Effect of Lactoferrin on the Expression Profiles of Long Non-coding RNA during Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells
Abstract
1. Introduction
2. Results
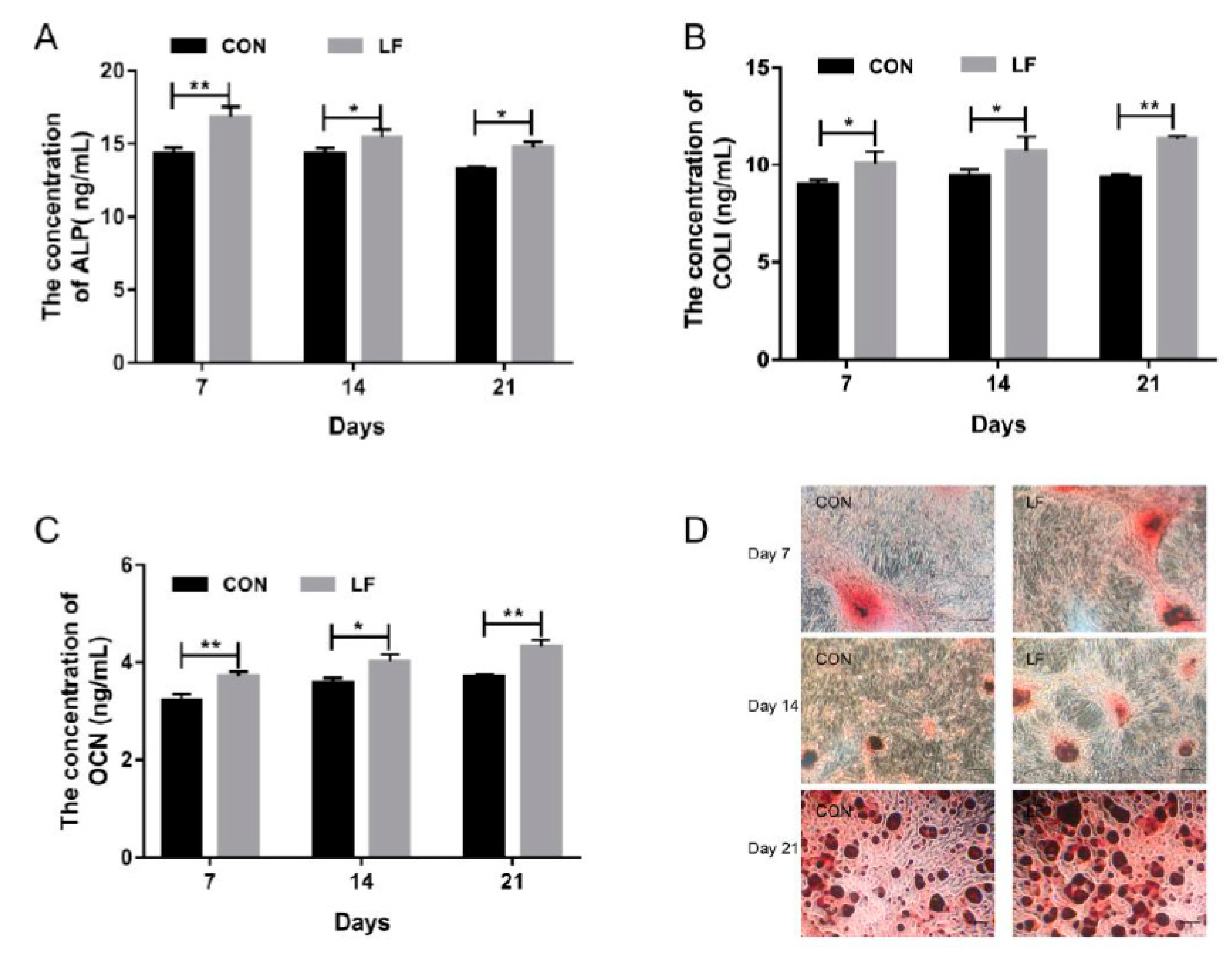
2.1. The Effect of LF on Osteogenic Differentiation Capacity of rBMSCs
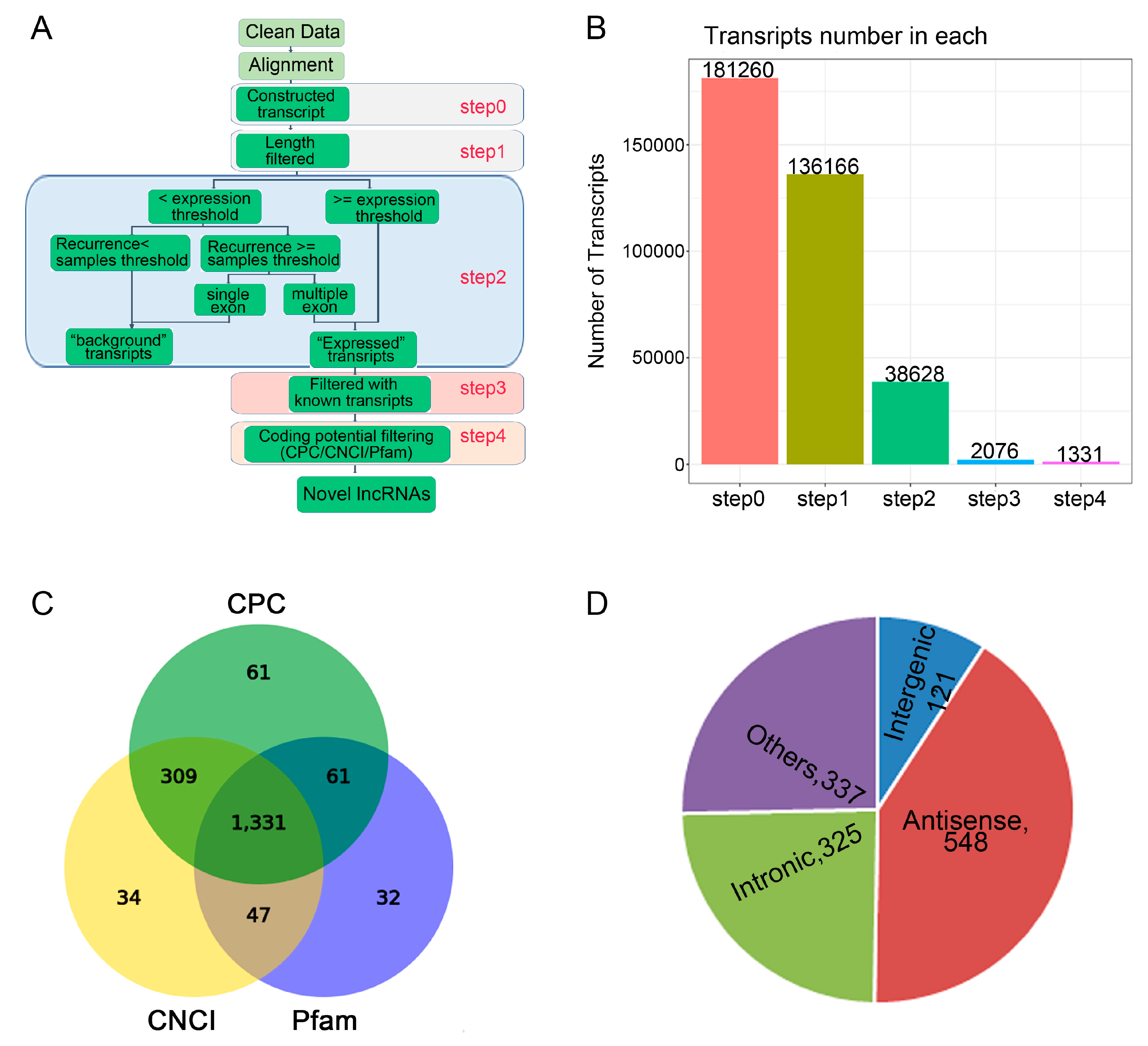
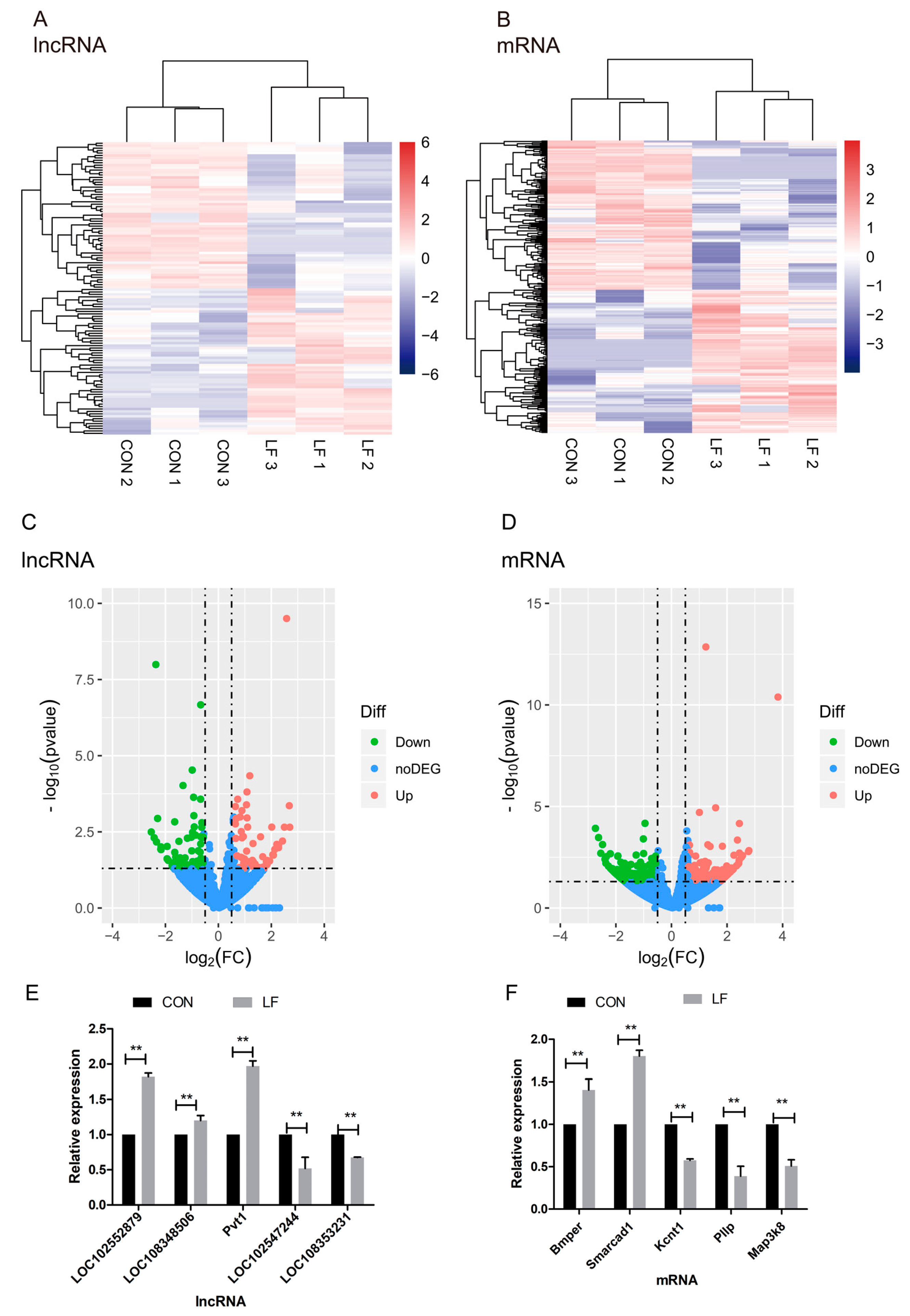
2.2. The Effect of LF on lncRNAs Profiles During Osteogenic Differentiation of rBMSCs
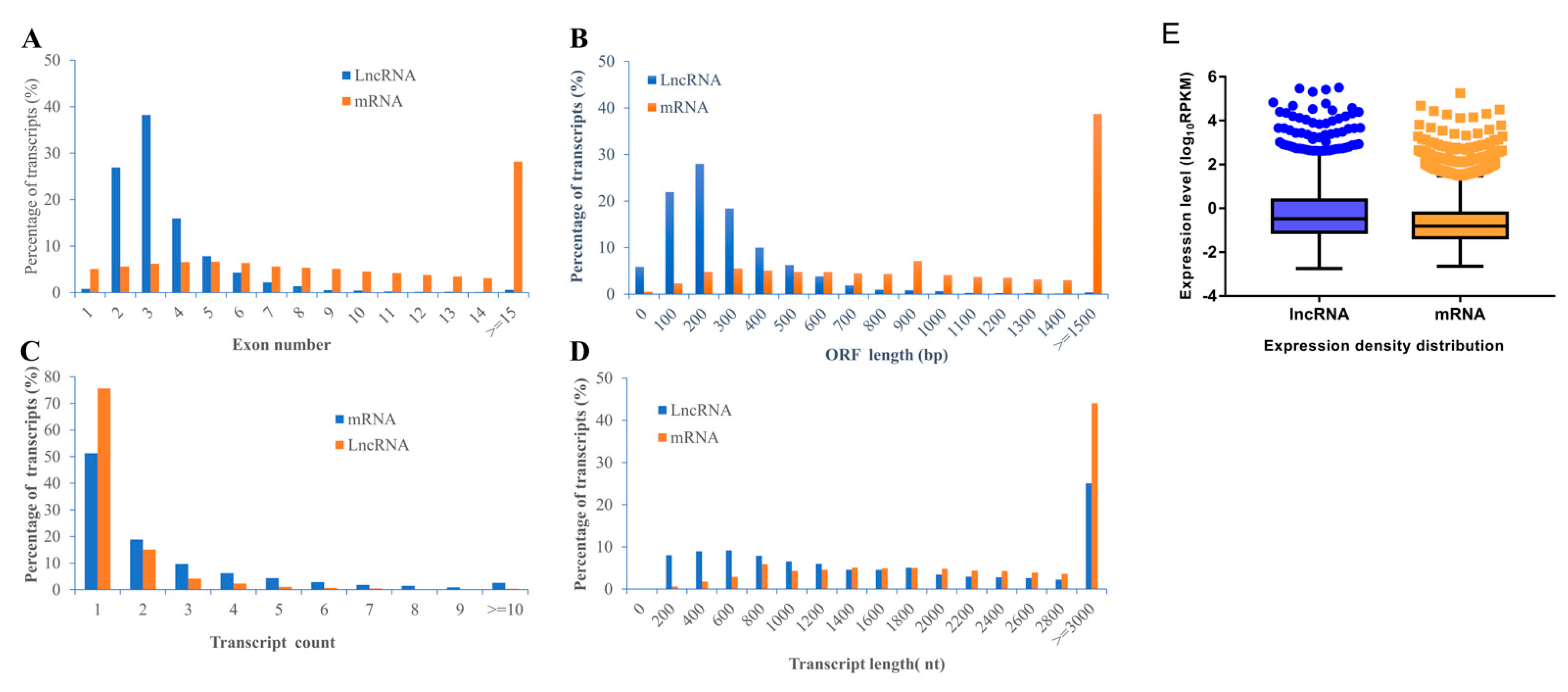
2.3. Characteristics of lncRNAs and mRNAs
2.4. The Effect of LF on Profiles of Differentially Expressed mRNAs and lncRNAs During Osteogenic Differentiation in rBMSCs
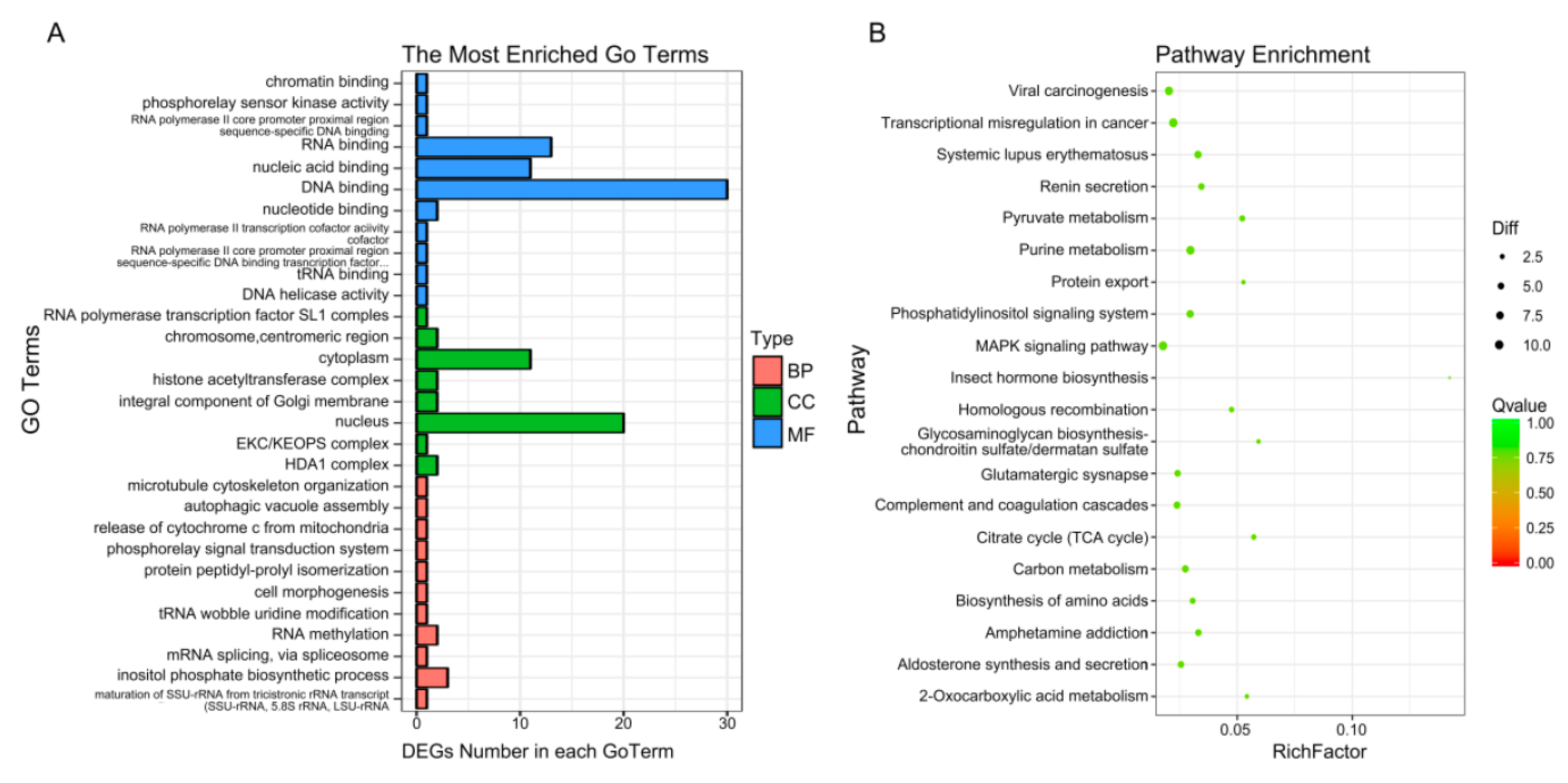
2.5. GO and KEGG Pathway Analysis of DEGs
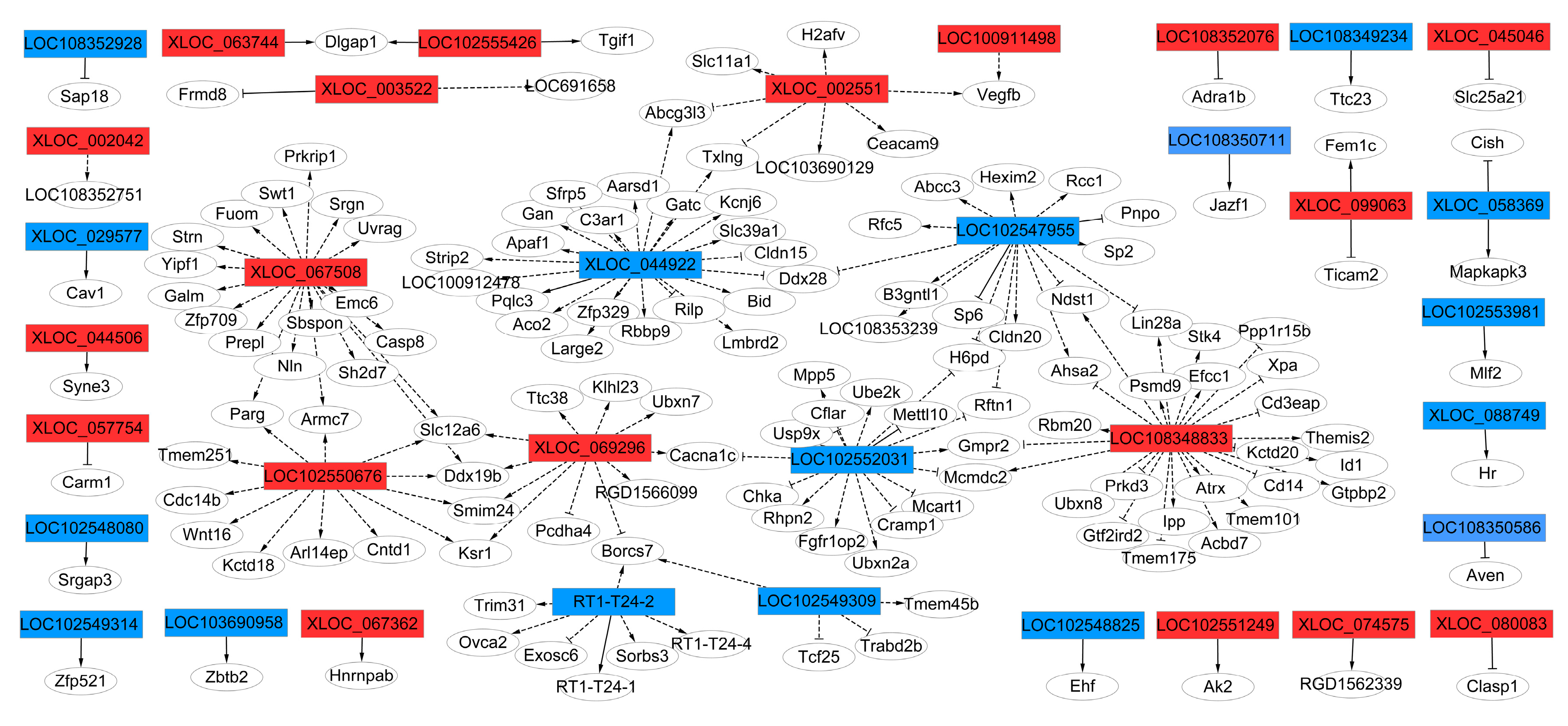
2.6. Target-gene Prediction of cis- and trans- lncRNA and lncRNA-mRNA Co-expression Networks
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Osteogenic Differentiation of rBMSCs
4.3. Alizarin Red Staining of rBMSCs
4.4. RNA Extraction, Library Construction, and High-throughput RNA Sequencing (RNA-Seq)
4.5. LncRNAs Identification and Classification
4.6. Differential Expression Genes Analysis of Transcripts
4.7. Bioinformatics Analysis
4.8. Target-Gene Prediction
4.9. Quantitative Real-Time PCR Validation
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| BMSCs | Bone marrow mesenchymal stem cells |
