomas, L.; Baumann, L.; Schiller, J.; Beck-Sickinger, A.G.; Huster, D.; Pisabarro, M.T. Characterization of the interaction of interleukin-8 with hyaluronan, chondroitin sulfate, dermatan sulfate and their sulfated derivatives by spectroscopy and molecular modeling. Glycobiology 2012, 22, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Penk, A.; Baumann, L.; Huster, D.; Samsonov, S.A. NMR and Molecular Modeling Reveal Specificity of the Interactions between CXCL14 and Glycosaminoglycans. Glycobiology 2019. [Google Scholar] [CrossRef] [PubMed]
- Bojarski, K.K.; Sieradzan, A.K.; Samsonov, S.A. Molecular Dynamics Insights into Protein-Glycosaminoglycan Systems from Microsecond-Scale Simulations. Biopolymers 2019. [Google Scholar] [CrossRef]
- Uciechowska-Kaczmarzyk, U.; Chauvot de Beauchene, I.; Samsonov, S.A. Docking software performance in protein-glycosaminoglycan systems. J. Mol. Graph. Mod. 2019, 90, 42–50. [Google Scholar] [CrossRef]
- Rother, S.; Samsonov, S.A.; Hofmann, T.; Blaszkiewicz, J.; Köhling, S.; Schnabelrauch, M.; Möller, S.; Rademann, J.; Kalkhof, S.; von Bergen, M.; et al. Structural and functional insights into the interaction of sulfated glycosaminoglycans with tissue inhibitor of metalloproteinase-3—A possible regulatory role on extracellular matrix homeostasis. Acta Biomater. 2016, 45, 143–154. [Google Scholar] [CrossRef]
- Vallet, S.D.; Miele, A.E.; Uciechowska-Kaczmarzyk, U.; Liwo, A.; Duclos, D.; Samsonov, S.A.; Ricard-Blum, S. Insights into the structure and dynamics of lysyl oxidase propeptide, a flexible protein with numerous partners. Sci. Rep. 2018, 8, 11768. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S.; Ruggiero, F.; van der Rest, M. The collagen superfamily. Top. Curr. Chem. 2005, 247, 35–84. [Google Scholar]
- Vadon-Le Goff, S.; Hulmes, D.J.S.; Moali, C. BMP-1/tolloid-like proteinases synchronize matrix assembly with growth factor activation to promote morphogenesis and tissue remodeling. Matrix Biol. 2015, 44–46, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Takahara, K.; Lyons, G.E.; Greenspan, D.S. Bone morphogenetic protein-1 and a mammalian tolloid homologue (mTld) are encoded by alternatively spliced transcripts which are differentially expressed in some tissues. J. Biol. Chem. 1994, 269, 32572–32578. [Google Scholar]
- Kessler, E.; Takahara, K.; Biniaminov, L.; Brusel, M.; Greenspan, D.S. Bone Morphogenetic Protein-1: The Type I Procollagen C-Proteinase. Science 1996, 271, 360–362. [Google Scholar] [CrossRef]
- Li, S.W.; Sieron, A.L.; Fertala, A.; Hojima, Y.; Arnold, W.V.; Prockop, D.J. The C-proteinase that processes procollagens to fibrillar collagens is identical to the protein previously identified as bone morphogenic protein-1. Proc. Natl. Acad. Sci. USA 1996, 93, 5127–5130. [Google Scholar] [CrossRef]
- Kessler, E.; Hassoun, E. Procollagen C-Proteinase Enhancer 1 (PCPE-1) in Liver Fibrosis. Methods Mol. Biol. 2019, 1944, 189–201. [Google Scholar]
