MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development
Abstract
1. Introduction
2. Results
2.1. Identification of the MYB Gene Family in Potato
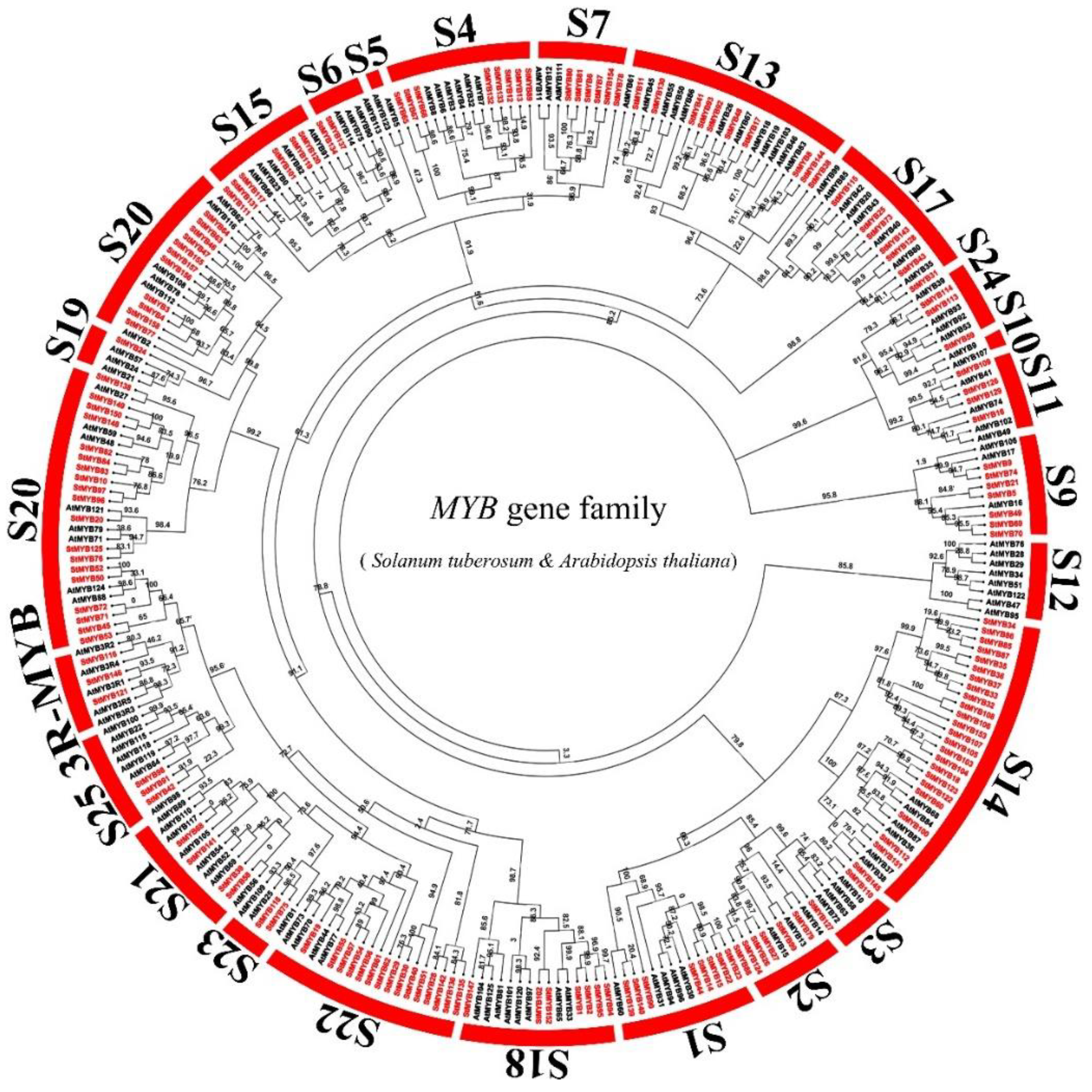
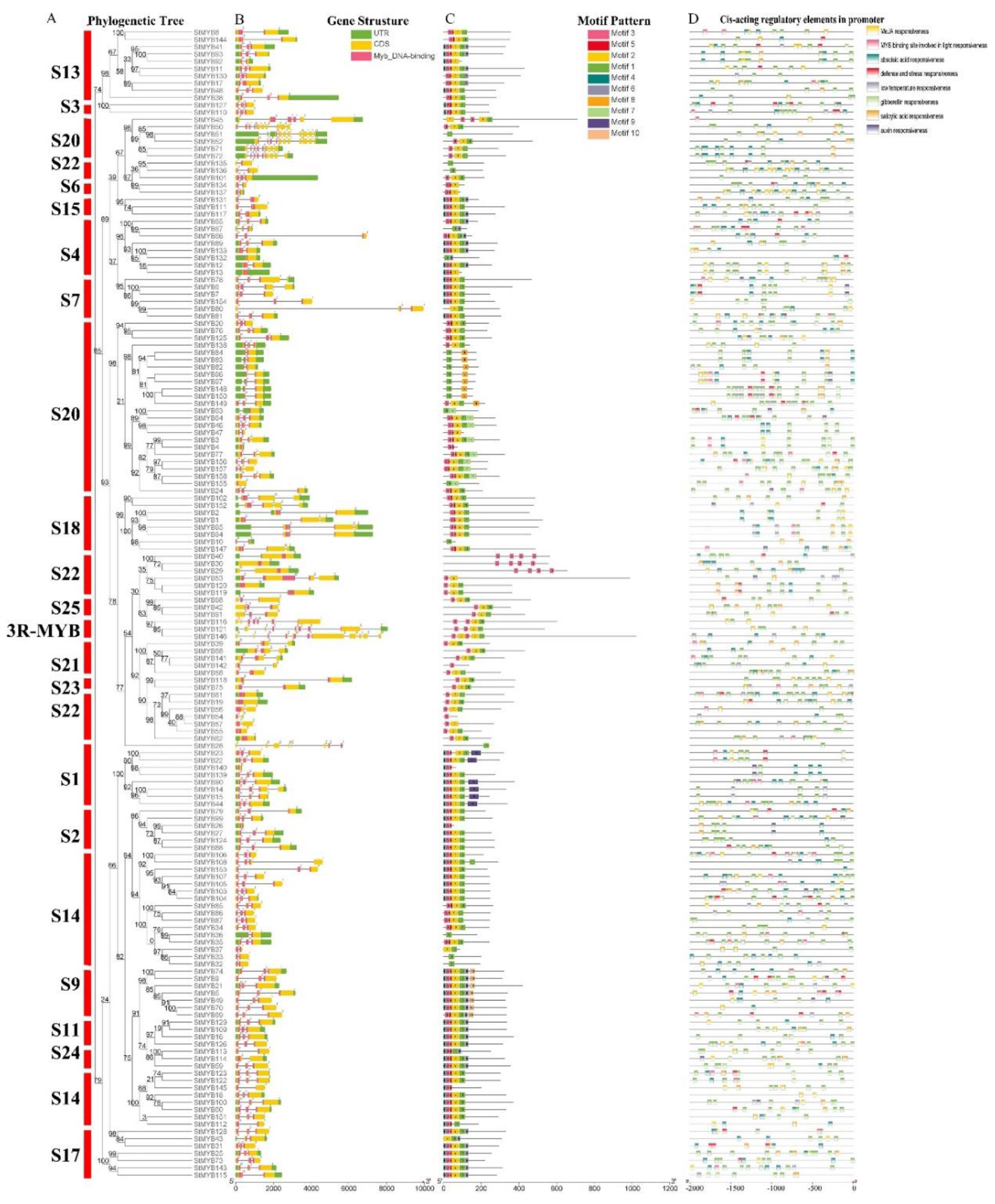
2.2. Phylogenetic Tree Analysis of MYB Genes in Potato and Arabidopsis thaliana (L.) Heynh
2.3. Gene Structure, Sequence Composition, and Promoter Cis-Acting Element Analysis
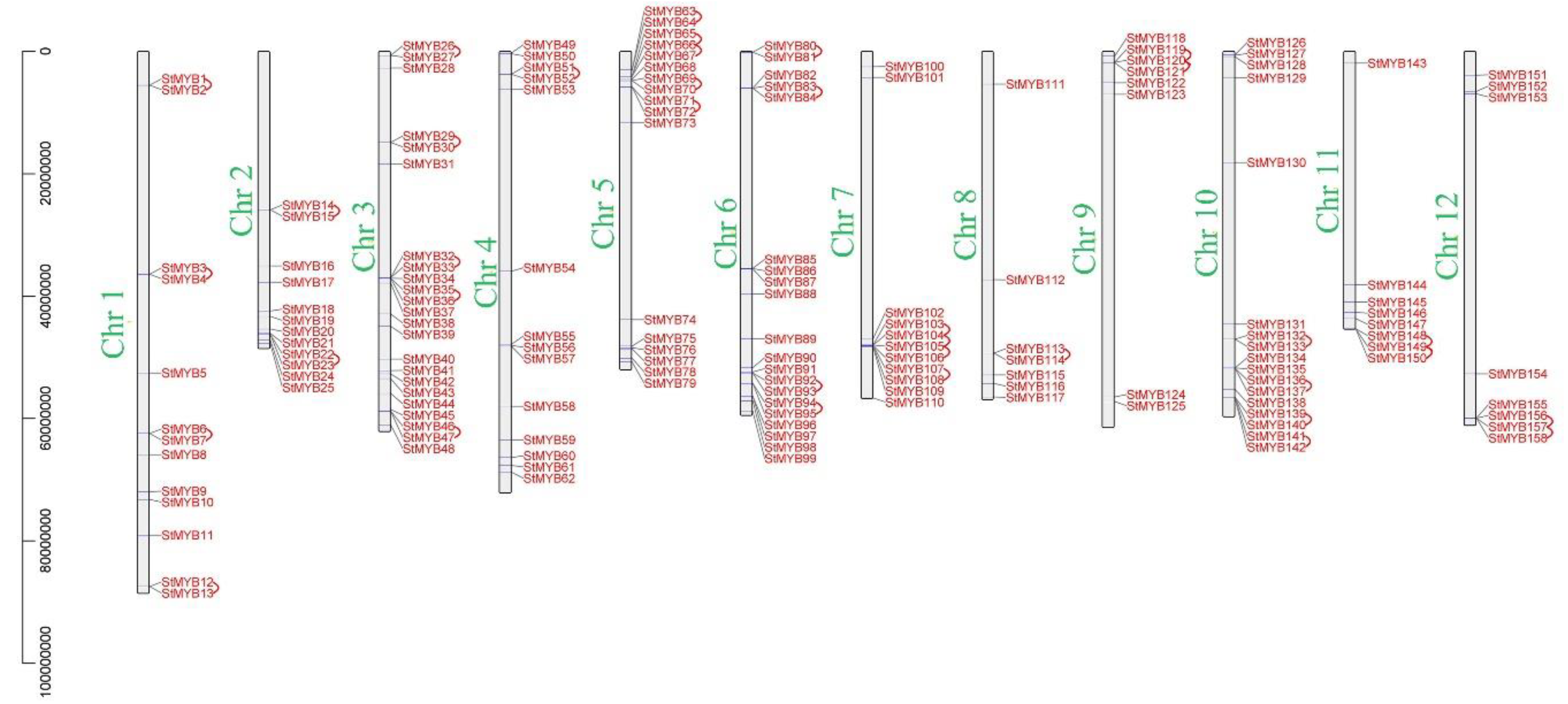
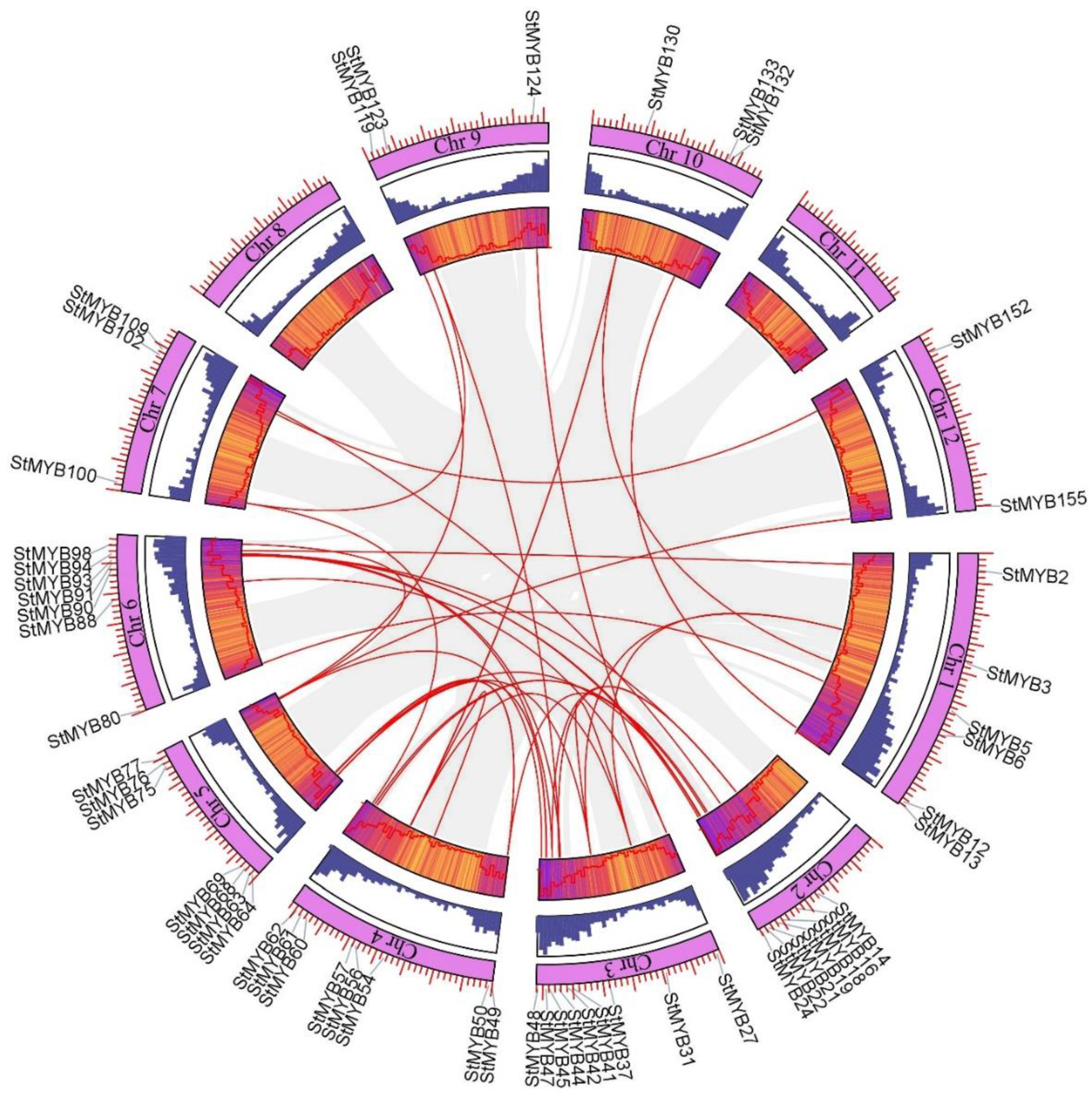
2.4. Analysis of Chromosome Distribution, Tandem Gene Duplication, and Segmental Gene Duplication of StMYB Genes
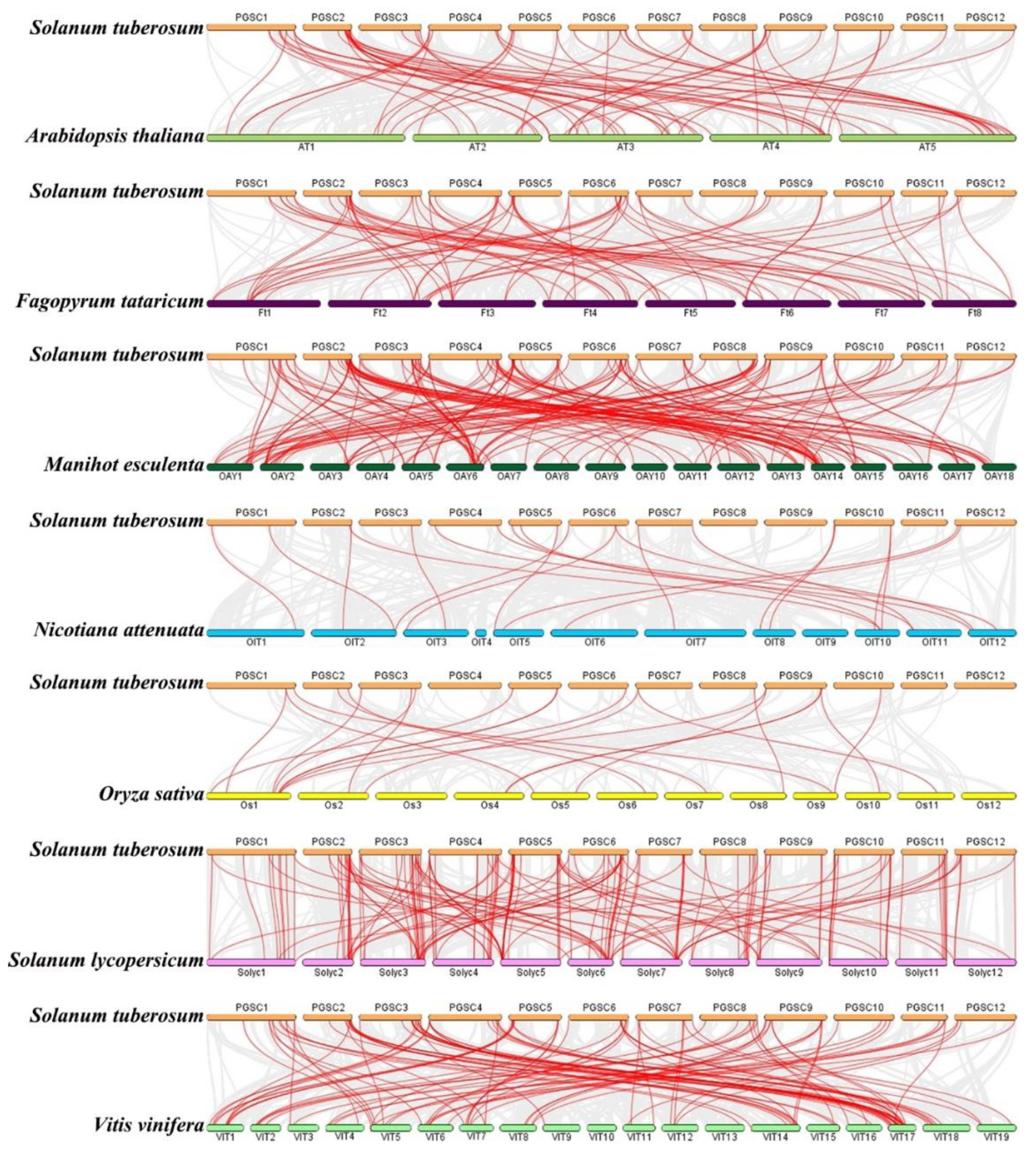
2.5. Evolutionary Analysis of StMYB Genes and the MYB Genes of Several Different Species
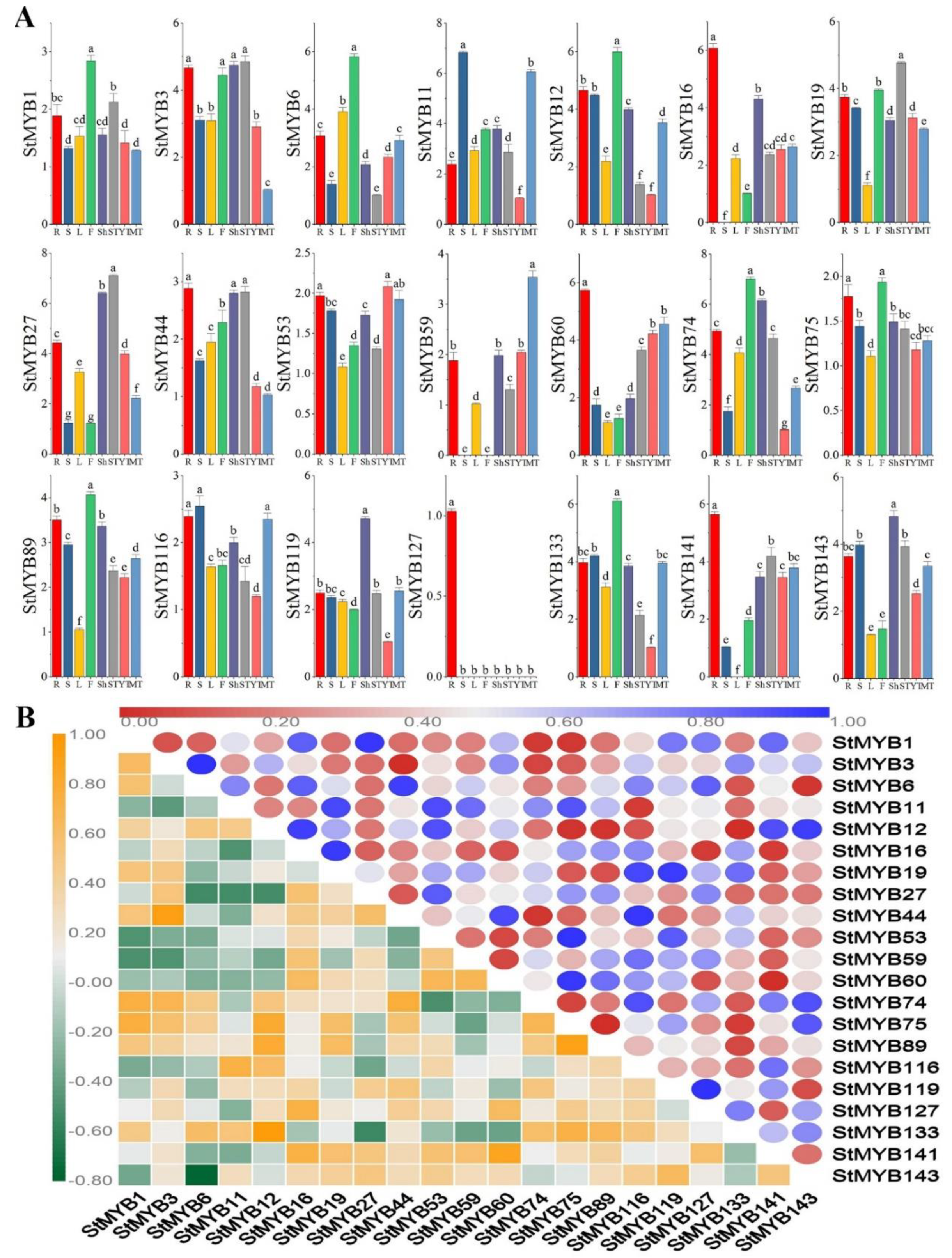
2.6. Expression Patterns of StMYB Genes in Different Plant Tissues
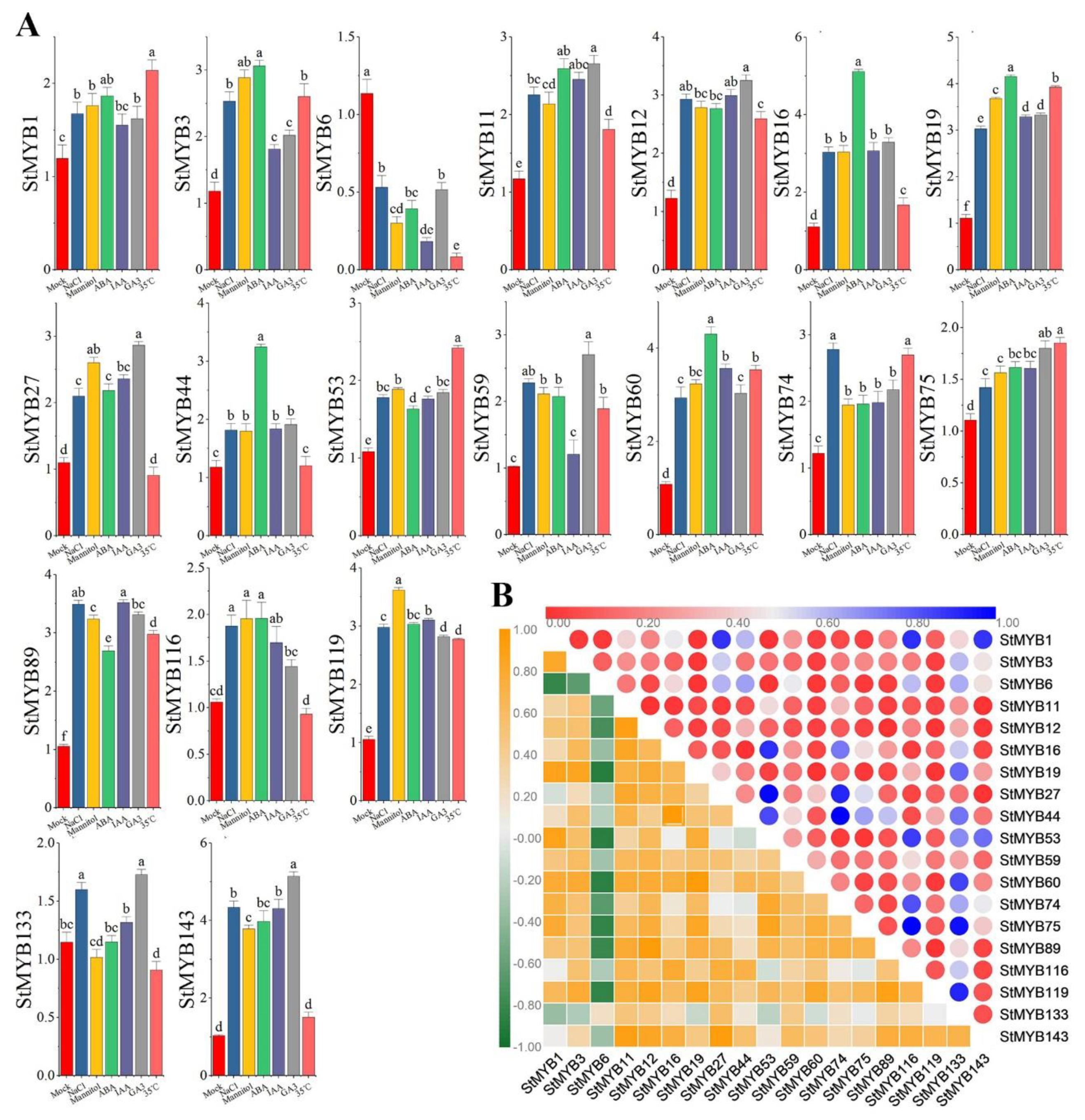
2.7. Expression Analysis of StMYB Genes under Phytohormone and Abiotic Stresses
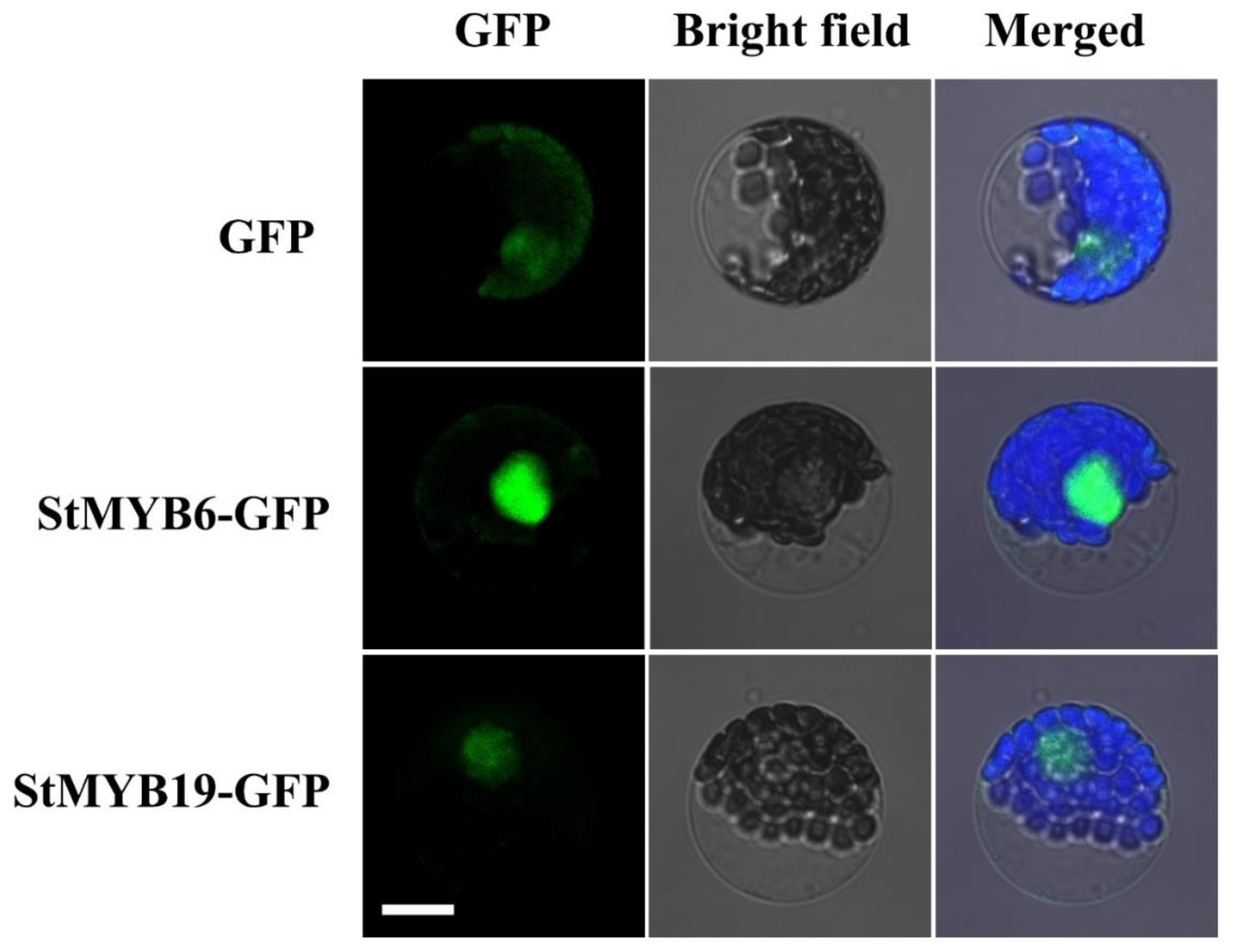
2.8. Subcellular Localization of StMYB6 and StMYB19
3. Discussion
3.1. Evolutionary Analysis of the StMYB Gene Family
3.2. StMYB Genes Play Crucial Roles in Potato Growth and Development and Response to Phytohormone and Abiotic Stresses