| CNCI | Coding non coding index |
| COL I | Collagen type I |
| CPC | Coding potential Calculator |
| DEG | Differentially expressed gene |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes. |
| LF | Lactoferrin |
| lncRNAs | Long non-coding RNAs |
| MAPK | Mitogen-activated protein kinase |
| ncRNAs | Non-coding RNAs |
| OCN | Osteocalcin |
| Pfam | Pfamscan |
| RNA-seq | RNA Sequencing |
| RPKM | Reads per kilo bases per million reads |
References
- Post, S.; Abdallah, B.M.; Bentzon, J.F.; Kassem, M. Demonstration of the presence of independent pre-osteoblastic and pre-adipocytic cell populations in bone marrow-derived mesenchymal stem cells. Bone 2008, 43, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.B.; Zhang, Y.; Wang, Y.Q.; He, Q.; Yu, Q. IGF-1 and BMP-7 synergistically stimulate articular cartilage repairing in the rabbit knees by improving chondrogenic differentiation of bone-marrow mesenchymal stem cells. J. Cell. Biochem. 2019, 120, 5570–5582. [Google Scholar] [CrossRef] [PubMed]
- Chanda, D.; Kumar, S.; Ponnazhagan, S. Therapeutic potential of adult bone marrow-derived mesenchymal stem cells in diseases of the skeleton. J. Cell. Biochem. 2010, 111, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Wang, F.; Li, Y.Y.; Dai, Y.F.; Liu, Y.J.; Wang, J.F.; Xue, C.H. Oil from Antarctic krill (Euphausia superba) facilitates bone formation in dexamethasone-treated mice. Food Sci. Biotechnol. 