- Hassoun, E.; Safrin, M.; Ziv, H.; Pri-Chen, S.; Kessler, E. Procollagen C-proteinase enhancer 1 (PCPE-1) as a plasma marker of muscle and liverf in mice. PLoS ONE 2016, 11, e0159606. [Google Scholar]
- Takahara, K.; Kessler, E.; Biniaminov, L.; Brusel, M.; Eddy, R.L.; Jani-Saitfl, S.; Shows, T.B.; Greenspan, D.S. Type I procollagen COOH-terminal proteinase enhancer protein: Identification, primary structure, and chromosomal localization of the cognate human gene (PCOLCE). J. Biol. Chem. 1994, 269, 26280–26285. [Google Scholar]
- Bányai, L.; Patthy, L. The NTR module: Domains of netrins, secreted frizzled related proteins, and type I procollagen C-proteinase enhancer protein are homologous with tissue inhibitors of metalloproteases. Protein Sci. 1999, 8, 1636–1642. [Google Scholar] [CrossRef]
- Liepinsh, E.; Banyai, L.; Pintacuda, G.; Trexler, M.; Patthy, L.; Otting, G. NMR structure of the netrin-like domain (NTR) of human type I procollagen C-proteinase enhancer defines structural consensus of NTR domains and assesses potential proteinase inhibitory activity and ligand binding. J. Biol. Chem. 2003, 278, 25982–25989. [Google Scholar] [CrossRef] [PubMed]
- Pulido, D.; Sharma, U.; Vadon-Le Goff, S.; Hussain, S.A.; Cordes, S.; Mariano, N.; Bettler, E.; Moali, C.; Aghajari, N.; Hohenester, E.; et al. Structural Basis for the Acceleration of Procollagen Processing by Procollagen C-Proteinase Enhancer-1. Structure 2018, 26, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Gaboriaud, C.; Gregory-Pauron, L.; Teillet, F.; Thielens, N.M.; Bally, I.; Arlaud, G.J. Structure and properties of the Ca2+-binding CUB domain, a widespread ligand-recognition unit involved in major biological functions. Biochem. J. 2011, 439, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Font, B.; Eichenberger, D.; Moreau, C.; Ricard-Blum, S.; Hulmes, D.J.S.; Moali, C. Insights into how CUB domains can exert specific functions while sharing a common fold. Conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J. Biol. Chem. 2007, 282, 16924–16933. [Google Scholar] [CrossRef]
- Bernocco, S.; Steiglitz, B.M.; Svergun, D.I.; Petoukhov, M.V.; Ruggiero, F.; Ricard-Blum, S.; Ebel, C.; Geourjon, C.; Deleage, G.; Font, B.; et al. Low resolution structure determination shows procollagen C-proteinase enhancer to be an elongated multidomain glycoprotein. J. Biol. Chem. 2003, 278, 7199–7205. [Google Scholar] [CrossRef]
- Moschcovich, L.; Bernocco, S.; Font, B.; Rivkin, H.; Eichenberger, D.; Chejanovsky, N.; Hulmes, D.J.; Kessler, E. Folding and activity of recombinant human procollagen C-proteinase enhancer. Eur. J. Biochem. 2001, 268, 2991–2996. [Google Scholar] [CrossRef]
- Weiss, T.; Ricard-Blum, S.; Moschcovich, L.; Wineman, E.; Mesilaty, S.; Kessler, E. Binding of procollagen C-proteinase enhancer-1 (PCPE-1) to heparin/heparan sulfate: Properties and role in PCPE-1 interaction with cells. J. Biol. Chem. 2010, 285, 33867–33874. [Google Scholar] [CrossRef]