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of the StMYB Genes in Potato
4.3. Phylogenetics, Intron–Exon Structure, Motif Composition, and Cis-Acting Elements
4.4. Chromosomal Mapping, Gene Duplication, and Synteny with Other Plants
4.5. Expression Analysis of the StMYB Genes by qRT-PCR
4.6. Subcellular Localization
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| GA3 | gibberellin acid 3 |
| GFP | green fluorescent protein |
| HTH | helix–turn–helix |
| IAA | indole-3-acetic acid |
| MEME | Multiple Em for Motif Elicitation |
| MWs | molecular weights |
| NCBI | National Center for Biotechnology Information |
| PIs | isoelectric points |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| R | repeat |
| SA | salicylic acid |
| SGs | subgroups |
| StMYB | Solanum tuberosum L. MYB |
| TFs | transcription factors |
References
- Goode, J.A.; Organizers, D.C. Improving Plant Drought, Salt and Freezing Tolerance by Gene Transfer of a Single Stress-Inducible Transcription Factor. Nat. Biotechnol. 2007, 17, 287–291. [Google Scholar]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, J.; Hu, R.; Wu, P.; Hou, X.L.; Song, X.M.; Xiong, A.S. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genom. 2015, 16, 17–38. [Google Scholar] [CrossRef]