2019, 28, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Weitzmann, M.N. The bone anabolic carotenoid p-hydroxycinnamic acid promotes osteoblast mineralization and suppresses osteoclast differentiation by antagonizing NF-κappa B activation. Int. J. Mol. Med. 2012, 30, 708–712. [Google Scholar] [CrossRef][Green Version]
- Li, M.; Zhang, C.; Li, X.H.; Lv, Z.H.; Chen, Y.; Zhao, J.Y. Isoquercitrin promotes the osteogenic differentiation of osteoblasts and BMSCs via the RUNX2 or BMP pathway. Connect. Tissue Res. 2019, 60, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Shen, Y.S.; He, M.G.; Yang, F.; Yang, P.; Pang, F.X.; He, W.; Cao, Y.M.; Wei, Q.S. Polydatin promotes the osteogenic differentiation of human bone mesenchymal stem cells by activating the BMP2-Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2019, 112, 108746. [Google Scholar] [CrossRef]
- Lootoing, L.; Davicco, M.J.; Lebecque, P.; Wittrant, Y.; Coxam, V. The flavonoid fisetin promotes osteoblasts differentiation through Runx2 transcriptional activity. Mol. Nutr. Food Res. 2014, 58, 1239–1248. [Google Scholar] [CrossRef]
- Fang, Y.Y.; Xue, Z.Y.; Zhao, L.G.; Yang, X.Y.; Yang, Y.F.; Zhou, X.L.; Feng, S.L.; Chen, K.M. Calycosin stimulates the osteogenic differentiation of rat calvarial osteoblasts by activating the IGF1R/PI3K/Akt signaling pathway. Cell Biol. Int. 2019, 43, 323–332. