Recent Advances in Our Understanding of the Link between the Intestinal Microbiota and Systemic Lupus Erythematosus
Abstract
1. Introduction
2. The Role of the Gut Microbiota in Human Health and Disease
3. The Gut Microbiome in Murine Lupus
4. The Gut Microbiome in Human Lupus
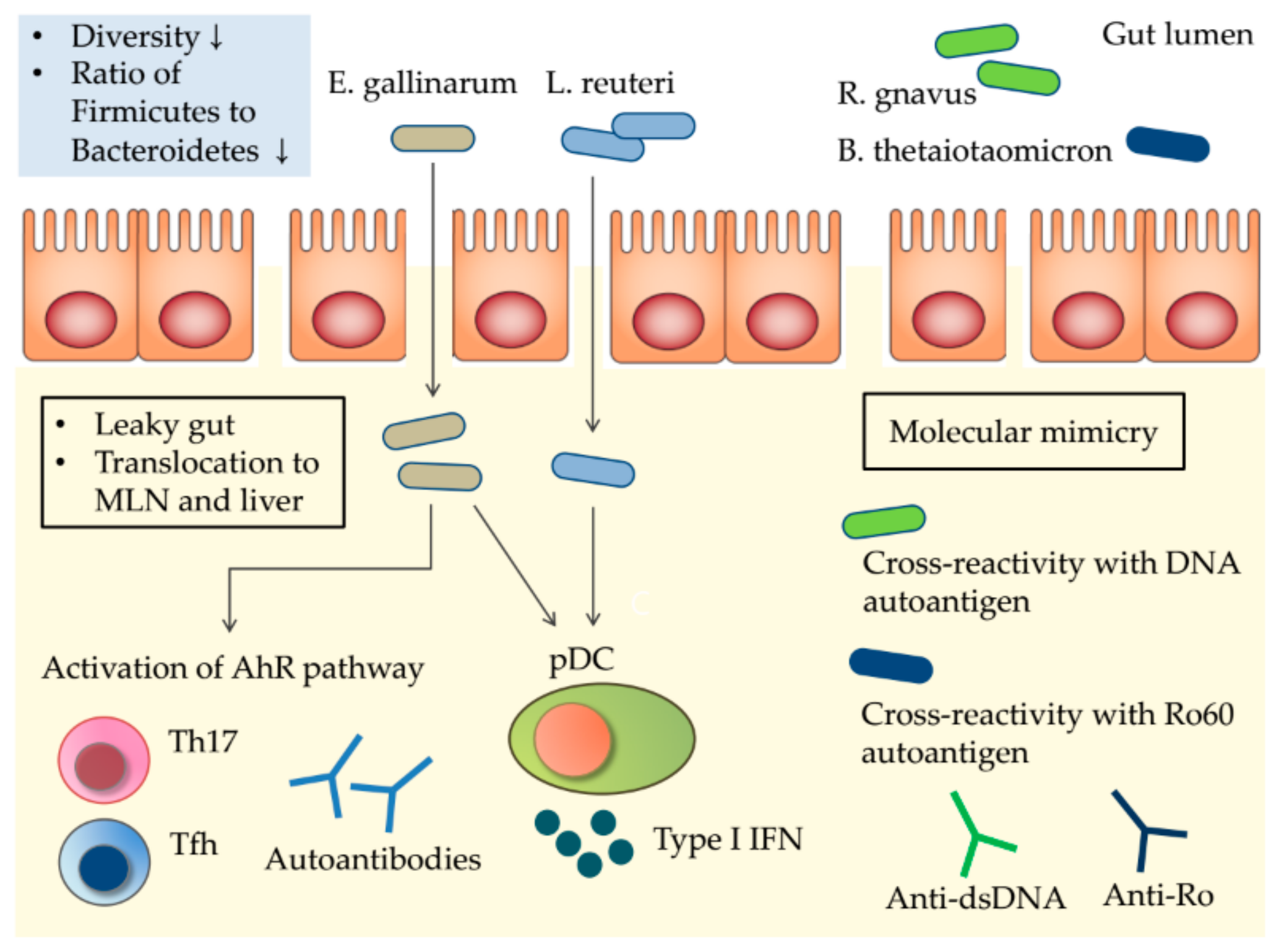
5. Potential Mechanisms Linking the Gut Microbiota to SLE
5.1. The Leaky Gut and Gut Microbiota Translocation
5.2. Molecular Mimicry
5.3. Sexual Dimorphism of the Gut Microbiota
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| rRNA | Ribosomal ribonucleic acid |
| SLEDAI | Systemic lupus erythematosus disease activity index |
References
- Tsokos, G.C.; Lo, M.S.; Costa Reis, P.; Sullivan, K.E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2016, 12, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Barrat, F.J.; Meeker, T.; Gregorio, J.; Chan, J.H.; Uematsu, S.; Akira, S.; Chang, B.; Duramad, O.; Coffman, R.L. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 2005, 202, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Lovgren, T.; Eloranta, M.L.; Bave, U.; Alm, G.V.; Ronnblom, L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004, 50, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Jego, G.; Palucka, A.K.; Blanck, J.P.; Chalouni, C.; Pascual, V.; Banchereau, J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity 2003, 19, 225–234. [Google Scholar] [CrossRef]
- Le Bon, A.; Schiavoni, G.; D’Agostino, G.; Gresser, I.; Belardelli, F.; Tough, D.F. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 2001, 14, 461–470. [Google Scholar] [CrossRef]
- Choi, J.Y.; Ho, J.H.; Pasoto, S.G.; Bunin, V.; Kim, S.T.; Carrasco, S.; Borba, E.F.; Goncalves, C.R.; Costa, P.R.; Kallas, E.G.; et al. Circulating follicular helper-like T cells in systemic lupus erythematosus: Association with disease activity. Arthritis Rheumatol. 2015, 67, 988–999. [Google Scholar] [CrossRef]
- Von Spee-Mayer, C.; Siegert, E.; Abdirama, D.; Rose, A.; Klaus, A.; Alexander, T.; Enghard, P.; Sawitzki, B.; Hiepe, F.; Radbruch, A.; et al. Low-dose interleukin-2 selectively corrects regulatory T cell defects in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2016, 75, 1407–1415. [Google Scholar] [CrossRef]
- Crispin, J.C.; Oukka, M.; Bayliss, G.; Cohen, R.A.; Van Beek, C.A.; Stillman, I.E.; Kyttaris, V.C.; Juang, Y.T.; Tsokos, G.C. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 2008, 181, 8761–8766. [Google Scholar] [CrossRef]
- Sieling, P.A.; Porcelli, S.A.; Duong, B.T.; Spada, F.; Bloom, B.R.; Diamond, B.; Hahn, B.H. Human double-negative T cells in systemic lupus erythematosus provide help for IgG and are restricted by CD1c. J. Immunol. 2000, 165, 5338–5344. [Google Scholar] [CrossRef]
- Berggren, O.; Hagberg, N.; Weber, G.; Alm, G.V.; Ronnblom, L.; Eloranta, M.L. B lymphocytes enhance interferon-alpha production by plasmacytoid dendritic cells. Arthritis Rheum. 2012, 64, 3409–3419. [Google Scholar] [CrossRef]
- Leonard, D.; Eloranta, M.L.; Hagberg, N.; Berggren, O.; Tandre, K.; Alm, G.; Ronnblom, L. Activated T cells enhance interferon-alpha production by plasmacytoid dendritic cells stimulated with RNA-containing immune complexes. Ann. Rheum. Dis. 2016, 75, 1728–1734. [Google Scholar] [CrossRef] [PubMed]
- Deapen, D.; Escalante, A.; Weinrib, L.; Horwitz, D.; Bachman, B.; Roy-Burman, P.; Walker, A.; Mack, T.M. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992, 35, 311–318. [Google Scholar] [PubMed]
