Differences in Expression of Genes Involved in Bone Development and Morphogenesis in the Walls of Internal Thoracic Artery and Saphenous Vein Conduits May Provide Markers Useful for Evaluation Graft Patency
Abstract
1. Introduction
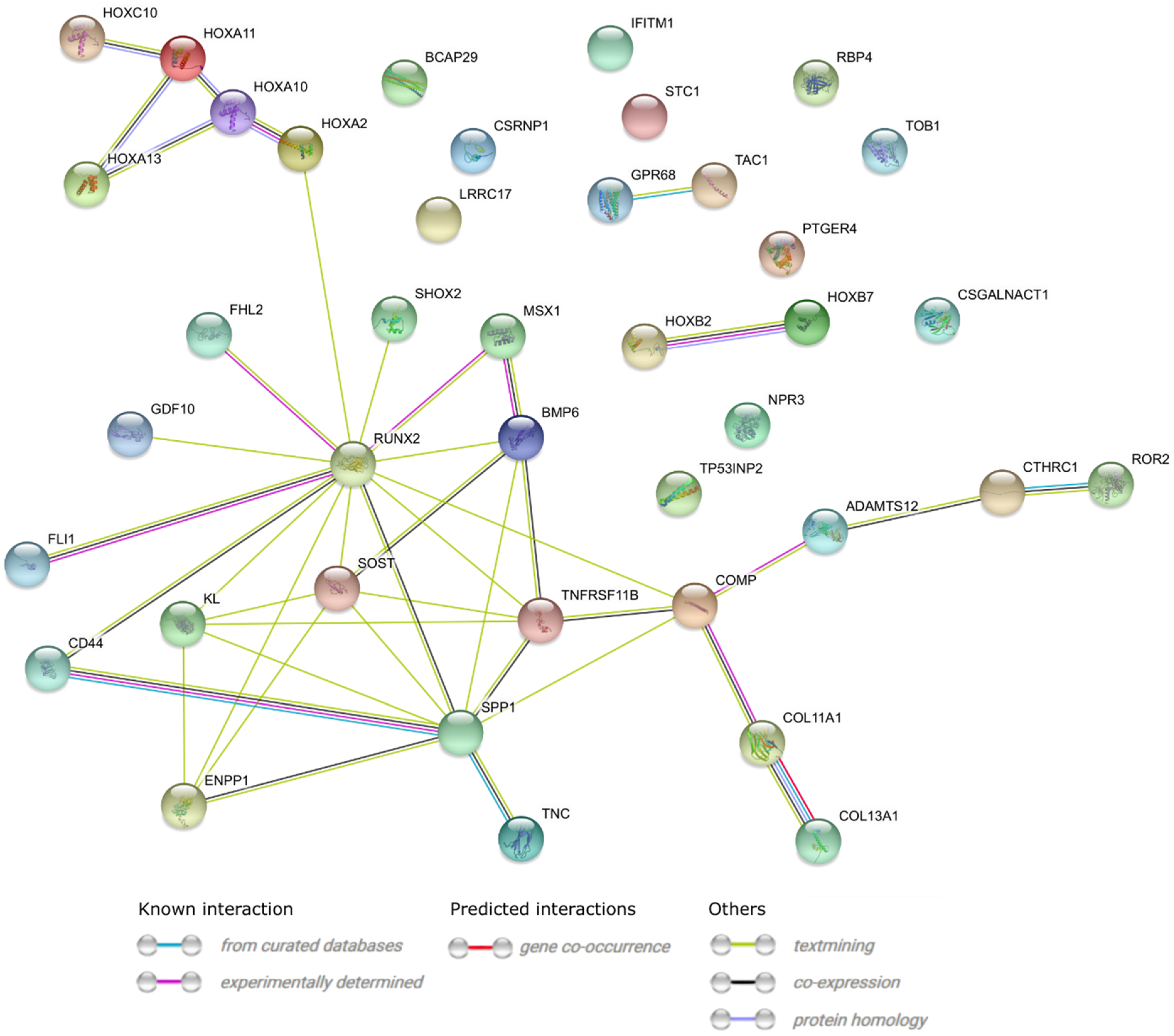
2. Results
3. Discussion
4. Materials and Methods
4.1. Human Subjects
4.2. Operation Procedure and Sample Collection
4.3. RNA Extraction and Reverse Transcription
4.4. Microarray Expression Study and Data Analysis
4.5. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
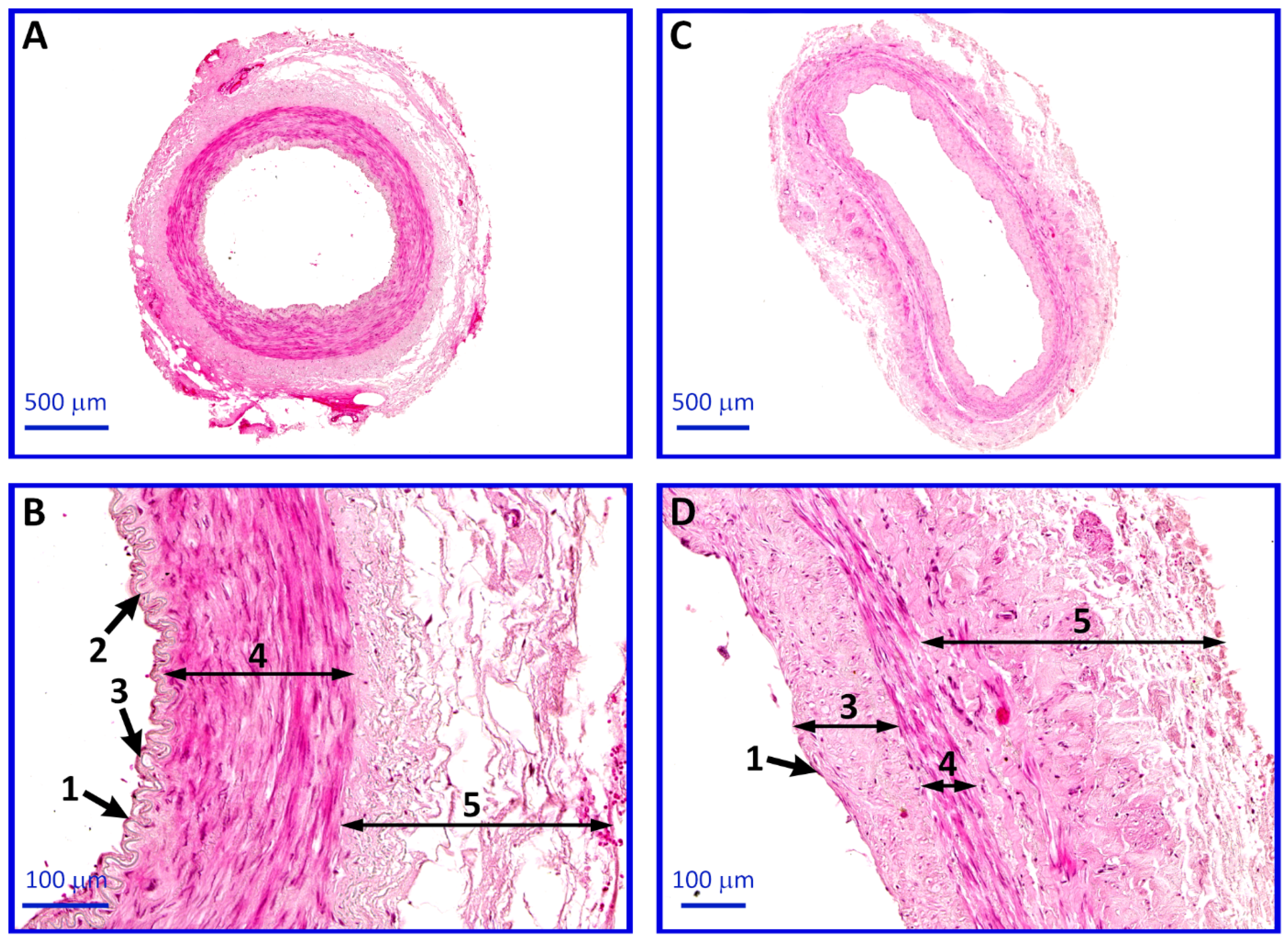
4.6. Histological Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Davierwala, P.M.; Mohr, F.W. Bilateral internal mammary artery grafting: Rationale and evidence. Int. J. Surg. 2015, 16, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Kappetein, A.P.; van Mieghem, N.M.; Head, S.J. Revascularization Options: Coronary artery bypass surgery and percutaneous coronary intervention. Cardiol. Clin. 2014, 32, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Al-Sabti, H.A.; Al Kindi, A.; Al-Rasadi, K.; Banerjee, Y.; Al-Hashmi, K.; Al-Hinai, A. Saphenous vein graft vs. radial artery graft searching for the best second coronary artery bypass graft. J. Saudi Hear. Assoc. 2013, 25, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Gansera, B.; Schmidtler, F.; Angelis, I.; Kiask, T.; Kemkes, B.; Botzenhardt, F. Patency of Internal Thoracic Artery Compared to Vein Grafts - Postoperative Angiographic Findings in 1189 Symptomatic Patients in 12 Years. Thorac. Cardiovasc. Surg. 2007, 55, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Taggart, D.P.; Altman, D.G.; Gray, A.M.; Lees, B.; Gerry, S.; Benedetto, U.; Flather, M.; ART Investigators. Randomized Trial of Bilateral versus Single Internal-Thoracic-Artery Grafts. N. Engl. J. Med. 2016, 375, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Taggart, D.P.; D’Amico, R.; Altman, D.G. Effect of arterial revascularisation on survival: A systematic review of studies comparing bilateral and single internal mammary arteries. Lancet 2001, 358, 870–875. [Google Scholar] [CrossRef]
- Weintraub, W.S.; Jones, E.L.; Craver, J.M.; Guyton, R.A. Frequency of repeat coronary bypass or coronary angioplasty after coronary artery bypass surgery using saphenous venous grafts. Am. J. Cardiol. 1994, 73, 103–112. [Google Scholar] [CrossRef]
- Harskamp, R.E.; Lopes, R.D.; Baisden, C.E.; de Winter, R.J.; Alexander, J.H. Saphenous Vein Graft Failure After Coronary Artery Bypass Surgery. Ann. Surg. 2013, 257, 824–833. [Google Scholar] [CrossRef]