- Bekhouche, M.; Kronenberg, D.; Vadon-Le Goff, S.; Bijakowski, C.; Lim, N.H.; Font, B.; Kessler, E.; Colige, A.; Nagase, H.; Murphy, G.; et al. Role of the netrin-like domain of procollagen C-proteinase enhancer-1 in the control of metalloproteinase activity. J. Biol. Chem. 2010, 285, 15950–15959. [Google Scholar] [CrossRef]
- Sankaranarayanan, N.V.; Nagarajan, B.; Desai, U.R. So you think computational approaches to understanding glycosaminoglycan-protein interactions are too dry and too rigid? Think again! Curr. Opin. Struct. Biol. 2018, 50, 91–100. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Pisabarro, M.T. Computational analysis of interactions in structurally available protein-glycosaminoglycan complexes. Glycobiology 2016, 26, 850–861. [Google Scholar] [CrossRef]
- Möbius, K.; Nordsieck, K.; Pichert, A.; Samsonov, S.A.; Thomas, L.; Schiller, J.; Kalkhof, S.; Pisabarro, M.T.; Beck-Sickinger, A.G.; Huster, D. Investigation of lysine side chain interactions of Interleukin-8 with Heparin and other glycosaminoglycans studied by a methylation-NMR approach. Glycobiology 2013, 23, 1260–1269. [Google Scholar] [CrossRef] [PubMed]
- Ester, M.; Kriegel, H.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Imberty, A.; Lortat-Jacob, H.; Pérez, S. Structural view of glycosaminoglycan-protein interactions. Carbohydr. Res. 2007, 342, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Capila, I.; Hernáiz, M.J.; Mo, Y.D.; Mealy, T.R.; Campos, B.; Dedman, J.R.; Linhardt, R.J.; Seaton, B.A. Annexin V—Heparin oligosaccharide complex suggests heparan sulfate—Mediated assembly on cell surfaces. Structure 2001, 9, 57–64. [Google Scholar] [CrossRef]
- Woodhead, N.E.; Long, W.F.; Willamson, F.B. Binding of zinc ions to heparin. Analysis by equilibrium dialysis suggests the occurrence of two, entropy-driven, processes. Biochem J. 1986, 237, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Stevic, I.; Parmar, N.; Paredes, N.; Berry, L.R.; Chan, A.K.C. Binding of heparin to metals. Cell Biochem. Biophys. 2011, 59, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Multhaup, G.; Bush, A.I.; Pollwein, P.; Masters, C.L. Binding of heparin to metals. FEBS Lett. 1994, 335, 151–154. [Google Scholar] [CrossRef]
- Eckert, R.; Ragg, H. Zinc ions promote the interaction between heparin and heparin cofactor II. FEBS Lett. 2003, 541, 121–125. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Féraud, O.; Lortat-Jacob, H.; Rencurosi, A.; Fukai, N.; Dkhissi, F.; Vittet, D.; Imberty, A.; Olsen, B.R.; van der Rest, M. Characterization of endostatin binding to heparin and heparan sulfate by surface plasmon resonance and molecular modeling: Role of divalent cations. J. Biol. Chem. 2004, 279, 2927–2936. [Google Scholar] [CrossRef]