- Li, C.; Ng, K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, C.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS ONE 2012, 7, e47576. [Google Scholar] [CrossRef]
- Nozomi, H.; Kiichi, K.; Masatake, M.; Satoshi, A.; Minoru, K.; Taku, D.; Kaoru, S.; Isabel, M.; Ute, V.; Gerd, J. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 2007, 134, 1101–1110. [Google Scholar]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- Jiao, D.; Ming, L.; Huang, L.; Mei, Y.; Yang, P. Genome-Wide Analysis of the R2R3 MYB Subfamily Genes in Lotus (Nelumbo nucifera). Plant Mol. Biol. Report. 2016, 34, 11. [Google Scholar]
- Rosinski, J.A.; Atchley, W.R. Molecular Evolution of the Myb Family of Transcription Factors: Evidence for Polyphyletic Origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Jiang, C.; Gu, J.; Chopra, S.; Gu, X.; Peterson, T. Ordered origin of the typical two- and three-repeat Myb genes. Gene 2004, 326, 13–22. [Google Scholar] [CrossRef]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83–98. [Google Scholar] [CrossRef]
- Dias, A.P.; Braun, E.L.; Mcmullen, M.D.; Erich, G. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 2003, 131, 610–620. [Google Scholar] [CrossRef]
- Martin, C.; Paz-Ares, J. MYB transcription factors in plants. Trends Genet. 1997, 13, 67–73. [Google Scholar] [CrossRef]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2010, 50, 660–677. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2010, 53, 814–827. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and Biochemistry of Seed Flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar]
- Lee, T.G.; Jang, C.S.; Kim, J.Y.; Kim, D.S.; Park, J.H.; Kim, D.Y.; Seo, Y.W. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: Roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Plant. 2010, 129, 375–385. [Google Scholar] [CrossRef]
- Mao, X.; Jia, D.; Li, A.; Zhang, H.; Tian, S.; Zhang, X.; Jia, J.; Jing, R. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct. Integr. Genom. 2011, 11, 445–465. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544–563. [Google Scholar] [CrossRef]