- Dar, M.I.; Dar, A.H.; Bilal, M.; Ahmad, M.; Haseeb, A. Association of Internal Mammary Artery Flow with Different Comorbidities and Post-Coronary Artery Bypass Graft Complications. Cureus 2017, 9, e1584. [Google Scholar] [CrossRef]
- Hegazy, Y.; Hassanein, W.; Ennker, J.; Keshk, N.; Bauer, S.; Sodian, R. The Use of Bilateral Internal Mammary Artery Grafting in Different Degrees of Obesity. Thorac. Cardiovasc. Surg. 2017, 65, 278–285. [Google Scholar] [CrossRef]
- Farkouh, M.E.; Domanski, M.; Fuster, V. Comparison of CABG versus PCI Outcomes in Diabetic, Mutivessel Coronary Artery Disease Patients With and Without Acute Coronary Syndromes. Circulation 2014, 130, A20356. [Google Scholar]
- Parolari, A.; Poggio, P.; Myasoedova, V.; Songia, P.; Bonalumi, G.; Pilozzi, A.; Pacini, D.; Alamanni, F.; Tremoli, E. Biomarkers in Coronary Artery Bypass Surgery: Ready for Prime Time and Outcome Prediction? Front. Cardiovasc. Med. 2015, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Tomizawa, Y.; Endo, M.; Nishida, H.; Niinami, H.; Tanaka, S.; Tomioka, H.; Ozawa, H.; Kikuchi, C.; Koyanagi, H. Use of arterial grafts for coronary revascularization. Experience of 2987 anastomoses. Jpn. J. Thorac. Cardiovasc. Surg. 1999, 47, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Malinska, A.; Perek, B.; Buczkowski, P.; Kowalska, K.; Ostalska-Nowicka, D.; Witkiewicz, W.; Nowicki, M. CD68 expression in aortocoronary saphenous vein bypass grafts. Histochem. Cell Biol. 2013, 140, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Perek, B.; Malińska, A.; Ostalska-Nowicka, D.; Puślecki, M.; Ligowski, M.; Misterski, M.; Zabel, M.; Jemielity, M.; Nowicki, M. Cytokeratin 8 in venous grafts: A factor of unfavorable long-term prognosis in coronary artery bypass grafting patients. Cardiol. J. 2013, 20, 583–591. [Google Scholar] [CrossRef]
- Perek, B.; Kowalska, K.; Kempisty, B.; Nowicki, A.; Jankowski, M.; Nawrocki, M.J.; Malińska, A. Role of macrophages in the pathogenesis of atherosclerosis and aortocoronary graft disease. J. Biol. Regul. Homeost. Agents 2018, 32, 1055–1059. [Google Scholar] [PubMed]
- Ozkan, S.; Akay, T.H.; Gultekin, B.; Aslim, E.; Arslan, A.; Ozdemir, B.H.; Becit, N.; Tasdelen, A. Atherosclerosis of Radial and Internal Thoracic Arteries Used in Coronary Bypass: Atherosclerosis in Arterial Grafts. J. Card. Surg. 2007, 22, 385–389. [Google Scholar] [CrossRef]
- Yanagawa, B.; Algarni, K.D.; Yau, T.M.; Rao, V.; Brister, S.J. Improving results for coronary artery bypass graft surgery in the elderly. Eur. J. Cardio-Thoracic. Surg. 2012, 42, 507–512. [Google Scholar] [CrossRef][Green Version]
- ElBardissi, A.W.; Aranki, S.F.; Sheng, S.; O’Brien, S.M.; Greenberg, C.C.; Gammie, J.S. Trends in isolated coronary artery bypass grafting: An analysis of the Society of Thoracic Surgeons adult cardiac surgery database. J. Thorac. Cardiovasc. Surg. 2012, 143, 273–281. [Google Scholar] [CrossRef]