- Han, Q.; Fu, Y.; Zhou, H.; He, Y.; Luo, Y. Contributions of Zn(II)-binding to the structural stability of endostatin. FEBS Lett. 2007, 581, 3027–3032. [Google Scholar] [CrossRef]
- Zhang, F.; Liang, X.; Beaudet, J.M.; Lee, Y.; Linhardt, R.J. The Effects of Metal Ions on Heparin/Heparin Sulfate-Protein Interactions. J. Biomed. Technol. Res. 2014, 1, 10. [Google Scholar] [CrossRef]
- Seo, Y.; Schenauer, M.R.; Leary, J.A. Biologically Relevant Metal-Cation Binding Induces Conformational Changes in Heparin Oligosaccharides as Measured by Ion Mobility Mass Spectrometry. Int. J. Mass Spectrom. 2011, 303, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Lerner, L.; Torchia, D.A. A multinuclear NMR study of the interactions of cations with proteoglycans, heparin, and Ficoll. J. Biol. Chem. 1986, 261, 12706–12714. [Google Scholar] [PubMed]
- Parrish, R.F.; Fair, W.R. Selective binding of zinc ions to heparin rather than to other glycosaminoglycans. Biochem. J. 1981, 193, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Grushka, E.; Cohen, A.S. The Binding of Cu(II) and Zn(II) Ions by Heparin. Anal. Lett. 1982, 15, 1277–1288. [Google Scholar] [CrossRef]
- Grant, D.; Long, W.F.; Williamson, F.B. Infrared spectroscopy of heparin-cation complexes. Biochem. J. 1987, 244, 143–149. [Google Scholar] [CrossRef]
- Sattelle, B.; Shakeri, J.; Almond, A. Does microsecond sugar ring flexing encode 3D-shape and bioactivity in the heparanome? Biomacromolecules 2013, 14, 1149–1159. [Google Scholar] [CrossRef]
- Samsonov, S.; Bichmann, L.; Pisabarro, M.T. Coarse-grained model of glycosaminglycans. J. Chem. Inf. Mod. 2015, 55, 114–124. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER16; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Rhee, Y.M.; Pande, V.S. Multiplexed-replica exchange molecular dynamics method for protein folding simulation. Biophys. J. 2003, 84, 775–786. [Google Scholar] [CrossRef]
- Hansmann, U.H.E.; Okamoto, Y. Comparative study of multicanonical and simulated annealing algorithms in the protein folding problem. Physica A 1994, 212, 415–437. [Google Scholar] [CrossRef]
- Liwo, A.; Baranowski, M.; Czaplewski, C.; Gołaś, E.; He, Y.; Jagieła, D.; Krupa, P.; Maciejczyk, M.; Makowski, M.; Mozolewska, M.A.; et al. A unified coarse-grained model of biological macromolecules based on mean-field multipole-multipole interactions. J. Mol. Model. 2014, 20, 2306. [Google Scholar] [CrossRef] [PubMed]
- Liwo, A.; Czaplewski, C.; Ołdziej, S.; Scheraga, H.A. Computational techniques for efficient conformational sampling of proteins. Curr. Opin. Struct. Biol. 2008, 18, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Karczyńska, A.S.; Mozolewska, M.A.; Krupa, P.; Giełdoń, A.; Liwo, A.; Czaplewski, C. Prediction of protein structure with the coarse-grained UNRES force field assisted by small X-ray scattering data and knowledge-based information. Proteins 2018, 86, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A.; Rosenberg, J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Murtagh, F.; Heck, A. Multivariate Data Analysis. In Astrophysics and Space Science Library; Kluwer Academic Publishers: Strasbourg, France, 1987; pp. 55–109. [Google Scholar]