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, H.; Wang, Z.Y.; Fan, F.J.; Shi, P.J.; Tu, M.L.; Du, M. Isolation and characterization of peptides from mytilus edulis with osteogenic activity in mouse MC3T3-E1 preosteoblast cells. J. Agric. Food Chem. 2019, 67, 1572–1584. [Google Scholar] [CrossRef]
- Ren, J.D.; Chakrabarti, S.; Wu, J.P. Phosvitin and its hydrolysate promote differentiation and inhibit TNF-alpha induced inflammation in MC3T3-E1 cells via ERK and AKT pathways. J. Funct. Foods 2019, 53, 259–265. [Google Scholar] [CrossRef]
- Gonzalez-Chavez, S.A.; Arevalo-Gallegos, S.; Rascon-Cruz, Q. Lactoferrin: Structure, function and applications. Int. J. Antimicrob. Agents 2009, 33, e1. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Watson, M.; Callon, K.E.; Tuari, D.; Dray, M.; Naot, D.; Amirapu, S.; Munro, J.T.; Cornish, J.; Musson, D.S. Local application of lactoferrin promotes bone regeneration in a rat critical-sized calvarial defect model as demonstrated by micro-CT and histological analysis. J. Tissue Eng. Regen. Med. 2018, 12, E620–E626. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Y.; Zhu, S.S.; Hu, J. Bone regeneration is promoted by orally administered bovine lactoferrin in a rabbit tibial distraction osteogenesis model. Clin. Orthop. Relat. Res. 2015, 473, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.J.; Shi, P.J.; Liu, M.; Chen, H.; Tu, M.L.; Lu, W.H.; Du, M. Lactoferrin preserves bone homeostasis by regulating the RANKL/RANK/OPG pathway of osteoimmunology. Food Funct. 