- Biglioli, P.; Cannata, A.; Alamanni, F.; Naliato, M.; Porqueddu, M.; Zanobini, M.; Tremoli, E.; Parolari, A. Biological effects of off-pump vs. on-pump coronary artery surgery: Focus on inflammation, hemostasis and oxidative stress. Eur. J. Cardiothorac. Surg. 2003, 24, 260–269. [Google Scholar] [CrossRef]
- Sanders, J.; Hawe, E.; Brull, D.J.; Hubbart, C.; Lowe, G.D.O.; Rumley, A.; Humphries, S.E.; Montgomery, H.E. Higher IL-6 levels but not IL6 −174G > C or −572G > C genotype are associated with post-operative complication following coronary artery bypass graft (CABG) surgery. Atherosclerosis 2009, 204, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Lobato, R.L.; White, W.D.; Mathew, J.P.; Newman, M.F.; Smith, P.K.; McCants, C.B.; Alexander, J.H.; Podgoreanu, M.V. Duke Perioperative Genetics and Safety Outcomes (PEGASUS) Investigative Team Thrombomodulin Gene Variants Are Associated With Increased Mortality After Coronary Artery Bypass Surgery in Replicated Analyses. Circulation 2011, 124, S143–S148. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hojo, Y.; Ikeda, U.; Takahashi, M.; Shimada, K. Interaction between monocytes and vascular smooth muscle cells enhances matrix metalloproteinase-1 production. J. Cardiovasc. Pharmacol. 2000, 36, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Ascione, R.; Lloyd, C.T.; Underwood, M.J.; Lotto, A.A.; Pitsis, A.A.; Angelini, G.D. Inflammatory response after coronary revascularization with or without cardiopulmonary bypass. Ann. Thorac. Surg. 2000, 69, 1198–1204. [Google Scholar] [CrossRef]
- Espinoza-Lewis, R.A.; Yu, L.; He, F.; Liu, H.; Tang, R.; Shi, J.; Sun, X.; Martin, J.F.; Wang, D.; Yang, J.; et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009, 327, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Song, Y.; Huang, Z.; Zhang, Y.; Chen, Y. Genetic Regulation of Sinoatrial Node Development and Pacemaker Program in the Venous Pole. J. Cardiovasc. Dev. Dis. 2015, 2, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Evans, S.M.; Sun, Y. Development of the cardiac pacemaker. Cell. Mol. Life Sci. 2017, 74, 1247. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Wang, J.; Song, Y.; Yu, D.; Sun, C.; Liu, C.; Chen, F.; Zhang, Y.; Wang, F.; Harvey, R.P.; et al. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 2015, 142, 2521–2532. [Google Scholar] [CrossRef]
- Hoffmann, S.; Clauss, S.; Berger, I.M.; Weiß, B.; Montalbano, A.; Röth, R.; Bucher, M.; Klier, I.; Wakili, R.; Seitz, H.; et al. Coding and non-coding variants in the SHOX2 gene in patients with early-onset atrial fibrillation. Basic Res. Cardiol. 2016, 111, 36. [Google Scholar] [CrossRef]