- Rotkiewicz, P.; Skolnick, J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J. Comput. Chem. 2008, 29, 1460–1465. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarcklan algorithm an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Samsonov, S.; Gehrcke, J.-P.; Pisabarro, M.T. Flexibility and explicit solvent in molecular dynamics-based docking of protein-glycosaminoglycan systems. J. Chem. Inf. Mod. 2014, 54, 582–592. [Google Scholar] [CrossRef]
- Joseph, R.R.; Mosier, P.D.; Desai, U.R.; Rajarathnam, K. Solution NMR characterization of chemokine CXCL8/IL-8 monomer and dimer binding to glycosaminoglycans: Structural plasticity mediates differential binding interactions. Biochem. J. 2015, 472, 121–133. [Google Scholar] [CrossRef]
- Nivedha, A.K.; Makeneni, S.; Foley, B.L.; Tessier, M.B.; Woods, R.J. Importance of ligand conformational energies in carbohydrate docking: Sorting the wheat from the chaff. J. Comput. Chem. 2014, 35, 526–539. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Zacharias, M.; Chauvot de Beauchene, I. Modeling large protein-glycosaminoglycan complexes using a fragment-based approach. J. Comput. Chem. 2019, 40, 1429–1439. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 4, 622–655. [Google Scholar] [CrossRef] [PubMed]
- Onufriev, A.; Case, D.A.; Bashford, D. Effective Born radii in the generalized Born approximation: The importance of being perfect. J. Comput. Chem. 2002, 23, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson-Boltzmann Surface Area Method. Mol. Inf. 2012, 31, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.S.; Mancera, R.L. Free energy calculations of glycosaminoglycan-protein interactions. Glycobiology 2009, 19, 1103–1115. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Schymkowitz, J.W.; Rousseau, F.; Martins, I.C.; Ferkinghoff-Borg, J.; Stricher, F.; Serrano, L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. USA 2005, 102, 10147–10152. [Google Scholar] [CrossRef]
- Hu, X.; Dong, Q.; Yang, J.; Zhang, Y. Recognizing metal and acid radical ion binding sites by integrating ab initio modeling with template-based transferals. Bioinformatics 2016, 32, 3260–3269. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 1.2r3pre; Schrödinger, LLC: New York, NY, USA, 2002.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. Available online: http://www.R-project.org/ (accessed on 1 May 2017).
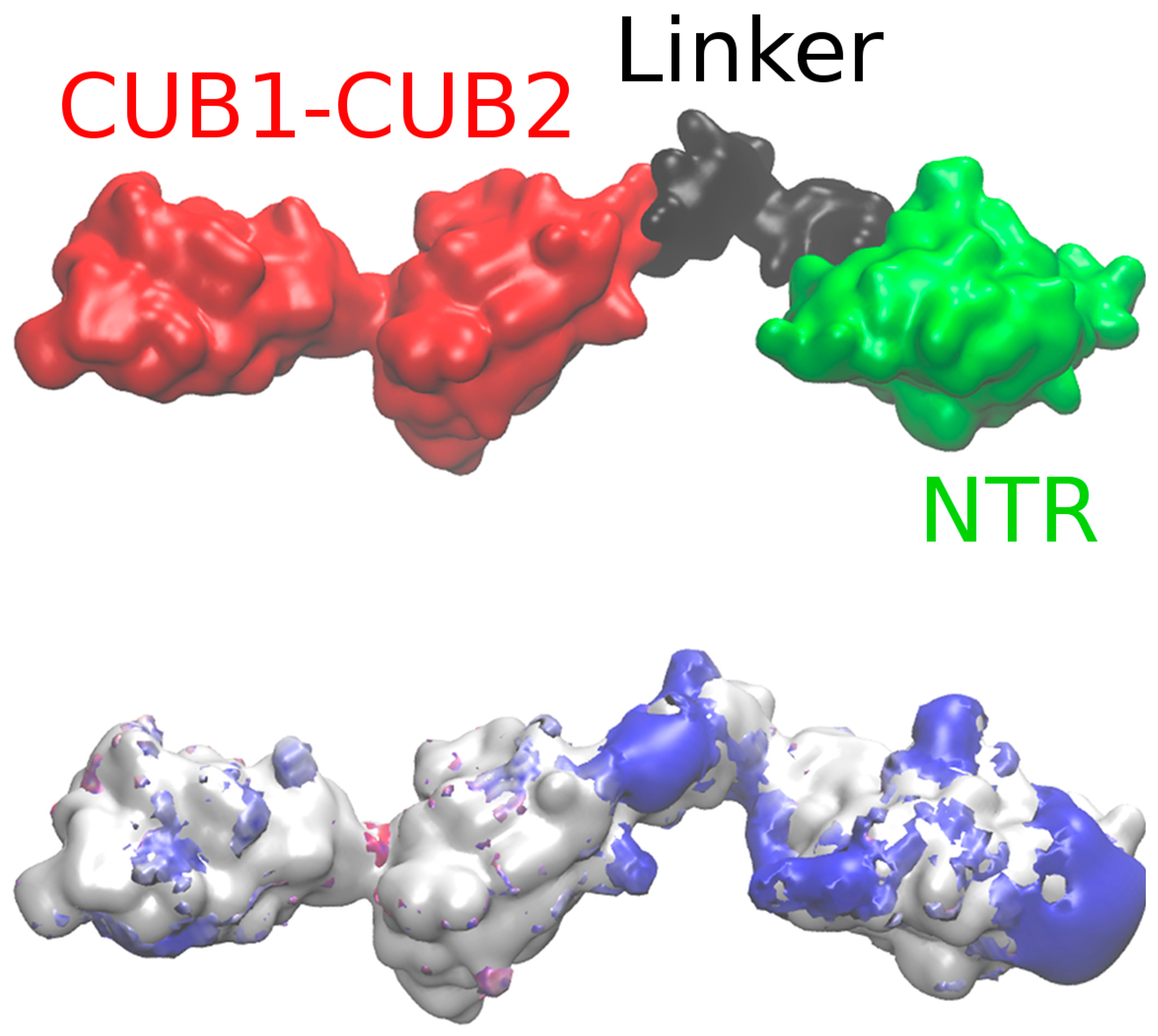

| Model | Restraints | Probability | Radius of Gyration (Å) |
|---|---|---|---|
| 1 | CUB1-CUB2, NTR domains | 34 | 22.2 |
| 2 | 32 | 24.8 | |
| 3 | 18 | 22.6 | |