2018, 9, 2653–2660. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, H.Y.; Jing, H.; Li, Y.X.; Wang, X.Y.; Zhang, H.; Jiang, L.; Ren, F.Z. Lactoferrin stimulates osteoblast differentiation through PKA and p38 pathways independent of Lactoferrin’s receptor LRP1. J. Bone Miner. Res. 2014, 29, 1232–1243. [Google Scholar] [CrossRef]
- Ying, X.Z.; Cheng, S.W.; Wang, W.; Lin, Z.Q.; Chen, Q.Y.; Zhang, W.; Kou, D.Q.; Shen, Y.; Cheng, X.J.; Peng, L.; et al. Effect of lactoferrin on osteogenic differentiation of human adipose stem cells. Int. Orthop. 2012, 36, 647–653. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Zhang, W.; Ren, F.; Guo, H. Activation of TGF-beta canonical and noncanonical signaling in bovine lactoferrin-induced osteogenic activity of C3H10T1/2 mesenchymal stem cells. Int. J. Mol. Sci. 2019, 20, 2880. [Google Scholar] [CrossRef]
- Grey, A.; Banovic, T.; Zhu, Q.; Watson, M.; Callon, K.; Palmano, K.; Ross, J.; Naot, D.; Reid, I.R.; Cornish, J. The low-density lipoprotein receptor-related protein 1 is a mitogenic receptor for lactoferrin in osteoblastic cells. Mol. Endocrinol. 2004, 18, 2268–2278. [Google Scholar] [CrossRef]
