PPP3CB Inhibits Migration of G401 Cells via Regulating Epithelial-to-Mesenchymal Transition and Promotes G401 Cells Growth
Abstract
1. Introduction
2. Results
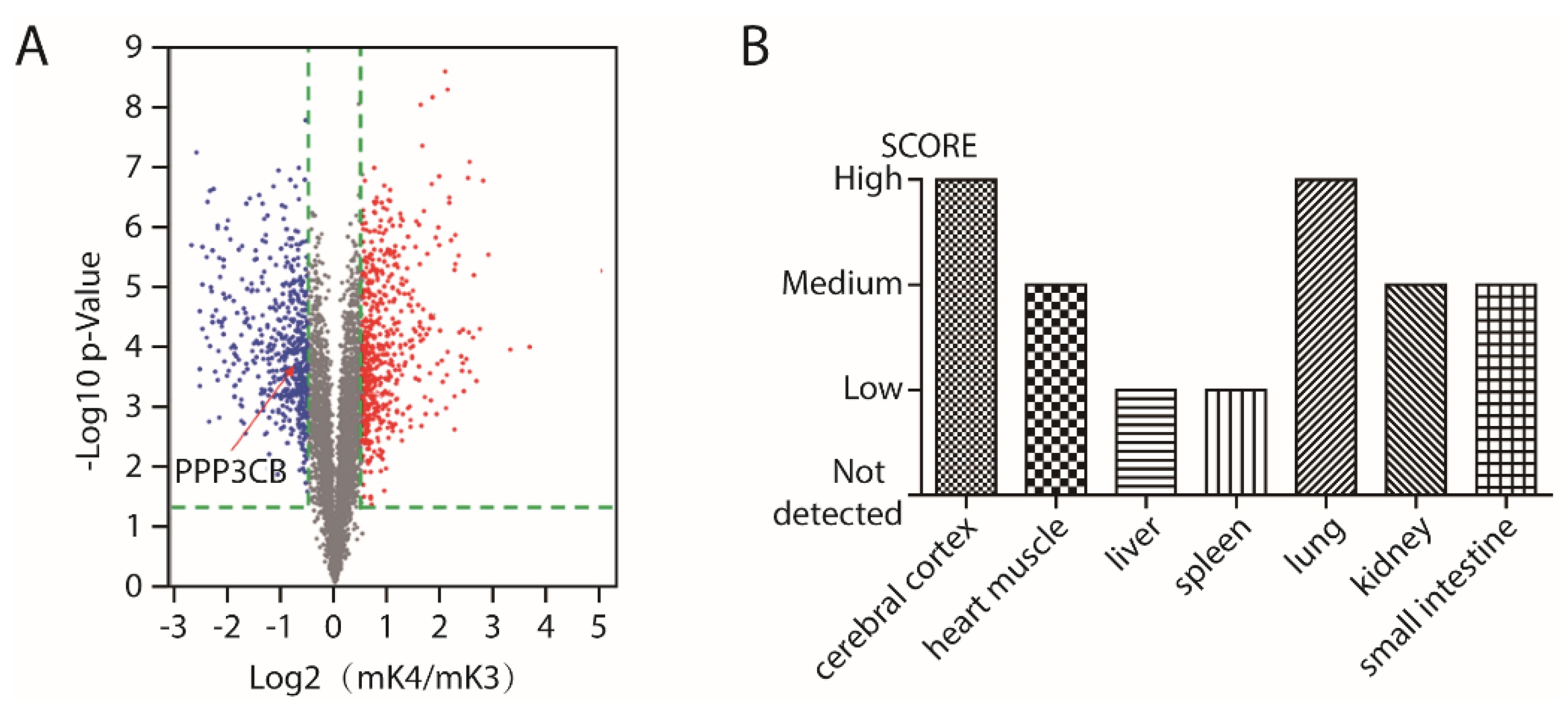
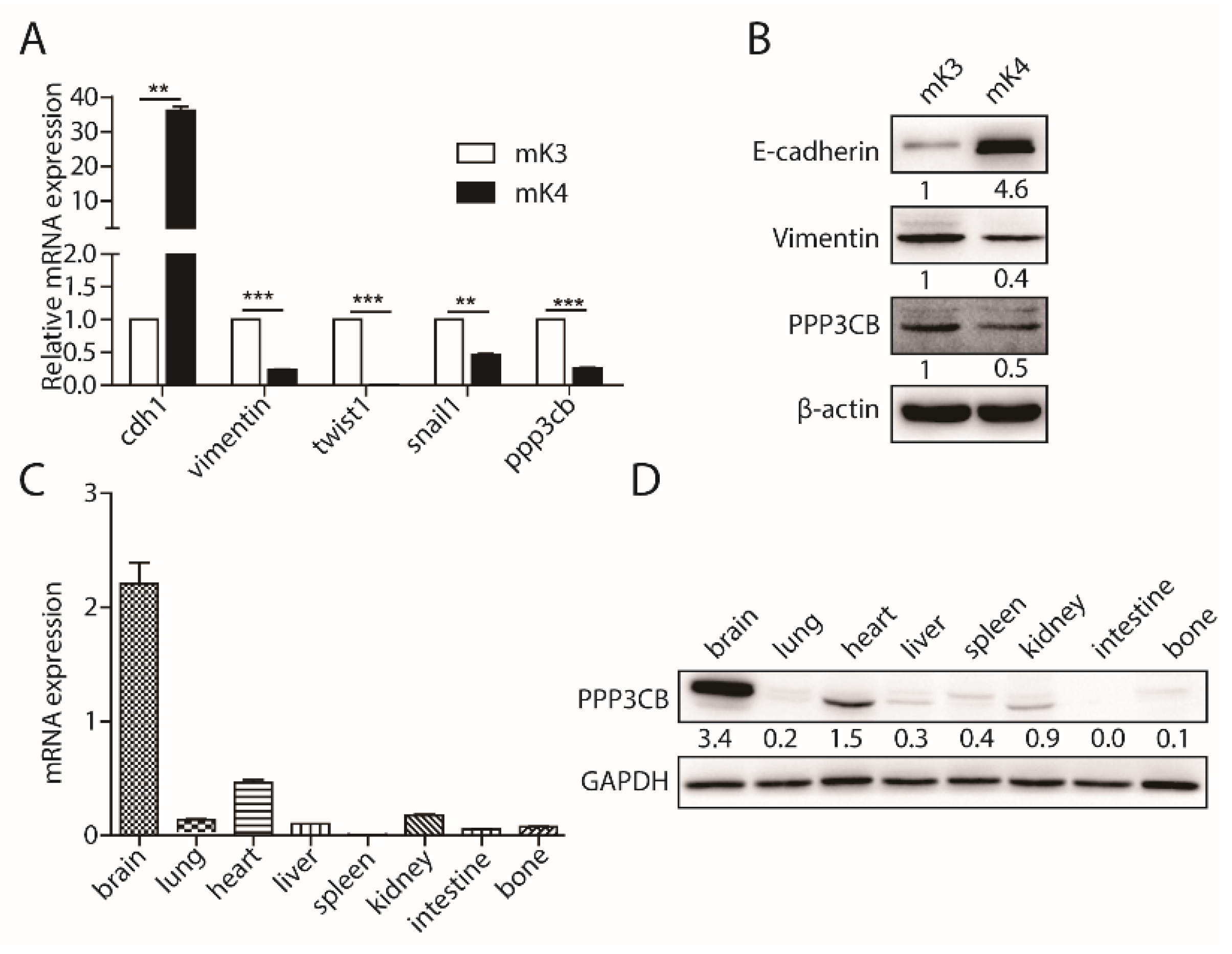
2.1. PPP3CB is an Important Regulator in Kidney
2.2. PPP3CB Suppresses EMT of G401 Cells
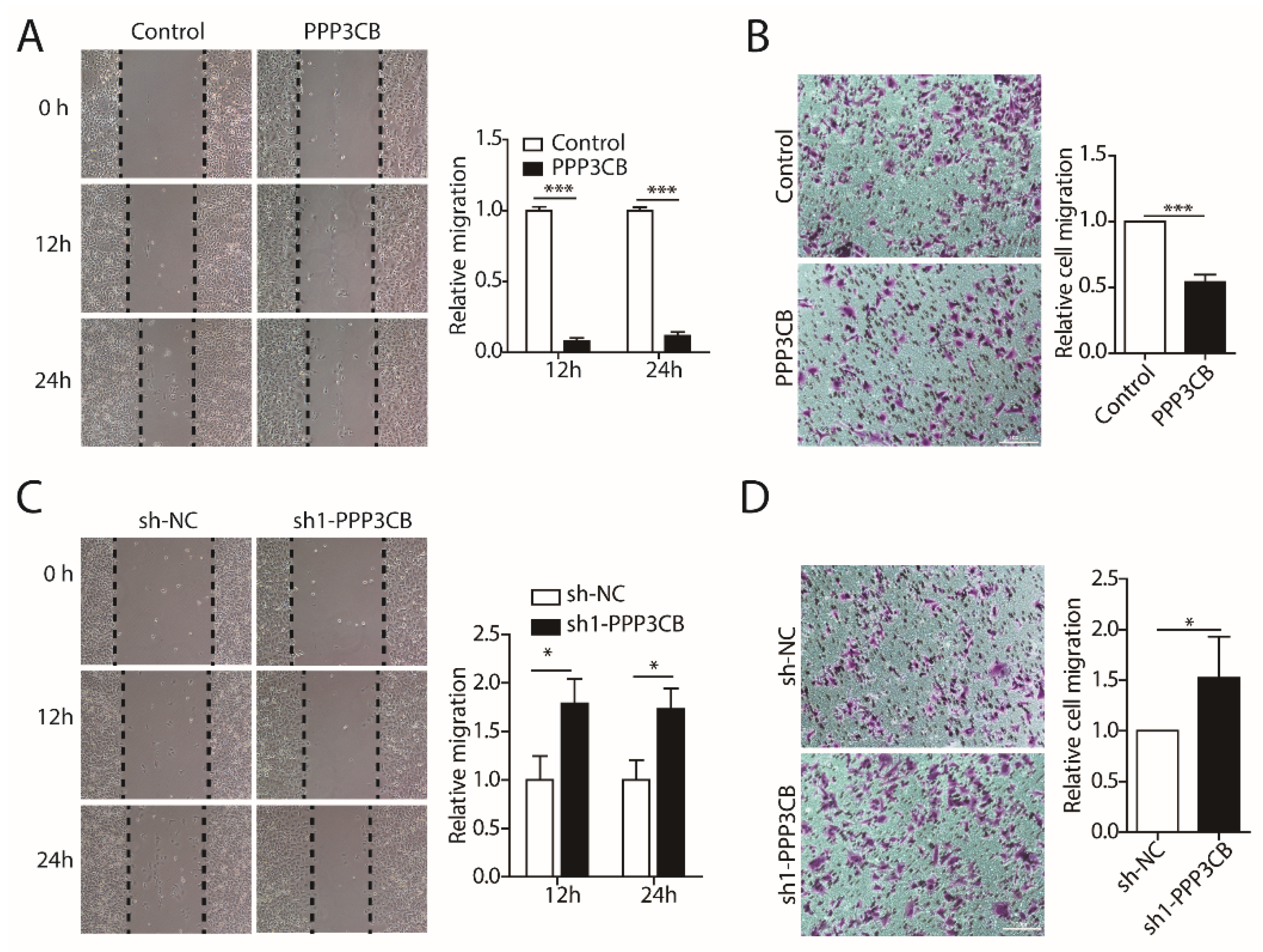
2.3. PPP3CB Inhibits Migration of G401 Cells
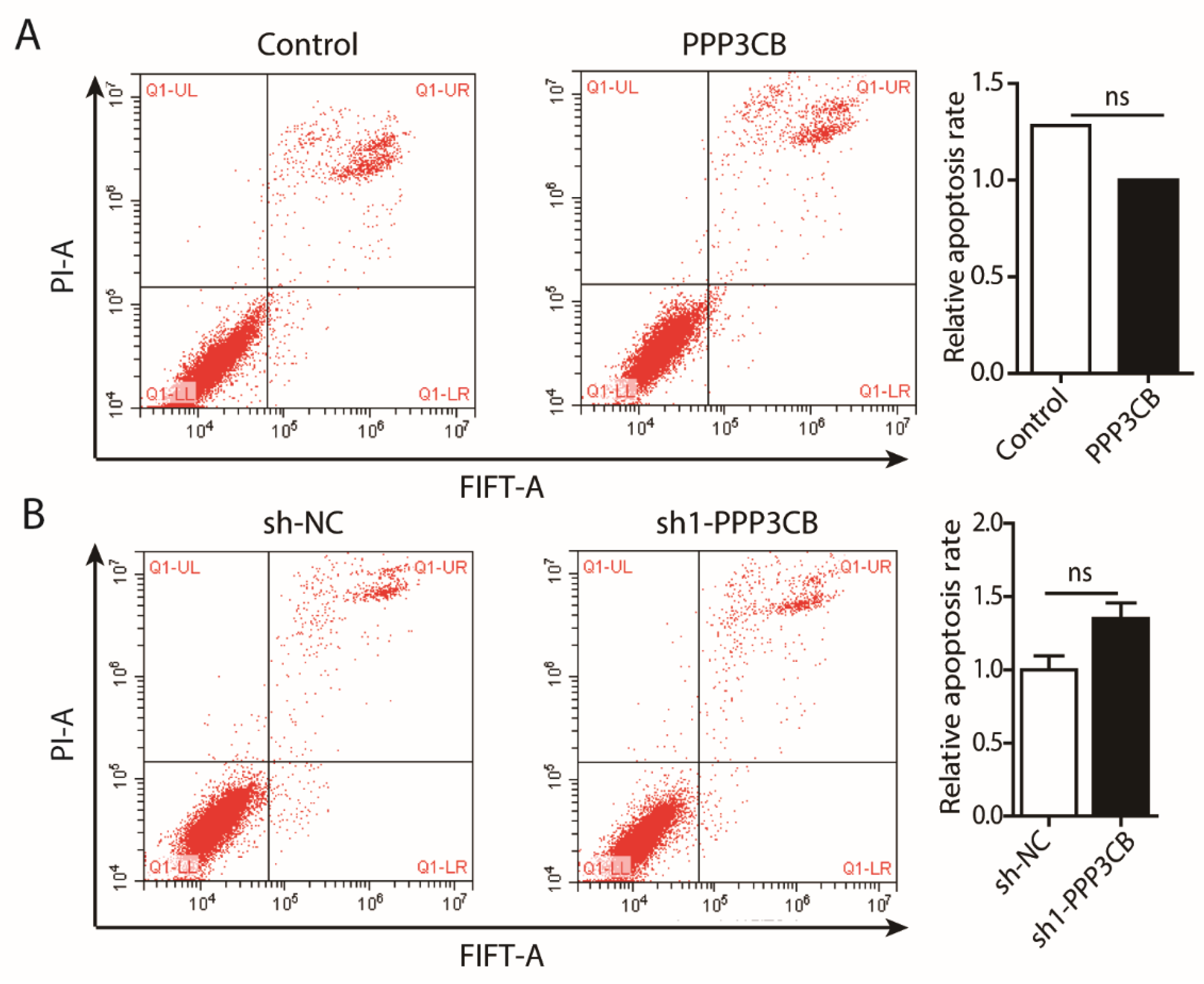
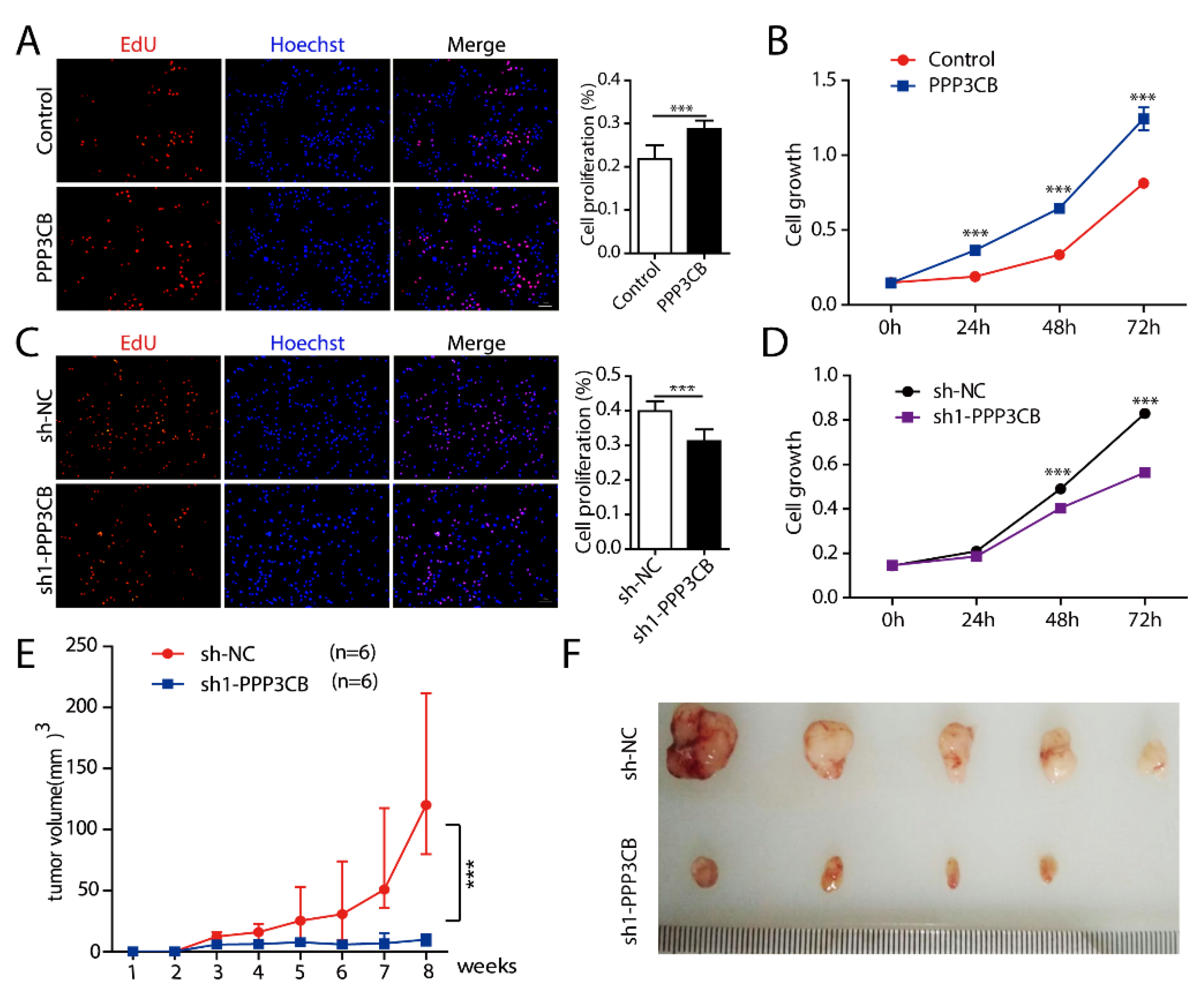
2.4. PPP3CB Promotes Cell Proliferation
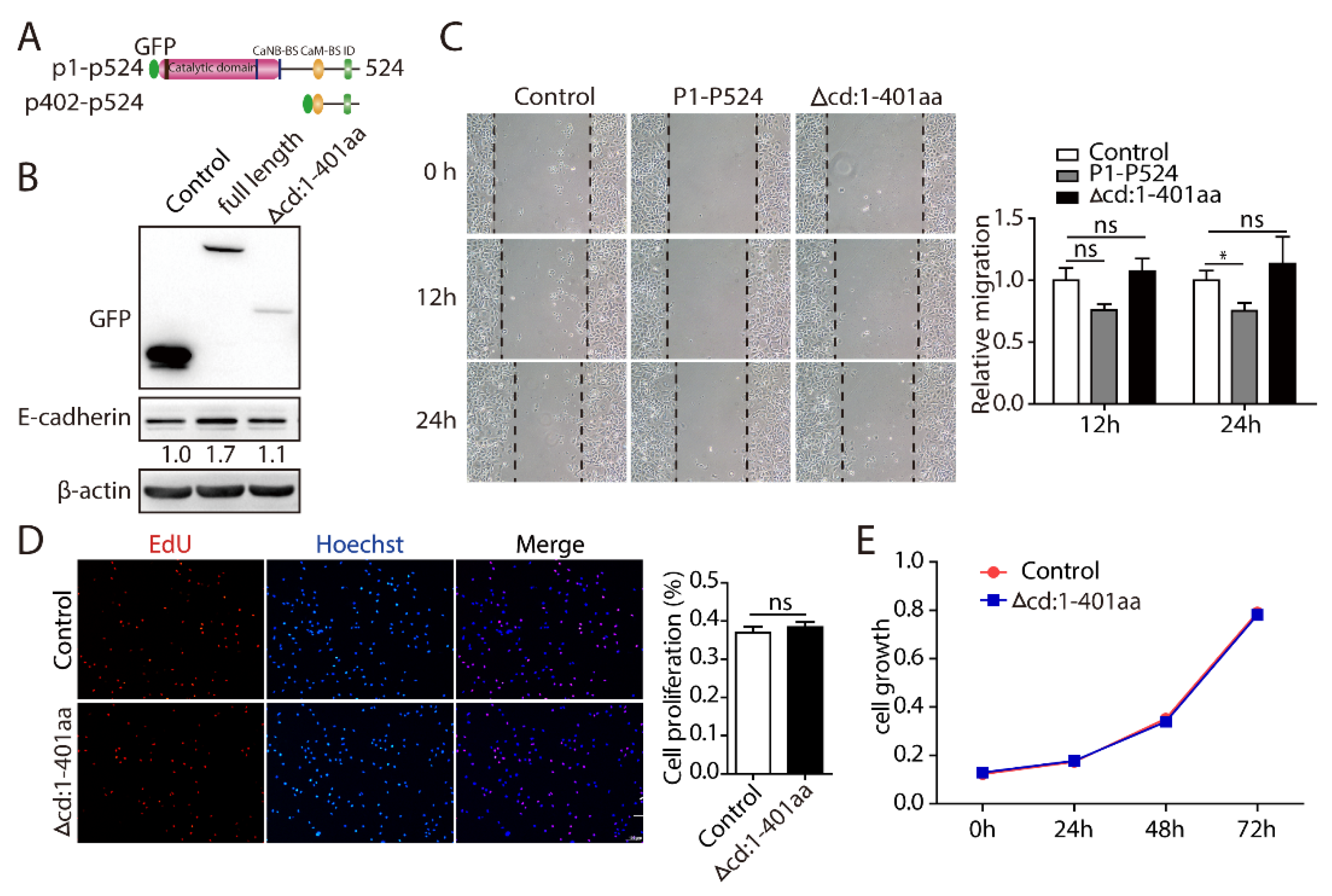
2.5. PPP3CB 1-401 Containing the Catalytic Domain Plays a Critical Role in EMT and Cell Proliferation
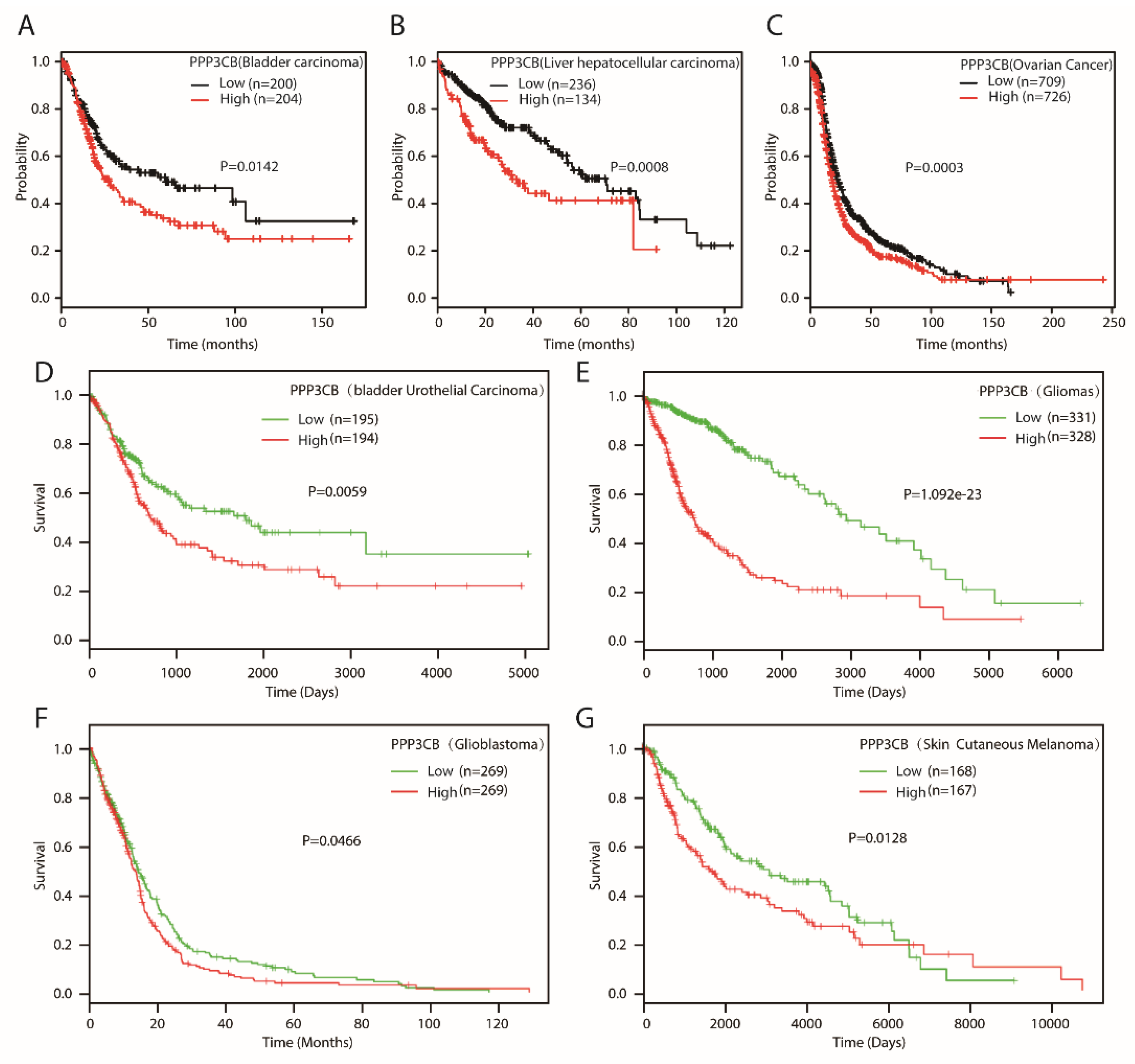
3. Discussion
4. Materials and Methods
4.1. Plasmids Construction
4.2. Cell Culture and Transfection