- Barbhaiya, M.; Costenbader, K.H. Environmental exposures and the development of systemic lupus erythematosus. Curr. Opin. Rheumatol. 2016, 28, 497–505. [Google Scholar] [CrossRef] [PubMed]
- James, J.A.; Neas, B.R.; Moser, K.L.; Hall, T.; Bruner, G.R.; Sestak, A.L.; Harley, J.B. Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum. 2001, 44, 1122–1126. [Google Scholar] [CrossRef]
- Wolf, S.J.; Estadt, S.N.; Gudjonsson, J.E.; Kahlenberg, J.M. Human and Murine Evidence for Mechanisms Driving Autoimmune Photosensitivity. Front. Immunol. 2018, 9, 2430. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.J.; Estadt, S.N.; Theros, J.; Moore, T.; Ellis, J.; Liu, J.; Reed, T.J.; Jacob, C.O.; Gudjonsson, J.E.; Kahlenberg, J.M. Ultraviolet light induces increased T cell activation in lupus-prone mice via type I IFN-dependent inhibition of T regulatory cells. J. Autoimmun. 2019. [Google Scholar] [CrossRef]
- Grimaldi, C.M. Sex and systemic lupus erythematosus: The role of the sex hormones estrogen and prolactin on the regulation of autoreactive B cells. Curr. Opin. Rheumatol. 2006, 18, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Vera-Lastra, O.; Jara, L.J.; Espinoza, L.R. Prolactin and autoimmunity. Autoimmun. Rev. 2002, 1, 360–364. [Google Scholar] [CrossRef]
- Bernier, M.O.; Mikaeloff, Y.; Hudson, M.; Suissa, S. Combined oral contraceptive use and the risk of systemic lupus erythematosus. Arthritis Rheum. 2009, 61, 476–481. [Google Scholar] [CrossRef]
- Costenbader, K.H.; Feskanich, D.; Stampfer, M.J.; Karlson, E.W. Reproductive and menopausal factors and risk of systemic lupus erythematosus in women. Arthritis Rheum. 2007, 56, 1251–1262. [Google Scholar] [CrossRef]
- Rosenbaum, J.T.; Silverman, G.J. The Microbiome and Systemic Lupus Erythematosus. New Engl. J. Med. 2018, 378, 2236–2237. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.J. The microbiome in SLE pathogenesis. Nat. Rev. Rheumatol. 2019, 15, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P.; De Paz, B.; Rodriguez-Carrio, J.; Hevia, A.; Sanchez, B.; Margolles, A.; Suarez, A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016, 6, 24072. [Google Scholar] [CrossRef] [PubMed]
- Van Praet, J.T.; Donovan, E.; Vanassche, I.; Drennan, M.B.; Windels, F.; Dendooven, A.; Allais, L.; Cuvelier, C.A.; Van de Loo, F.; Norris, P.S.; et al. Commensal microbiota influence systemic autoimmune responses. EMBO J. 2015, 34, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Neish, A.S. Microbes in gastrointestinal health and disease. Gastroenterology 2009, 136, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Baumler, A.J. The germ-organ theory of non-communicable diseases. Nat. Rev. Microbiol. 2018, 16, 103–110. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chaudhary, N.; Yang, N.; Granato, A.; Turner, J.A.; Howard, S.L.; Devereaux, C.; Zuo, T.; Shrestha, A.; Goel, R.R.; et al. Microbial symbionts regulate the primary Ig repertoire. J. Exp. Med. 2018, 215, 1397–1415. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108, 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Garrett, W.S.; Gallini, C.A.; Yatsunenko, T.; Michaud, M.; DuBois, A.; Delaney, M.L.; Punit, S.; Karlsson, M.; Bry, L.; Glickman, J.N.; et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010, 8, 292–300. [Google Scholar] [CrossRef]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008, 455, 1109–1113. [Google Scholar] [CrossRef]