- Liu, M.; Fan, F.J.; Shi, P.J.; Tu, M.L.; Yu, C.P.; Yu, C.X.; Du, M. Lactoferrin promotes MC3T3-E1 osteoblast cells proliferation via MAPK signaling pathways. Int. J. Biol. Macromol. 2018, 107, 137–143. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Ren, F.; Zhang, W.; Zhang, H.; Zhao, L.; Zhang, M.; Cui, W.; Wang, X.; Guo, H. Lactoferrin Promotes Osteogenesis through TGF-β Receptor II Binding in Osteoblasts and Activation of Canonical TGF-β Signaling in MC3T3-E1 Cells and C57BL/6J Mice. J. Nutr. 2018, 148, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.P.; Cao, L.H.; He, S.W.; Zhong, Y.C.; Ma, H.T.; Zhang, Y.R.; Shuai, C.J. An overview of long noncoding RNAs involved in bone regeneration from mesenchymal stem cells. Stem Cells Int. 2018, 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.B.; Zhou, Y.C.; He, X.Y.; Wan, M.; Du, W.; Xu, X.; Ye, L.; Zhou, X.D.; Zheng, L.W. Insight into the role of long non-coding RNAs during osteogenesis in mesenchymal stem cells. Curr. Stem Cell Res. Ther. 2018, 13, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Jia, L.F.; Li, X.B.; Guo, R.Z.; Huang, Y.P.; Zheng, Y.F.; Li, W.R. Long noncoding RNAs: New players in the osteogenic differentiation of bone marrow- and adipose-derived mesenchymal stem cells. Stem Cell Rev. Rep. 2018, 14, 297–308. [Google Scholar] [CrossRef]
- Lammens, T.; D’Hont, I.; D’Herde, K.; Benoit, Y.; Diez-Fraile, A. Long non-coding RNAs in pluripotent stem cell biology. Vet. Q. 2013, 33, 202–206. [Google Scholar] [CrossRef][Green Version]