- Soshnikova, N. Hox genes regulation in vertebrates. Dev. Dyn. 2014, 243, 49–58. [Google Scholar] [CrossRef]
- Pruett, N.D.; Visconti, R.P.; Jacobs, D.F.; Scholz, D.; McQuinn, T.; Sundberg, J.P.; Awgulewitsch, A. Evidence for Hox-specified positional identities in adult vasculature. BMC Dev. Biol. 2008, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Pruett, N.D.; Hajdu, Z.; Zhang, J.; Visconti, R.P.; Kern, M.J.; Wellik, D.M.; Majesky, M.W.; Awgulewitsch, A. Changing topographic Hox expression in blood vessels results in regionally distinct vessel wall remodeling. Biol. Open 2012, 1, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Roles of Runx2 in Skeletal Development. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 962, pp. 83–93. [Google Scholar]
- Kendrick, J.; Chonchol, M. The Role of Phosphorus in the Development and Progression of Vascular Calcification. Am. J. Kidney Dis. 2011, 58, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Román-García, P.; Carrillo-López, N.; Fernández-Martín, J.L.; Naves-Díaz, M.; Ruiz-Torres, M.P.; Cannata-Andía, J.B. High phosphorus diet induces vascular calcification, a related decrease in bone mass and changes in the aortic gene expression. Bone 2010, 46, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Moreno, J.M.; Muñoz-Castañeda, J.R.; Herencia, C.; de Oca, A.M.; Estepa, J.C.; Canalejo, R.; Rodríguez-Ortiz, M.E.; Perez-Martinez, P.; Aguilera-Tejero, E.; Canalejo, A.; et al. In vascular smooth muscle cells paricalcitol prevents phosphate-induced Wnt/β-catenin activation. Am. J. Physiol. Physiol. 2012, 303, F1136–F1144. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-E.; Chen, T.; Leaf, E.M.; Speer, M.Y.; Giachelli, C.M. Runx2 Expression in Smooth Muscle Cells Is Required for Arterial Medial Calcification in Mice. Am. J. Pathol. 2015, 185, 1958. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Sun, D.; Duan, Y.; Wen, P.; Dai, C.; Yang, J.; He, W. WNT/β-catenin signaling promotes VSMCs to osteogenic transdifferentiation and calcification through directly modulating Runx2 gene expression. Exp. Cell Res. 2016. [Google Scholar] [CrossRef]
- Langenfeld, E.M.; Langenfeld, J. Bone morphogenetic protein-2 stimulates angiogenesis in developing tumors. Mol. Cancer Res. 2004, 2, 141–149. [Google Scholar]
- Wiley, D.M.; Kim, J.-D.; Hao, J.; Hong, C.C.; Bautch, V.L.; Jin, S.-W. Distinct signalling pathways regulate sprouting angiogenesis from the dorsal aorta and the axial vein. Nat. Cell Biol. 2011, 13, 686–692. [Google Scholar] [CrossRef]