- Mathis, D.; Benoist, C. The influence of the microbiota on type-1 diabetes: On the threshold of a leap forward in our understanding. Immunol. Rev. 2012, 245, 239–249. [Google Scholar] [CrossRef]
- Bjorksten, B.; Naaber, P.; Sepp, E.; Mikelsaar, M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 1999, 29, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Demoor, T.; Rauch, M.; Faruqi, A.A.; Jang, S.; Johnson, C.C.; Boushey, H.A.; Zoratti, E.; Ownby, D.; Lukacs, N.W.; et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Van Nimwegen, F.A.; Penders, J.; Stobberingh, E.E.; Postma, D.S.; Koppelman, G.H.; Kerkhof, M.; Reijmerink, N.E.; Dompeling, E.; Van den Brandt, P.A.; Ferreira, I.; et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 2011, 128, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Ivanov, I.I.; Darce, J.; Hattori, K.; Shima, T.; Umesaki, Y.; Littman, D.R.; Benoist, C.; Mathis, D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010, 32, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, J.W.; You, H.J.; Park, S.J.; Lee, J.; Ju, J.H.; Park, M.S.; Jin, H.; Cho, M.L.; Kwon, B.; et al. Gut microbial composition and function are altered in patients with early rheumatoid arthritis. J. Clin. Med. 2019, 8, 693. [Google Scholar] [CrossRef]
- Berer, K.; Mues, M.; Koutrolos, M.; Rasbi, Z.A.; Boziki, M.; Johner, C.; Wekerle, H.; Krishnamoorthy, G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–541. [Google Scholar] [CrossRef]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108, 4615–4622. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nature reviews. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Fukunaga, R.; Brannan, C.I.; Copeland, N.G.; Jenkins, N.A.; Nagata, S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature 1992, 356, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liao, X.; Sparks, J.B.; Luo, X.M. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 2014, 80, 7551–7560. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R.; Kleerebezem, M.; Kopp, M.V.; Rescigno, M. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 2012, 12, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef]
- Mu, Q.; Zhang, H.; Liao, X.; Lin, K.; Liu, H.; Edwards, M.R.; Ahmed, S.A.; Yuan, R.; Li, L.; Cecere, T.E.; et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.S.; Eisenberg, R.A.; Theofilopoulos, A.N.; Izui, S.; Wilson, C.B.; McConahey, P.J.; Murphy, E.D.; Roths, J.B.; Dixon, F.J. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J. Exp. Med. 1978, 148, 1198–1215. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.M.; Edwards, M.R.; Mu, Q.; Yu, Y.; Vieson, M.D.; Reilly, C.M.; Ahmed, S.A.; Bankole, A.A. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Zegarra-Ruiz, D.F.; El Beidaq, A.; Iniguez, A.J.; Lubrano Di Ricco, M.; Manfredo Vieira, S.; Ruff, W.E.; Mubiru, D.; Fine, R.L.; Sterpka, J.; Greiling, T.M.; et al. A diet-Sensitive commensal lactobacillus strain mediates TLR7-Dependent Systemic Autoimmunity. Cell Host Microbe 2019, 25, 113–127. [Google Scholar] [CrossRef]
- Richard, M.L.; Gilkeson, G. Mouse models of lupus: What they tell us and what they don’t. Lupus Sci. Med. 2018, 5, e000199. [Google Scholar] [CrossRef]