4.3. Construction of Stable Cell Lines
4.4. RNA Extraction and Real-Time PCR
4.5. Western Blotting
4.6. Scratch Wound Healing Assay
4.7. Migration Assay
4.8. MTT Assay
4.9. 5-Ethynyl-21-Deoxyuridine (EdU) Assay
4.10. Flow Cytometry Assay
4.11. Nude Mice Tumorigenesis
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EMT | Epithelial-mesenchymal transition |
| PPP | Phosphoprotein phosphatase |
| PP1 | Protein Phosphatases1 |
| PP2A | Protein Phosphatases2A |
| PP2B | Protein Phosphatases2B |
| CsA | cyclosporineA |
| QPCR | Quantitative polymerase chain reaction |
| NB | neuroblastoma |
| FITC | fluoresceine isothiocyanate |
| PI | propidium iodide |
Appendix A

References
- Yang, M.H.; Hsu, D.S.; Wang, H.W.; Wang, H.J.; Lan, H.Y.; Yang, W.H.; Huang, C.H.; Kao, S.Y.; Tzeng, C.H.; Tai, S.K.; et al. Bmi1 is essential in Twist1-induced epithelial–mesenchymal transition. Nat. Cell Biol. 2010, 12, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Charles, J.D.; Han, Y.H.; Mo, C. TGF-b Tumor Suppression through a Lethal EMT. Cell 2016, 164, 1015–1030. [Google Scholar]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nature reviews. Clin. Oncol. 2017, 14, 611–629. [Google Scholar]
- Lyons, S.P.; Jenkins, N.P.; Nasa, I.; Choy, M.S.; Adamo, M.E.; Page, R.; Peti, W.; Moorhead, G.B.; Kettenbach, A.N. A Quantitative Chemical Proteomic Strategy for Profiling Phosphoprotein Phosphatases from Yeast to Humans. Mol. Cell. Proteom. 2018, 17, 2448–2461. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Singh, S.; Srivastava, S.K.; Arora, S.; Hyde, S.J.; Andrews, J.; Grizzle, W.E.; Singh, A.P. Restoration of PPP2CA