| 4 | 8 | 22.8 | |
| 5 | 8 | 22.6 | |
| 1 | CUB1-CUB2, NTR domains + SAXS-based | 39 | 43.5 |
| 2 | 21 | 44.5 | |
| 3 | 17 | 43.5 | |
| 4 | 14 | 43.3 | |
| 5 | 9 | 43.9 |
| GAG | 1 m, ε | 2 # | 3 Size | 4 ΔG (kcal/mol) | 5 Polarity |
|---|---|---|---|---|---|
| CS6, dp4 | 3, 2 | 1 | 19 | −42.0 ± 6.6; −48.3 ± 7.7; −41.3 ± 6.6 | 17/2 |
| 2 | 6 | −30.1 ± 16.0; −63.3 ± 7.1 | 6/0 | ||
| 3 | 4 | −34.4 ± 9.6; −38.4 ± 8.6 | 2/2 | ||
| 4 | 3 | −46.7 ± 10.5 | 3/0 | ||
| CS6, dp6 | 3, 2 | 1 | 3 | −56.6 ± 9.0 | 3/0 |
| 2 | 3 | −33.9 ± 9.2 | 3/0 | ||
| 3 | 3 | −36.8 ± 7.1; −64.2 ± 11.8 | 3/0 | ||
| DS, dp6 | 3, 2 | 1 | 6 | −35.5 ± 6.3; −41.5 ± 6.8 | 5/1 |
| 2 | 4 | −36.7 ± 6.6 | 4/0 | ||
| 3 | 3 | −63.7 ± 8.3 | 3/0 | ||
| 4 | 3 | −37.8 ± 8.2 | 2/1 | ||
| HP, dp2 | 3, 2 | 1 | 25 | −44.9 ± 9.3; −41.1 ± 7.3; −23.1 ± 7.6 | 25/0 |
| 2 | 12 | −27.9 ± 9.2 | 12/0 | ||
| 3 | 9 | −42.0 ± 9.0 | 9/0 | ||
| 4 | 3 | −27.7 ± 8.9; −28.7 ± 5.6 | 3/0 | ||
| HP, dp4 | 3, 2 | 1 | 32 | −39.0 ± 7.2; −29.4 ± 10.4 | 21/11 |
| 2 | 3 | −53.9 ± 7.2 | 3/0 | ||
| 3 | 3 | −50.6 ± 11.5; −57.4 ± 8.6 | 2/1 | ||
| HP, dp6 | 3, 2 | 1 | 15 | −69.5 ± 7.8; −56.7 ± 7.4; −43.5 ± 9.7; −54.0 ± 14.8; −80.5 ± 10.7 | 9/6 |
| 2 | 7 | −68.3 ± 11.0; −44.7 ± 7.5; −57.1 ± 10.6; −55.4 ± 9.1 | 4/3 | ||
| 3 | 6 | −50.1 ± 10.0; −44.6 ± 9.5; −65.7 ± 11.5; −61.8 ± 13.6 | 4/2 |
| GAG | CS6, dp4 | CS6, dp6 | DS, dp6 | HP, dp2 | HP, dp4 | HP, dp6 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CS6, dp4 | 10 | 7 | 7 | 6 | 6 | 4 | 7 | 5 | 6 | 6 | 4 | 7 | 5 | 5 | 5 | 6 | 6 | 6 | 7 | 7 | 7 |
| 7 | 10 | 6 | 9 | 3 | 2 | 6 | 6 | 6 | 7 | 5 | 6 | 7 | 6 | 6 | 5 | 8 | 7 | 8 | 7 | 9 | |
| 7 | 6 | 10 | 5 | 4 | 3 | 5 | 6 | 5 | 7 | 4 | 7 | 4 | 6 | 5 | 4 | 5 | 7 | 7 | 7 | 6 | |
| 6 | 9 | 5 | 10 | 3 | 2 | 5 | 6 | 6 | 6 | 4 | 5 | 7 | 5 | 5 | 6 | 9 | 6 | 7 | 6 | 8 | |
| CS6, dp6 | 6 | 3 | 4 | 3 | 10 | 7 | 5 | 2 | 5 | 3 | 1 | 4 | 3 | 1 | 1 | 5 | 3 | 2 | 5 | 5 | 4 |
| 4 | 2 | 3 | 2 | 7 | 10 | 5 | 2 | 4 | 2 | 1 | 5 | 2 | 1 | 1 | 4 | 2 | 1 | 4 | 3 | 3 | |
| 7 | 6 | 5 | 5 | 5 | 5 | 10 | 5 | 5 | 4 | 5 | 6 | 5 | 4 | 4 | 7 | 5 | 4 | 7 | 6 | 7 | |
| DS, dp6 | 5 | 6 | 6 | 6 | 2 | 2 | 5 | 10 | 5 | 6 | 6 | 6 | 6 | 7 | 6 | 5 | 6 | 8 | 7 | 7 | 7 |
| 6 | 6 | 5 | 6 | 5 | 4 | 5 | 5 | 10 | 5 | 5 | 4 | 6 | 4 | 4 | 6 | 7 | 5 | 7 | 6 | 6 | |
| 6 | 7 | 7 | 6 | 3 | 2 | 4 | 6 | 5 | 10 | 6 | 6 | 6 | 6 | 6 | 3 | 5 | 7 | 7 | 8 | 6 | |
| 4 | 5 | 4 | 4 | 1 | 1 | 5 | 6 | 5 | 6 | 10 | 4 | 4 | 5 | 5 | 3 | 4 | 6 | 5 | 5 | 5 | |
| HP, dp2 | 7 | 6 | 7 | 5 | 4 | 5 | 6 | 6 | 4 | 6 | 4 | 10 | 5 | 5 | 5 | 5 | 5 | 6 | 7 | 7 | 7 |
| 5 | 7 | 4 | 7 | 3 | 2 | 5 | 6 | 6 | 6 | 4 | 5 | 10 | 5 | 5 | 7 | 7 | 5 | 7 | 7 | 7 | |
| 5 | 6 | 6 | 5 | 1 | 1 | 4 | 7 | 4 | 6 | 5 | 5 | 5 | 10 | 6 | 3 | 5 | 7 | 6 | 6 | 6 | |
| 5 | 6 | 5 | 5 | 1 | 1 | 4 | 6 | 4 | 6 | 5 | 5 | 5 | 6 | 10 | 3 | 5 | 6 | 6 | 6 | 6 | |
| HP, dp4 | 6 | 5 | 4 | 6 | 5 | 4 | 7 | 5 | 6 | 3 | 3 | 5 | 7 | 3 | 3 | 10 | 7 | 3 | 6 | 5 | 6 |