- Mouillesseaux, K.P.; Wiley, D.S.; Saunders, L.M.; Wylie, L.A.; Kushner, E.J.; Chong, D.C.; Citrin, K.M.; Barber, A.T.; Park, Y.; Kim, J.-D.; et al. Notch regulates BMP responsiveness and lateral branching in vessel networks via SMAD6. Nat. Commun. 2016, 7, 13247. [Google Scholar] [CrossRef]
- Uemura, M.T.; Ihara, M.; Maki, T.; Nakagomi, T.; Kaji, S.; Uemura, K.; Matsuyama, T.; Kalaria, R.N.; Kinoshita, A.; Takahashi, R. Pericyte-derived bone morphogenetic protein 4 underlies white matter damage after chronic hypoperfusion. Brain Pathol. 2018, 28, 521–535. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, T.K.; Aronow, B.J.; Tong, W.S.; Manka, D.; Tang, Y.; Bogdanov, V.Y.; Unruh, D.; Blomkalns, A.L.; Piegore, M.G.; Weintraub, D.S.; et al. Human coronary artery perivascular adipocytes overexpress genes responsible for regulating vascular morphology, inflammation, and hemostasis. Physiol. Genom. 2013, 45, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.M.; Yokota, T.; Maekawa, S.; Lü, P.; Zheng, Y.-W.; Tei, I.; Taniguchi, H.; Yokoyama, U.; Kato, T.; Minamisawa, S. Transcription profiles of endothelial cells in the rat ductus arteriosus during a perinatal period. PLoS ONE 2013, 8, e73685. [Google Scholar] [CrossRef] [PubMed]
- Deol, H.K.; Varghese, R.; Wagner, G.F.; DiMattia, G.E. Dynamic regulation of mouse ovarian stanniocalcin expression during gestation and lactation. Endocrinology 2000, 141, 3412–3421. [Google Scholar] [CrossRef] [PubMed]
- Yeung, B.H.Y.; Law, A.Y.S.; Wong, C.K.C. Evolution and roles of stanniocalcin. Mol. Cell. Endocrinol. 2012, 349, 272–280. [Google Scholar] [CrossRef]
- Johnston, J.; Ramos-Valdes, Y.; Stanton, L.-A.; Ladhani, S.; Beier, F.; Dimattia, G.E. Human stanniocalcin-1 or -2 expressed in mice reduces bone size and severely inhibits cranial intramembranous bone growth. Transgenic Res. 2010, 19, 1017–1039. [Google Scholar] [CrossRef] [PubMed]
- Law, A.Y.S.; Wong, C.K.C. Stanniocalcin-1 and -2 promote angiogenic sprouting in HUVECs via VEGF/VEGFR2 and angiopoietin signaling pathways. Mol. Cell. Endocrinol. 2013, 374, 73–81. [Google Scholar] [CrossRef]
- Li, K.; Dong, D.; Yao, L.; Dai, D.; Gu, X.; Guo, L. Identification of STC1 as an β-amyloid activated gene in human brain microvascular endothelial cells using cDNA microarray. Biochem. Biophys. Res. Commun. 2008, 376, 399–403. [Google Scholar] [CrossRef]
- Hägg, P.; Rehn, M.; Huhtala, P.; Väisänen, T.; Tamminen, M.; Pihlajaniemi, T. Type XIII Collagen Is Identified as a Plasma Membrane Protein. J. Biol. Chem. 1998, 273, 15590–15597. [Google Scholar] [CrossRef]