- Feng, S.; Xu, Y.; Long, Y.; Sun, S.; Wang, D.; Chen, X. Genome-wide identification and characterization of R2R3-MYB transcription factors in pear. Sci. Hortic. 2015, 197, 176–182. [Google Scholar] [CrossRef]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet ( Beta vulgaris ). BMC Plant Biol. 2014, 14, 249–266. [Google Scholar] [CrossRef]
- Salih, H.; Gong, W.; He, S.; Sun, G.; Sun, J.; Du, X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016, 17, 129–141. [Google Scholar] [CrossRef]
- Hai, D.; Bo-Run, F.; Si-Si, Y.; Yu-Bi, H.; Yi-Xiong, T. The R2R3-MYB transcription factor gene family in maize. PLoS ONE 2012, 7, e37463–e37475. [Google Scholar]
- Dongjin, S.; Seok-Jun, M.; Seyoun, H.; Beom-Gi, K.; Ryeol, P.S.; Seong-Kon, L.; Hye-Jin, Y.; Hye Eun, L.; Hawk-Bin, K.; Dongwon, B. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011, 155, 421–432. [Google Scholar]
- Cheng, Y.J.; Kim, M.D.; Deng, X.P.; Kwak, S.S.; Chen, W. Enhanced Salt Stress Tolerance in Transgenic Potato Plants Expressing IbMYB1, a Sweet Potato Transcription Factor. J. Microbiol. Biotechnol. 2013, 23, 1737–1746. [Google Scholar] [CrossRef]
- Li, X.; Guo, C.; Ahmad, S.; Wang, Q.; Yu, J.; Liu, C.; Guo, Y. Systematic Analysis of MYB Family Genes in Potato and Their Multiple Roles in Development and Stress Responses. Biomolecules 2019, 9, 317. [Google Scholar] [CrossRef]