expression reverses epithelial-to-mesenchymal transition and suppresses prostate tumour growth and metastasis in an orthotopic mouse model. Br. J. Cancer 2014, 110, 2000–2010. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, L.; Yang, A.; Lin, J.; Tang, F.; Jin, S.; Wei, Z.; Li, J.; Jin, Y. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell 2011, 8, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Srinivasan, S.; Ruthel, G.; Kashina, A.K.; Carstens, R.P.; Mendoza, A.; Khanna, C.; Van Winkle, T.; Avadhani, N.G. Mitochondrial retrograde signaling induces epithelial–mesenchymal transition and generates breast cancer stem cells. Oncogene 2014, 33, 5238–5250. [Google Scholar] [CrossRef]
- Li, H.; Rao, A.; Hogan, P.G. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011, 21, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Balakrishnan-Renuka, A.; Hagemann, N.; Theiss, C.; Chankiewitz, V.; Chen, J.; Pu, Q.; Erdmann, K.S.; Brand-Saberi, B. A novel interaction between ATOH8 and PPP3CB. Histochem. Cell Biol. 2016, 145, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.E.; Bai, Y. Differential expression of cyclosporine A-Induced calcineurin isoform-specific matrix metalloproteinase 9 (MMP-9) in renal fibroblasts. Biochem. Biophys. Res. Commun. 2018, 503, 2549–2554. [Google Scholar] [CrossRef] [PubMed]
- Pena, J.A.; Titus, L.; Jackson, J.; Kirk, A.D.; Gooch, J.L. Differential Regulation of Calcineurin Isoforms in Transplant Patients: A New Look at an Old Problem. Transplantation 2013, 96, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zheng, C.; Wang, X.; Yun, S.; Zhao, Y.; Liu, L.; Lu, Y.; Ye, Y.; Zhu, X.; Zhang, C.; et