- Hägg, P.; Väisänen, T.; Tuomisto, A.; Rehn, M.; Tu, H.; Huhtala, P.; Eskelinen, S.; Pihlajaniemi, T. Type XIII collagen: A novel cell adhesion component present in a range of cell-matrix adhesions and in the intercalated discs between cardiac muscle cells. Matrix Biol. 2001, 19, 727–742. [Google Scholar] [CrossRef]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Toritsuka, M.; Ohnishi, S.; Shimada, K.; Furuya, H.; Khadka, V.S.; Deng, Y.; et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017, 8, 36099–36114. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Kato, K.; Yokoi, K.; Oguri, M.; Watanabe, S.; Metoki, N.; Yoshida, H.; Satoh, K.; Aoyagi, Y.; Nozawa, Y.; et al. Association of genetic variants with hemorrhagic stroke in Japanese individuals. Int. J. Mol. Med. 2010, 25, 649–656. [Google Scholar] [PubMed]
- Gimbrone, M.A.; García-Cardeña, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Doherty, T.M.; Asotra, K.; Fitzpatrick, L.A.; Qiao, J.-H.; Wilkin, D.J.; Detrano, R.C.; Dunstan, C.R.; Shah, P.K.; Rajavashisth, T.B. Calcification in atherosclerosis: Bone biology and chronic inflammation at the arterial crossroads. Proc. Natl. Acad. Sci. USA 2003, 100, 11201–11206. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, M.V.; Tarigopula, M.; Mintz, G.S.; Maehara, A.; Stone, G.W.; Généreux, P. Coronary Artery Calcification. J. Am. Coll. Cardiol. 2014, 63, 1703–1714. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, F.; Sakakura, K.; Yahagi, K.; Joner, M.; Virmani, R. Has Our Understanding of Calcification in Human Coronary Atherosclerosis Progressed? Arterioscler. Thromb. Vasc. Biol. 2014, 34, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Motwani, J.G.; Topol, E.J. Aortocoronary saphenous vein graft disease: Pathogenesis, predisposition, and prevention. Circulation 1998, 97, 916–931. [Google Scholar] [CrossRef]
- Lardenoye, J.H.P.; de Vries, M.R.; Löwik, C.W.G.M.; Xu, Q.; Dhore, C.R.; Cleutjens, J.P.M.; van Hinsbergh, V.W.M.; van Bockel, J.H.; Quax, P.H.A. Accelerated atherosclerosis and calcification in vein grafts: A study in APOE*3 Leiden transgenic mice. Circ. Res. 2002, 91, 577–584. [Google Scholar] [CrossRef]
- Nasso, G.; Anselmi, A.; Modugno, P.; Alessandrini, F. Minimally invasive saphenous vein harvesting guided by preoperative echotomography: Results of a prospective randomized study. Interact. Cardiovasc. Thorac. Surg. 2005, 4, 464–468. [Google Scholar] [CrossRef][Green Version]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Nawrocki, M.J.; Celichowski, P.; Jankowski, M.; Kranc, W.; Bryja, A.; Borys-Wójcik, S.; Jeseta, M.; Antosik, P.; Bukowska, D.; Bruska, M.; et al. Ontology groups representing angiogenesis and blood vessels development are highly up-regulated during porcine oviductal epithelial cells long-term real-time proliferation—A primary cell culture approach. Med. J. Cell Biol. 2018, 6, 186–194. [Google Scholar] [CrossRef]