- Wei, B.F.; Wei, W.; Zhao, B.X.; Guo, X.X.; Liu, S. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in nontraumatic osteonecrosis of femoral head. PLoS ONE 2017, 12, e0169097. [Google Scholar]
- Jin, C.Y.; Jia, L.F.; Huang, Y.P.; Zheng, Y.F.; Du, N.; Liu, Y.S.; Zhou, Y.S. Inhibition of lncRNA MIR31HG Promotes Osteogenic Differentiation of Human Adipose-Derived Stem Cells. Stem Cells 2016, 34, 2707–2720. [Google Scholar] [CrossRef]
- Yoshioka, H.; Yoshiko, Y. The Roles of long non-protein-coding RNAs in osteo-adipogenic lineage commitment. Int. J. Mol. Sci. 2017, 18, 1239. [Google Scholar]
- Ju, C.; Liu, R.F.; Zhang, Y.W.; Zhang, Y.; Zhou, R.H.; Sun, J.; Lv, X.B.; Zhang, Z.P. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed. Pharmacother. 2019, 115, 108912. [Google Scholar] [CrossRef]
- Xie, Z.Y.; Wang, P.; Wu, Y.F.; Shen, H.Y. Long non-coding RNA: The functional regulator of mesenchymal stem cells. World J. Stem Cells 2019, 11, 167–179. [Google Scholar] [CrossRef]
- Zhuang, W.Z.; Ge, X.P.; Yang, S.J.; Huang, M.L.; Zhuang, W.Y.; Chen, P.; Zhang, X.H.; Fu, J.X.; Qu, J.; Li, B.Z. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells 2015, 33, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Liao, Z.; Xiao, H.; Liu, H.; Hu, Y.H.; Liao, Q.D.; Zhong, D. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp. Mol. Pathol. 