| 6 | 8 | 5 | 9 | 3 | 2 | 5 | 6 | 7 | 5 | 4 | 5 | 7 | 5 | 5 | 7 | 10 | 6 | 7 | 6 | 8 | |
| 6 | 7 | 7 | 6 | 2 | 1 | 4 | 8 | 5 | 7 | 6 | 6 | 5 | 7 | 6 | 3 | 6 | 10 | 6 | 7 | 7 | |
| HP, dp6 | 7 | 8 | 7 | 7 | 5 | 4 | 7 | 7 | 7 | 7 | 5 | 7 | 7 | 6 | 6 | 6 | 7 | 6 | 10 | 9 | 9 |
| 7 | 7 | 7 | 6 | 5 | 3 | 6 | 7 | 6 | 8 | 5 | 7 | 7 | 6 | 6 | 5 | 6 | 7 | 9 | 10 | 8 | |
| 7 | 9 | 6 | 8 | 4 | 3 | 7 | 7 | 6 | 6 | 5 | 7 | 7 | 6 | 6 | 6 | 8 | 7 | 9 | 8 | 10 | |
| GAG | CS6, dp4 | CS6, dp6 | DS, dp6 | HP, dp2 | HP, dp4 | HP, dp6 |
|---|---|---|---|---|---|---|
| CS6, dp4 | 10 | 5 | 7 | 6 | 9 | 8 |
| CS6, dp6 | 5 | 10 | 4 | 4 | 6 | 6 |
| DS, dp6 | 7 | 4 | 10 | 6 | 7 | 7 |
| HP, dp2 | 6 | 4 | 6 | 10 | 7 | 7 |
| HP, dp4 | 9 | 6 | 7 | 7 | 10 | 9 |
| HP, dp6 | 8 | 6 | 7 | 7 | 9 | 10 |
| 1 m, ε | 2 # | 3 Size | 4 ΔG, kcal/mol | 5 TopMM-GBSA 10 Residues for GAG Binding | 6 Polarity |
|---|---|---|---|---|---|
| 2, 2.64 | 1 | 4 | −62.4.8 ± 19.0; −54.9 ± 9.1 −49.6 ± 18.6 | R435, K436, R275, R288, K279, K299, K365, K434, N331, K295 | 4/0 |
| 2 | 3 | −50.1 ± 9.7; −79.0 ± 17.0; −38.1 ± 9.4 | K436, R435, K365, K299, K434, K271, K295, R288, K165, K279 | 3/0 | |
| 3 | 3 | −30.8 ± 10.7; −36.0 ± 7.8; −42.3 ± 10.6 | K299, K436, K279, K365, K271, K434, K295, K165, Q282, R435 | 2/1 |
| Protein | PDB ID | Experimental Structure | Method | ||
|---|---|---|---|---|---|
| FoldX | IonCom | 1 MD | |||
| Annexin V | 1G5N | 9 | 6 | 8 | 6 |
| NTR | 1UAP | 0 | 0 | 0 | 2 |
| 3 | |||||
| 1 | |||||
| 1 | |||||
| 1 | |||||
| CUB1-CUB2 | 6FZV | 2 | 2 | 2 | 2 |
| 2 | |||||
| 1 | |||||
| 2 | |||||
| 1 | |||||
| Ca2+ Number (X-Ray) | 1 ΔG, kcal/mol | 2 ΔGele, kcal/mol | 3 FoldX | 3 IonCom | 3 MD |
|---|---|---|---|---|---|
| 319 | 57.2 ± 4.7 | −310.4 ± 10.3 | + | + | + |
| 320 | 47.5 ± 4.8 | −264.8 ± 15.7 | + | + | – |
| 321 | 36.5 ± 3.5 | −296.0 ± 10.9 | – | + | + |
| 322 | 59.7 ± 4.9 | −380.5 ± 9.5 | + | + | – |
| 323 | 36.4 ± 3.5 | −332.4 ± 7.9 | – | – | + |
| 324 | 62.4 ± 4.4 | −376.6 ± 8.1 | + | + | + |
| 325 | 47.5 ± 6.1 | −413.2 ± 13.0 | + | + | + |
| 326 | 39.3 ± 3.7 | −312.2 ± 9.2 | – | + | + |
| 327 | 59.2 ± 4.7 | −302.3 ± 8.6 | + | + | – |
| PCPE-1 Domain | Ca2+ Site | 2 ΔGele, kcal/mol | Site Occupancy, ns |
|---|---|---|---|
| NTR, MD1 | E405, E406, N407 G367, D370 | −116.5 ± 20.4 −58.3 ± 15.7 | 65 40 |
| NTR, MD2 | E405, E406, N407 D314/N-terminus of NTR G367, D370 | −125.8 ± 14.7 −38.6 ± 19.0 −49.6 ± 14.2 | 85 35 90 |
| NTR, MD3 | E405, E406, N407 | −120.3 ± 15.9 | 75 |
| NTR, MD4 | E405, E406, N407 | −123.4 ± 12.2 | 45 |
| NTR, MD5 | E405, E406, N407 1 E405, E406, N407/G367, D370 | −51.7 ± 13.7; −156.2 ± 33.6 | 65 25 |
| CUB1-CUB2 (X-ray, PDB ID: 6FZV) | E85, Y92, D93, D134 Y180, E208, D216, D258 | −363.2 ± 10.8 −466.5 ± 12.5 | 100 100 |
| CUB1-CUB2, MD1 | E85, Y92, D93, D134 Y180, E208, D216, D258 | −389.9 ± 19.3 −302.9 ± 24.3 | 25 90 |