- Budna, J.; Celichowski, P.; Bryja, A.; Jeseta, M.; Jankowski, M.; Bukowska, D.; Antosik, P.; Nowicki, A.; Brüssow, K.P.; Bruska, M.; et al. Expression Changes in Fatty acid Metabolic Processrelated Genes in Porcine Oocytes During in Vitro Maturation. Med. J. Cell Biol. 2018, 6, 48–54. [Google Scholar] [CrossRef]
- Borys-Wójcik, S.; Kocherova, I.; Celichowski, P.; Popis, M.; Jeseta, M.; Bukowska, D.; Antosik, P.; Nowicki, M.; Kempisty, B. Protein oligomerization is the biochemical process highly up-regulated in porcine oocytes before in vitro maturation (IVM). Med. J. Cell Biol. 2018, 6, 155–162. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis: Figure 1. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
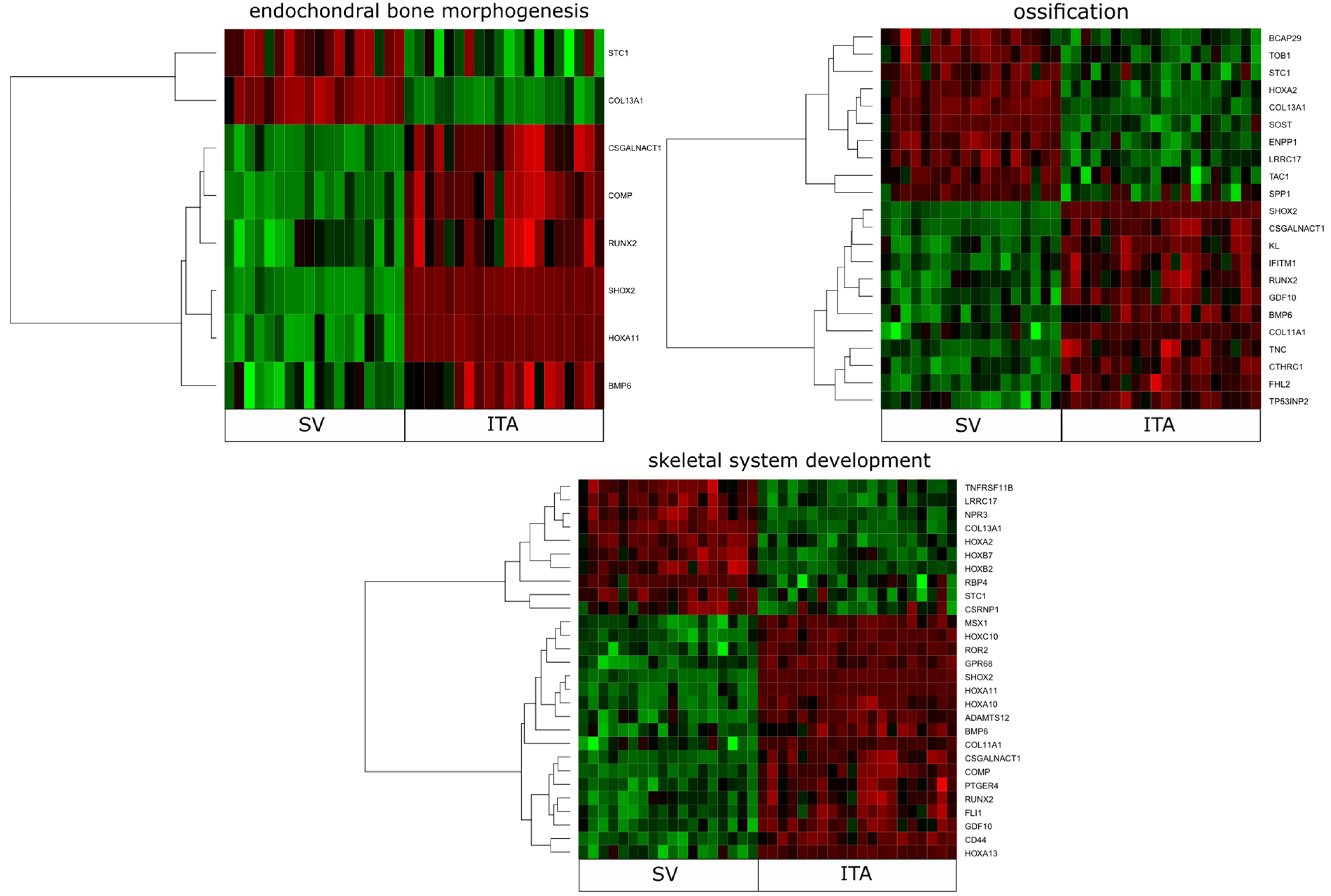
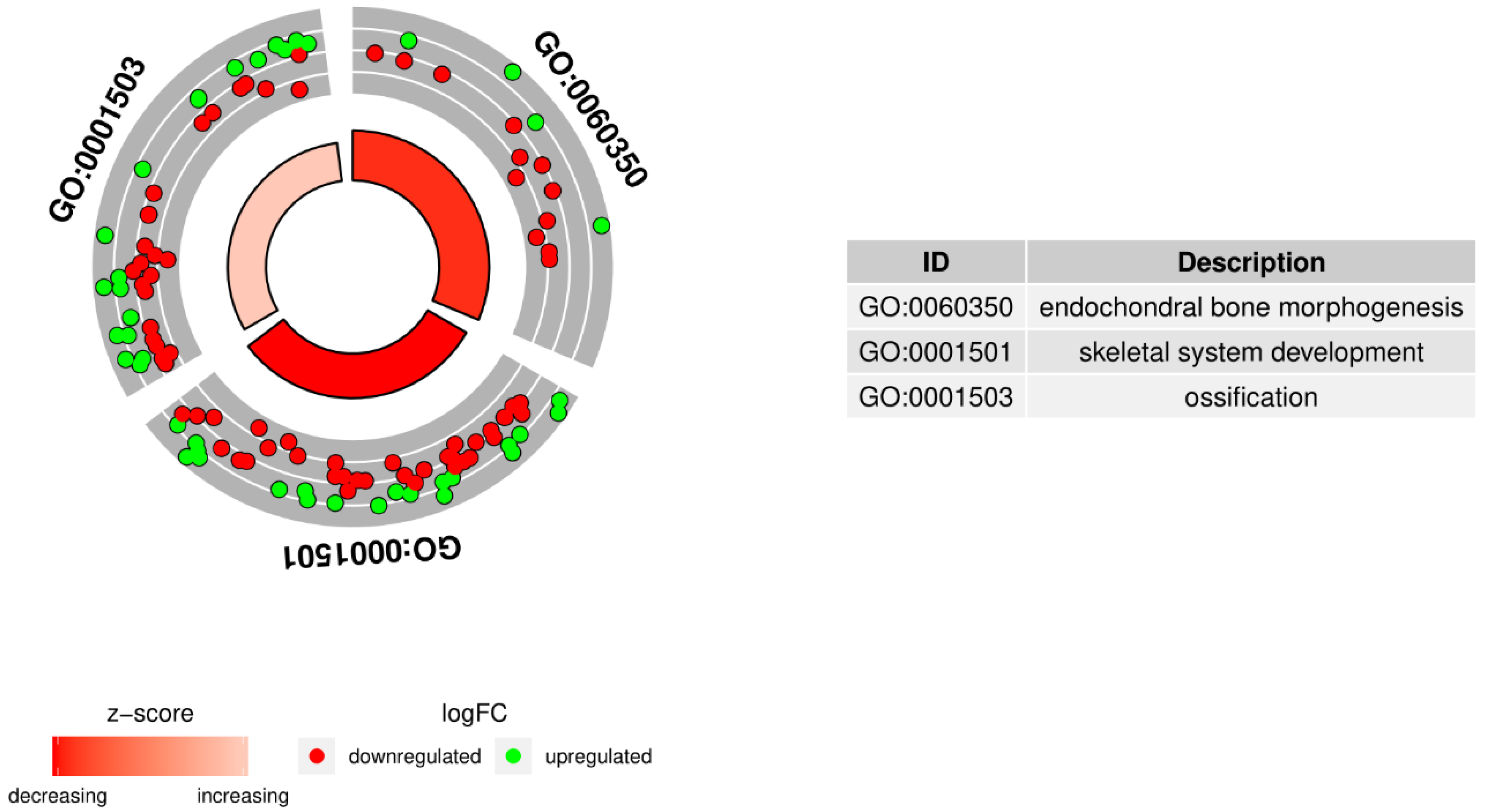
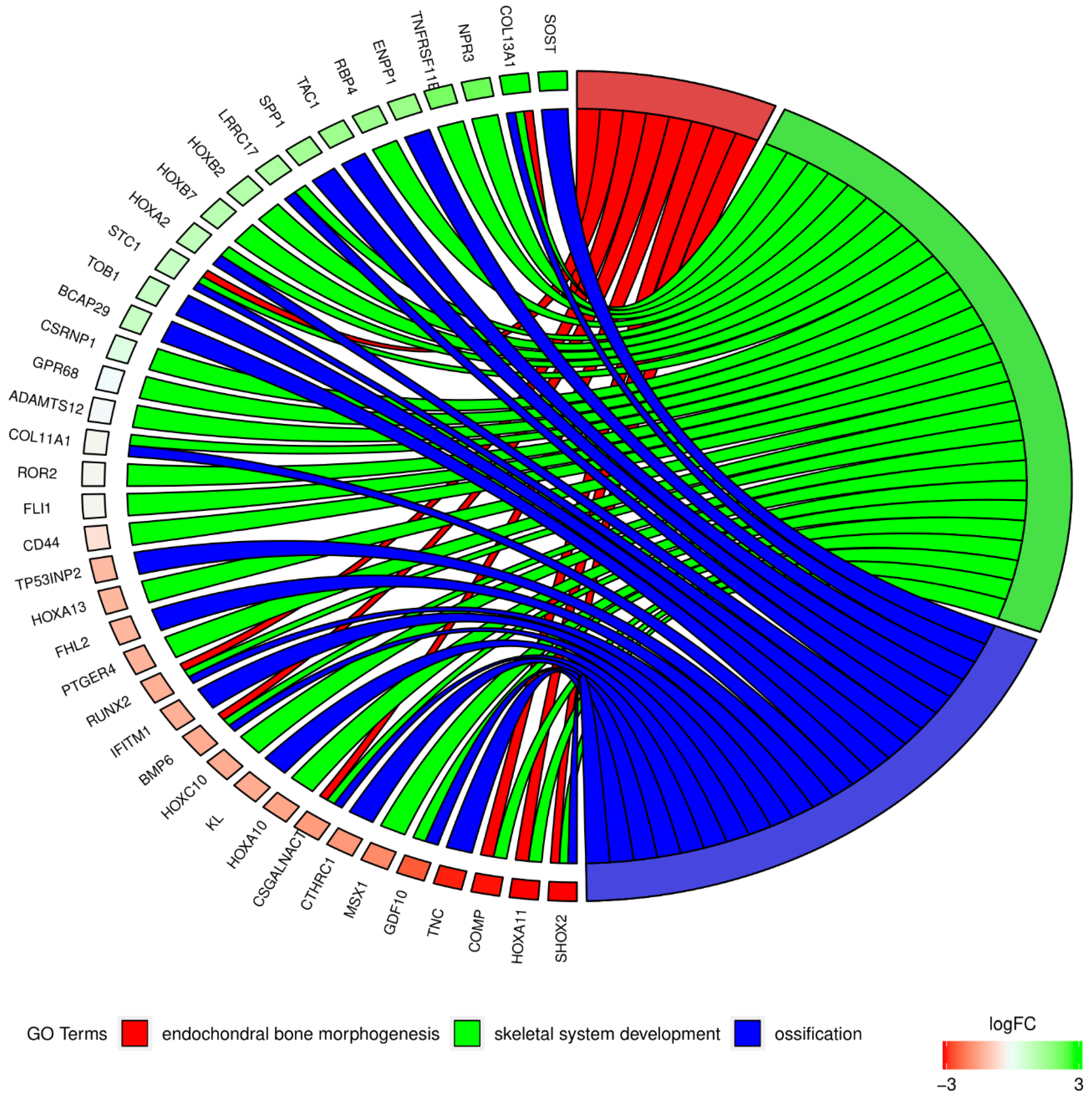
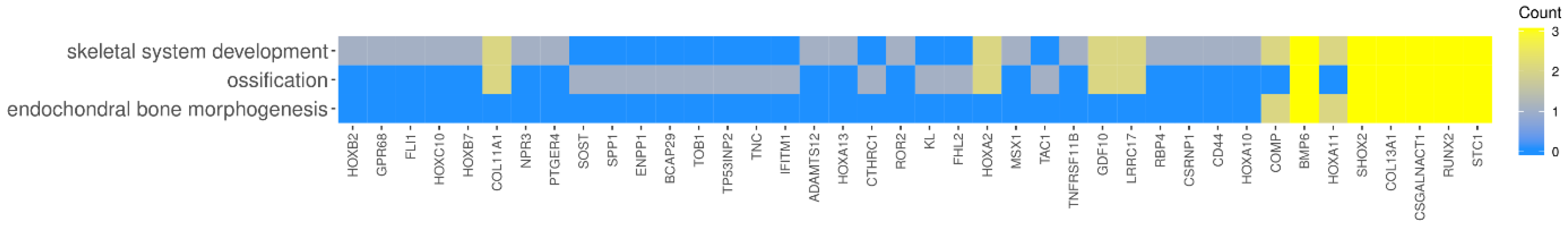


| Gene Symbol | Ratio | Adjusted p Value | Entrez Gene ID |
|---|---|---|---|
| SHOX2 | −22.8240429 | 1.03 × 10−27 | 6474 |
| HOXA11 | −9.830934198 | 7.60 × 10−17 | 3207 |
| COMP | −7.773021124 | 3.33 × 10−13 | 1311 |
| TNC | −7.430294969 | 1.88 × 10−08 | 3371 |
| GDF10 | −5.309648509 | 2.16 × 10−08 | 2662 |
| MSX1 | −3.455950944 | 9.17 × 10−13 | 4487 |
| CTHRC1 | −3.018066571 | 5.20 × 10−11 | 115908 |
| CSGALNACT1 | −2.881617301 | 2.10 × 10−12 | 55790 |
| HOXA10 | −2.63199568 | 1.36 × 10−12 | 3206 |
| KL | −2.458105612 | 2.59 × 10−08 | 9365 |
| HOXC10 | −2.457042843 | 7.09 × 10−15 | 3226 |
| BMP6 | −2.45216404 | 2.98 × 10−07 | 654 |
| IFITM1 | −2.349841846 | 2.37 × 10−08 | 8519 |
| RUNX2 | −2.331169268 | 5.34 × 10−07 | 860 |
| PTGER4 | −2.280369849 | 7.12 × 10−09 | 5734 |
| FHL2 | −2.216349206 | 6.35 × 10−10 | 2274 |
| HOXA13 | −2.166453813 | 2.16 × 10−10 | 3209 |
| TP53INP2 | −2.127154082 | 3.09 × 10−08 | 58476 |
| CD44 | −1.387817152 | 0.006540227 | 960 |
| FLI1 | –1.131519079 | 0.002636506 | 2313 |
| ROR2 | −1.128865754 | 0.004346143 | 4920 |
| COL11A1 | −1.12582207 | 0.000190115 | 1301 |
| ADAMTS12 | −1.054115576 | 0.188720272 | 81792 |
| GPR68 | −1.028071249 | 0.512393039 | 8111 |
| CSRNP1 | 1.338139052 | 0.026644084 | 64651 |
| BCAP29 | 1.823380549 | 3.52 × 10−07 | 55973 |
| TOB1 | 1.841885719 | 1.99 × 10−08 | 10140 |
| STC1 | 1.845964521 | 0.000109973 | 6781 |
| HOXA2 | 2.01745347 | 5.20 × 10−11 | 3199 |
| HOXB7 | 2.230080809 | 3.31 × 10−09 | 3217 |
| HOXB2 | 2.433099517 | 5.86 × 10−11 | 3212 |
| LRRC17 | 2.543411445 | 2.26 × 10−10 | 10234 |
| SPP1 | 2.96640349 | 0.001152397 | 6696 |
| TAC1 | 3.167584851 | 4.92 × 10−05 | 6863 |
| RBP4 | 3.228449588 | 3.61 × 10−05 | 5950 |