- Qin, Z.; Hao, R.; Xu, Z.; Yang, W.; Jia, W.; Cheng, T.; Pan, H.; Zhang, Q. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell Tissue Organ Cult. 2017, 131, 13. [Google Scholar]
- Gao, F.; Zhou, J.; Deng, R.Y.; Zhao, H.X.; Li, C.L.; Chen, H.; Suzuki, T.; Park, S.U.; Wu, Q. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J. Plant Physiol. 2017, 214, 81–90. [Google Scholar] [CrossRef]
- Meng, D.; He, M.; Bai, Y.; Xu, H.; Dandekar, A.M.; Fei, Z.; Cheng, L. Decreased sorbitol synthesis leads to abnormal stamen development and reduced pollen tube growth via an MYB transcription factor, MdMYB39L, in apple (Malus domestica). New Phytol. 2017, 217, 641–656. [Google Scholar] [CrossRef]
- Fang, Q.; Jiang, T.; Xu, L.; Liu, H.; Mao, H.; Wang, X.; Jiao, B.; Duan, Y.; Wang, Q.; Dong, Q. A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2017, 114, 100–110. [Google Scholar] [CrossRef]
- Ruan, M.B.; Guo, X.; Wang, B.; Yang, Y.L.; Li, W.Q.; Yu, X.L.; Zhang, P.; Peng, M. Genome-wide characterization and expression analysis enables identification of abiotic stress-responsive MYB transcription factors in cassava (Manihot esculenta). J. Exp. Bot. 2017, 68, 3657–3672. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef]