| CUB1-CUB2, MD2 | E85, Y92, D93, D134 Y180, E208, D216, D258 | −368.3 ± 18.7 −371.9 ± 11.8 | 85 90 |
| CUB1-CUB2, MD3 | Y180, E208, D216, D258 | −389.2 ± 18.6 | 85 |
| CUB1-CUB2, MD4 | E85, Y92, D93, D134 Y180, E208, D216, D258 | −293.7 ± 9.0 −374.0 ± 16.1 | 75 95 |
| CUB1-CUB2, MD5 | Y180, E208, D216, D258 | −521.4 ± 10.8 | 95 |
| 1 m, ε | 2 # | 3 Size | 4 ΔG, kcal/mol | 5 TopMM-GBSA 10 Residues for GAG Binding | 6 Polarity |
|---|---|---|---|---|---|
| 2, 2.8 | 1 | 6 | −58.8 ± 12.2; −56.0 ± 19.7; −58.6 ± 13.7 | R435, K436, K434, K365, K299, K279, K295, R288, P438, K271 | 5/1 |
| 2 | 4 | −79.5 ± 15.6; −32.6 ± 11.0; −42.8 ± 10.0 | K436, R435, K279, R288, K365, K299, K434, Q282, G281, K287 | 3/1 | |
| 3 | 3 | −30.8 ± 10.7; −70.7 ± 13.2; −56.6 ± 18.4 | R435, K299, K436, K365, K434, R275, K279, K295, K271, K305 | 3/0 |
| 1 # | 2 Ca2+ | 3 ΔG, kcal/mol | 4 TopMM-GBSA 10 Residues for GAG Binding |
|---|---|---|---|
| 1 | – | −65.6 ± 12.3 | K299, R288, R435, K436, K295, K293, K305, K287, K365, P298 |
| 2 | – | −64.2 ± 11.2 | K436, K434, R275, K279, K295, R435, K365, R288, K299, K287 |
| 3 | – | −94.7 ± 12.4 | K295, K436, R435, K365, K434, K293, K299, R288, K305, V294 |
| 1 | + | −93.3 ± 12.3 | R435, K279, K295, K436, K305, K299, K434, N331, K271, R324 |
| 2 | + | −73.8 ± 11.9 | R435, K434, K436, K299, K295, K279, P298, K293, K287, K305 |
| 3 | + | −102.5 ± 14.7 | K436, K299, K295, R435, R275, K293, K279, K434, K305, P441 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potthoff, J.; Bojarski, K.K.; Kohut, G.; Lipska, A.G.; Liwo, A.; Kessler, E.; Ricard-Blum, S.; Samsonov, S.A. Analysis of Procollagen C-Proteinase Enhancer-1/Glycosaminoglycan Binding Sites and of the Potential Role of Calcium Ions in the Interaction. Int. J. Mol. Sci. 2019, 20, 5021. https://doi.org/10.3390/ijms20205021
Potthoff J, Bojarski KK, Kohut G, Lipska AG, Liwo A, Kessler E, Ricard-Blum S, Samsonov SA. Analysis of Procollagen C-Proteinase Enhancer-1/Glycosaminoglycan Binding Sites and of the Potential Role of Calcium Ions in the Interaction. International Journal of Molecular Sciences. 2019; 20(20):5021. https://doi.org/10.3390/ijms20205021
Chicago/Turabian StylePotthoff, Jan, Krzysztof K. Bojarski, Gergely Kohut, Agnieszka G. Lipska, Adam Liwo, Efrat Kessler, Sylvie Ricard-Blum, and Sergey A. Samsonov. 2019. "Analysis of Procollagen C-Proteinase Enhancer-1/Glycosaminoglycan Binding Sites and of the Potential Role of Calcium Ions in the Interaction" International Journal of Molecular Sciences 20, no. 20: 5021. https://doi.org/10.3390/ijms20205021
APA StylePotthoff, J., Bojarski, K. K., Kohut, G., Lipska, A. G., Liwo, A., Kessler, E., Ricard-Blum, S., & Samsonov, S. A. (2019). Analysis of Procollagen C-Proteinase Enhancer-1/Glycosaminoglycan Binding Sites and of the Potential Role of Calcium Ions in the Interaction. International Journal of Molecular Sciences, 20(20), 5021. https://doi.org/10.3390/ijms20205021