| ENPP1 | 3.320786599 | 4.83 × 10−10 | 5167 |
| TNFRSF11B | 4.741562441 | 1.54 × 10−10 | 4982 |
| NPR3 | 5.295452922 | 2.59 × 10−12 | 4883 |
| COL13A1 | 8.475105942 | 8.83 × 10−18 | 1305 |
| SOST | 12.36428646 | 1.01 × 10−10 | 50964 |
| Gene | Primer Sequence (5′–3′) | Product Size (bp) | |
|---|---|---|---|
| SHOX2 | F | GAAGGCCAGACCAAAATCAA | 234 |
| R | GGCCCCTATGAGAACACCTT | ||
| RUNX2 | F | GGACGAGGCAAGAGTTTCAC | 165 |
| R | GAGGCGGTCAGAGAACAAAC | ||
| BMP6 | F | AAGAAGGCTGGCTGGAATTT | 170 |
| R | GAAGGGCTGCTTGTCGTAAG | ||
| STC1 | F | TGATCAGTGCTTCTGCAACC | 242 |
| R | GACGAATGCTTTTCCCTGAG | ||
| CSGALNACT1 | F | CAGAAAGGGACAAAGGGACA | 243 |
| R | TGAGATGGACTCTCCCATCC | ||
| COL13A1 | F | CAAAGGGAGAAGCAGGTGTC | 175 |
| R | TCCTGGAGAGCCTCATTGAT | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawrocki, M.J.; Perek, B.; Sujka-Kordowska, P.; Konwerska, A.; Kałużna, S.; Zawierucha, P.; Bruska, M.; Zabel, M.; Jemielity, M.; Nowicki, M.; et al. Differences in Expression of Genes Involved in Bone Development and Morphogenesis in the Walls of Internal Thoracic Artery and Saphenous Vein Conduits May Provide Markers Useful for Evaluation Graft Patency. Int. J. Mol. Sci. 2019, 20, 4890. https://doi.org/10.3390/ijms20194890
Nawrocki MJ, Perek B, Sujka-Kordowska P, Konwerska A, Kałużna S, Zawierucha P, Bruska M, Zabel M, Jemielity M, Nowicki M, et al. Differences in Expression of Genes Involved in Bone Development and Morphogenesis in the Walls of Internal Thoracic Artery and Saphenous Vein Conduits May Provide Markers Useful for Evaluation Graft Patency. International Journal of Molecular Sciences. 2019; 20(19):4890. https://doi.org/10.3390/ijms20194890
Chicago/Turabian StyleNawrocki, Mariusz J., Bartłomiej Perek, Patrycja Sujka-Kordowska, Aneta Konwerska, Sandra Kałużna, Piotr Zawierucha, Małgorzata Bruska, Maciej Zabel, Marek Jemielity, Michał Nowicki, and et al. 2019. "Differences in Expression of Genes Involved in Bone Development and Morphogenesis in the Walls of Internal Thoracic Artery and Saphenous Vein Conduits May Provide Markers Useful for Evaluation Graft Patency" International Journal of Molecular Sciences 20, no. 19: 4890. https://doi.org/10.3390/ijms20194890
APA StyleNawrocki, M. J., Perek, B., Sujka-Kordowska, P., Konwerska, A., Kałużna, S., Zawierucha, P., Bruska, M., Zabel, M., Jemielity, M., Nowicki, M., Kempisty, B., & Malińska, A. (2019). Differences in Expression of Genes Involved in Bone Development and Morphogenesis in the Walls of Internal Thoracic Artery and Saphenous Vein Conduits May Provide Markers Useful for Evaluation Graft Patency. International Journal of Molecular Sciences, 20(19), 4890. https://doi.org/10.3390/ijms20194890