- Hajiebrahimi, A.; Owji, H.; Hemmati, S. Genome-wide identification, functional prediction and evolutionary analysis of R2R3-MYB superfamily in Brassica napus. Genome 2017, 60, 797. [Google Scholar] [CrossRef]
- Du, H.; Yang, S.S.; Liang, Z.; Feng, B.R.; Liu, L.; Huang, Y.B.; Tang, Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106–128. [Google Scholar] [CrossRef]
- Wang, W.; Feng, B.; Xiao, J.; Xia, Z.; Zhou, X.; Li, P.; Zhang, W.; Wang, Y.; Møller, B.L.; Zhang, P. Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 2014, 5, 5110–5119. [Google Scholar] [CrossRef]
- Aoki, K.; Yano, K.; Suzuki, A.; Kawamura, S.; Sakurai, N.; Suda, K.; Kurabayashi, A.; Suzuki, T.; Tsugane, T.; Watanabe, M. Large-scale analysis of full-length cDNAs from the tomato ( Solanum lycopersicum ) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genom. 2010, 11, 210–226. [Google Scholar] [CrossRef]
- Boulos, C.; France, D.; Shengyi, L.; Parkin, I.A.P.; Haibao, T.; Xiyin, W.; Julien, C.; Harry, B.; Chaobo, T.; Birgit, S. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953.