al. MicroRNA-30 family members regulate calcium/calcineurin signaling in podocytes. J. Clin. Investig. 2015, 125, 4091–4106. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, S.; Stebbing, J.; Frampton, A.E.; Zagorac, S.; Krell, J.; de Giorgio, A.; Trabulo, S.M.; Nguyen, V.M.; Magnani, L.; Feng, H.; et al. TGF-β induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression. Nat. Commun. 2018, 9, 1845. [Google Scholar] [CrossRef] [PubMed]
- Shakhova, I.; Li, Y.; Yu, F.; Kaneko, Y.; Nakamura, Y.; Ohira, M.; Izumi, H.; Mae, T.; Varfolomeeva, S.R.; Rumyantsev, A.G.; et al. PPP3CB contributes to poor prognosis through activating nuclear factor of activated T-cells signaling in neuroblastoma. Mol. Carcinog. 2018. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Mancini, M.; Toker, A. NFAT Proteins: Emerging Roles in Cancer Progression. Nat. Rev. Cancer 2009, 9, 810–820. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, X.; Wang, S.; Yuan, X.; Pang, Y.; Chen, J.; Wang, J. Cancer Stem Cells are Regulated by STAT3 Signaling in Wilms Tumor. J. Cancer 2018, 9, 1486–1499. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; He, Q.; Liu, Y.; Wu, Y.; Ni, D.; Liu, J.; Hu, Y.; Gu, Y.; Xie, Y.; Zhou, Q.; et al. PPP3CB Inhibits Migration of G401 Cells via Regulating Epithelial-to-Mesenchymal Transition and Promotes G401 Cells Growth. Int. J. Mol. Sci. 2019, 20, 275. https://doi.org/10.3390/ijms20020275
Chen L, He Q, Liu Y, Wu Y, Ni D, Liu J, Hu Y, Gu Y, Xie Y, Zhou Q, et al. PPP3CB Inhibits Migration of G401 Cells via Regulating Epithelial-to-Mesenchymal Transition and Promotes G401 Cells Growth. International Journal of Molecular Sciences. 2019; 20(2):275. https://doi.org/10.3390/ijms20020275
Chicago/Turabian StyleChen, Lei, Qingling He, Yamin Liu, Yafei Wu, Dongsheng Ni, Jianing Liu, Yanxia Hu, Yuping Gu, Yajun Xie, Qin Zhou, and et al. 2019. "PPP3CB Inhibits Migration of G401 Cells via Regulating Epithelial-to-Mesenchymal Transition and Promotes G401 Cells Growth" International Journal of Molecular Sciences 20, no. 2: 275. https://doi.org/10.3390/ijms20020275
APA StyleChen, L., He, Q., Liu, Y., Wu, Y., Ni, D., Liu, J., Hu, Y., Gu, Y., Xie, Y., Zhou, Q., & Li, Q. (2019). PPP3CB Inhibits Migration of G401 Cells via Regulating Epithelial-to-Mesenchymal Transition and Promotes G401 Cells Growth. International Journal of Molecular Sciences, 20(2), 275. https://doi.org/10.3390/ijms20020275