- Maeda, Y.; Takeda, K. Role of Gut Microbiota in Rheumatoid Arthritis. J. Clin. Med. 2017, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.M.; Gaudreau, M.C.; Al-Gadban, M.M.; Gudi, R.; Vasu, C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin. Exp. Immunol. 2015, 181, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Hevia, A.; Milani, C.; Lopez, P.; Cuervo, A.; Arboleya, S.; Duranti, S.; Turroni, F.; Gonzalez, S.; Suarez, A.; Gueimonde, M.; et al. Intestinal dysbiosis associated with systemic lupus erythematosus. Microbiology 2014, 5, e01548-14. [Google Scholar] [CrossRef]
- He, Z.; Shao, T.; Li, H.; Xie, Z.; Wen, C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016, 8, 64. [Google Scholar] [CrossRef]
- Greiling, T.M.; Dehner, C.; Chen, X.; Hughes, K.; Iniguez, A.J.; Boccitto, M.; Ruiz, D.Z.; Renfroe, S.C.; Vieira, S.M.; Ruff, W.E.; et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Van der Meulen, T.A.; Harmsen, H.J.M.; Vila, A.V.; Kurilshikov, A.; Liefers, S.C.; Zhernakova, A.; Fu, J.; Wijmenga, C.; Weersma, R.K.; De Leeuw, K.; et al. Shared gut, but distinct oral microbiota composition in primary Sjogren’s syndrome and systemic lupus erythematosus. J. Autoimmun. 2019, 97, 77–87. [Google Scholar] [CrossRef]
- Azzouz, D.; Omarbekova, A.; Heguy, A.; Schwudke, D.; Gisch, N.; Rovin, B.H.; Caricchio, R.; Buyon, J.P.; Alekseyenko, A.V.; Silverman, G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019, 78, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Breban, M.; Tap, J.; Leboime, A.; Said-Nahal, R.; Langella, P.; Chiocchia, G.; Furet, J.P.; Sokol, H. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann. Rheum. Dis. 2017, 76, 1614–1622. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Hirano, T. Molecular regulation of B lymphocyte response. Annual review of immunology 1988, 6, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Kishimoto, T. IL-6: Regulator of Treg/Th17 balance. Eur. J. Immunol. 2010, 40, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- McClain, M.T.; Heinlen, L.D.; Dennis, G.J.; Roebuck, J.; Harley, J.B.; James, J.A. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat. Med. 2005, 11, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Szymula, A.; Rosenthal, J.; Szczerba, B.M.; Bagavant, H.; Fu, S.M.; Deshmukh, U.S. T cell epitope mimicry between Sjogren’s syndrome Antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin. Immunol. 2014, 152, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; Von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [PubMed]
| Study | Mice | Results |
|---|---|---|
| Zhang et al. 2014 [53] | MRL/lpr mice | Lactobacillaceae ↓, Lachnospiraceae ↑, Ruminococcaceae ↑, Rikenellaceae (genus Alistipes) ↑ in lupus-prone mice compared to control mice. Gut microbiota of lupus-prone mice distinct between the sexes. Higher abundance of Lachnospiraceae and lower abundance of Lactobacillaceae associated with the lupus disease indices of lymphadenopathy and glomerulonephritis. |
| Mu et al. 2017 [56] | MRL/lpr mice | Lactobacillales ↓ and a leaky gut evident in lupus-prone mice. Lactobacillus treatment enhanced gut mucosal barrier, suppressed gut inflammation, and attenuated lupus nephritis. |
| Luo et al. 2018 [58] | NZB/W F1 mice | Gut microbiota changed before and after disease onset in lupus-prone mice. Species in the genera Clostridium, Dehalobacterium, Lactobacillus, Oscillospira, Dorea, Bilophila, AF12 and an unnamed genus within the family Ruminococcaceae increased during lupus progression. The relative abundance of Lactobacillus tended to be associated with poorer renal function and more extensive systemic autoimmunity. |