- Yu, J.; Wang, J.; Lin, W.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C. The Genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38–e54. [Google Scholar] [CrossRef] [PubMed]
- Zapata, L.; Jia, D.; Willing, E.M.; Hartwig, B.; Bezdan, D.; Jiao, W.B.; Patel, V.; James, G.V.; Koornneef, M.; Ossowski, S. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA 2016, 113, E4052–E4060. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490–508. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome Wide Analysis of the Apple MYB Transcription Factor Family Allows the Identification of MdoMYB121 Gene Confering Abiotic Stress Tolerance in Plants. PLoS ONE 2013, 8, e69955–e69968. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Otto, S.P.; Yong, P. The evolution of gene duplicates. Adv. Genet. 2002, 46, 451–483. [Google Scholar]
- Duarte, J.M.; Liying, C.; P Kerr, W.; Qing, Z.; Xiaohong, Z.; Jim, L.M.; Hong, M.; Naomi, A.; Depamphilis, C.W. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 2006, 23, 469–478. [Google Scholar] [CrossRef]
- Hou, D.; Zhanchao, C.; Lihua, X.; Xiangyu, L.; Juan, L.; Shaohua, M.; Jian, G. The R2R3MYB Gene Family in Phyllostachys edulis: Genome-Wide Analysis and Identification of Stress or Development-Related R2R3MYBs. Front. Plant Sci. 2018, 9, 738–760. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Veilleux, R.E. Adaptation of potato to high temperatures and salinity-a review. Am. J. Potato Res. 2007, 84, 487–506. [Google Scholar] [CrossRef]
- Arvin, M.J.; Donnelly, D.J. Screening potato cultivars and wild species to abiotic stresses using an electrolyte leakage bioassay. J. Agric. Sci. Technol. 2008, 10, 33–42. [Google Scholar]
- Trudi, H.; Feng, L.S.; Parish, R.W. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2010, 35, 177–192. [Google Scholar]
- Rumi, T.; Mineko, I.; Kiyotaka, O.; Takuji, W. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 2007, 19, 2264–2277. [Google Scholar]
- Frank, M.; Harald, K.; Pawel, B.; Bernd, W. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar]
- Teng, S.; Keurentjes, J.; Bentsink, L.N.; Koornneef, M.; Smeekens, S. Sucrose-Specific Induction of Anthocyanin Biosynthesis in Arabidopsis Requires the MYB75/PAP1 Gene. Plant physiol. 2006, 139, 1840–1852. [Google Scholar] [CrossRef]
- Yuan-Yuan, L.; Ke, M.; Cheng, Z.; Xian-Yan, Z.; Hua-Lei, Z.; Huai-Rui, S.; Yu-Jin, H. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar]
- Hiroshi, A.; Takeshi, U.; Takuya, I.; Motoaki, S.; Kazuo, S.; Kazuko, Y.S. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a MYB transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef]
- Yao, L.; Jiang, Y.; Lu, X.; Wang, B.; Zhou, P.; Wu, T. A R2R3-MYB transcription factor from Lablab purpureus induced by drought increases tolerance to abiotic stress in Arabidopsis. Mol. Biol. Rep. 2016, 43, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2014, 19, 6150–6161. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.; Wheeler, J.J.; Li, S.; Parish, R. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2010, 40, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Silvia, F.; Eric, L.; Salazar-Henao, J.E.; Pedro, F.N.; Joan, R.; David, C.R. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A Guard-Cell-Specific MYB Transcription Factor Regulates Stomatal Movements and Plant Drought Tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.; Chua, N. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2010, 49, 592–606. [Google Scholar] [CrossRef]
- Manu, A.; Yujin, H.; Avnish, K.; Chun-Hai, D.; Hiroaki, F.; Xianwu, Z.; Jian-Kang, Z. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 Gene Encodes an R2R3MYB Transcription Factor Protein That Is Required for Biotic and Abiotic Stress Responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.I. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Reference gene selection for RT-qPCRnormalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2012, 25, 402–408. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Ma, Z.; Chen, H.; Liu, M. MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development. Int. J. Mol. Sci. 2019, 20, 4847. https://doi.org/10.3390/ijms20194847
Sun W, Ma Z, Chen H, Liu M. MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development. International Journal of Molecular Sciences. 2019; 20(19):4847. https://doi.org/10.3390/ijms20194847
Chicago/Turabian StyleSun, Wenjun, Zhaotang Ma, Hui Chen, and Moyang Liu. 2019. "MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development" International Journal of Molecular Sciences 20, no. 19: 4847. https://doi.org/10.3390/ijms20194847
APA StyleSun, W., Ma, Z., Chen, H., & Liu, M. (2019). MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development. International Journal of Molecular Sciences, 20(19), 4847. https://doi.org/10.3390/ijms20194847