| Johnson et al. 2015 [62] | SNF1 mice | Abundant Rikenellaceae associated with more rapid lupus progression. Giving mice acidic water delayed lupus development compared to intake of neutral water. |
| Manfredo Vieira et al. 2018 [63] | (NZW × BXSB) F1 mice | Enterococcus gallinarum detected in feces, the small intestine, and the liver via culture-based PCR. Translocation of the gut commensal E. gallinarum to the liver triggered type I interferon expression and anti-dsDNA antibody production. |
| Zegarra-Ruiz et al. 2019 [59] | TLR7-dependent spontaneous and induced mice | Enrichment of fecal Lactobacillus reuteri and translocation of L. reuteri in lupus-prone mice. L. reuteri exacerbated lupus by inducing the pDC activation, type I interferon expression, and enhancing glomerulonephritis. |
| Study | Patients [F:M] | Results |
|---|---|---|
| Hevia et al. 2014 [67] | 20 SLE patients [20:0] | Bacteriodetes ↑ and Firmicutes/Bacteroidetes ratio ↓ in SLE patients compared to healthy controls. |
| He et al. 2016 [68] | 45 SLE patients [45:0] | Firmicutes ↓, Bacteriodetes ↑, and Firmicutes/Bacteroidetes ratio ↓ in SLE patients compared to healthy controls. Rhodococcus, Eggerthella, Klebsiella, Prevotella, Eubacterium, Flavonifractor, and incertae sedis enriched and Dialister and Pseudobutyrivibrio depleted in SLE patients. |
| Luo et al. 2018 [58] | 14 SLE patients [10:4] | Firmicutes/Bacteroidetes ratio not significantly different between SLE patients and healthy controls. Proteobacteria ↑, Odoribacter ↓, and Blautia ↑ in SLE patients. |
| Greiling et al. 2018 [69] | 16 SLE and 2 SCLE patients [17:1] | Firmicutes/Bacteroidetes ratio ↓ in SLE patients compared to healthy controls. Commensal bacteria containing Ro60 orthologs common in humans, including SLE patients. |
| van der Meulen et al. 2019 [70] | 30 SLE patients [28:2] | Firmicutes/Bacteroidetes ratio ↓, Bacteroidetes ↑, Bacteroides ↑, Alistipes ↑, Proteobacteria ↑ in SLE patients compared to healthy controls. |
| Zegarra-Ruiz et al. 2019 [59] | 28 SLE patients | Lactobacillus spp. enriched in SLE patients compared to healthy controls. |
| Azzouz et al. 2019 [71] | 61 SLE patients [61:0] | The abundance of Ruminococcus gnavus associated with lupus disease activity and lupus nephritis. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-W.; Kwok, S.-K.; Choe, J.-Y.; Park, S.-H. Recent Advances in Our Understanding of the Link between the Intestinal Microbiota and Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2019, 20, 4871. https://doi.org/10.3390/ijms20194871
Kim J-W, Kwok S-K, Choe J-Y, Park S-H. Recent Advances in Our Understanding of the Link between the Intestinal Microbiota and Systemic Lupus Erythematosus. International Journal of Molecular Sciences. 2019; 20(19):4871. https://doi.org/10.3390/ijms20194871
Chicago/Turabian StyleKim, Ji-Won, Seung-Ki Kwok, Jung-Yoon Choe, and Sung-Hwan Park. 2019. "Recent Advances in Our Understanding of the Link between the Intestinal Microbiota and Systemic Lupus Erythematosus" International Journal of Molecular Sciences 20, no. 19: 4871. https://doi.org/10.3390/ijms20194871
APA StyleKim, J.-W., Kwok, S.-K., Choe, J.-Y., & Park, S.-H. (2019). Recent Advances in Our Understanding of the Link between the Intestinal Microbiota and Systemic Lupus Erythematosus. International Journal of Molecular Sciences, 20(19), 4871. https://doi.org/10.3390/ijms20194871