2019, 107, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Tao, Z.W.; Wang, Y.L. Long non-coding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int. J. Mol. Med. 2018, 41, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.X.; Yu, J.J.; Du, J.; Zhong, G.S.; Qiao, L.; Lin, J.T. LncRNA HOXA-AS2 positively regulates osteogenesis of mesenchymal stem cells through inactivating NF-κappa B signalling. J. Cell. Mol. Med. 2019, 23, 1325–1332. [Google Scholar]
- Zheng, X.; Ning, C.; Zhao, P.; Feng, W.; Jin, Y.; Zhou, L.; Yu, Y.; Liu, J. Integrated analysis of long noncoding RNA and mRNA expression profiles reveals the potential role of long noncoding RNA in different bovine lactation stages. J. Dairy Sci. 2018, 101, 11061–11073. [Google Scholar] [CrossRef]
- Ruszkowska, M.; Nynca, A.; Paukszto, L.; Sadowska, A.; Swigonska, S.; Orlowska, K.; Molcan, T.; Jastrzebski, J.P.; Ciereszko, R.E. Identification and characterization of long non-coding RNAs in porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Anim. Sci. Biotechnol. 2018, 9, 72. [Google Scholar] [CrossRef]
- Ma, Q.M.; Li, L.Y.; Tang, Y.; Fu, Q.; Liu, S.; Hu, S.W.; Qiao, J.; Chen, C.F.; Ni, W. Analyses of long non-coding RNAs and mRNA profiling through RNA sequencing of MDBK cells at different stages of bovine viral diarrhea virus infection. Res. Vet. Sci. 2017, 115, 508–516. [Google Scholar] [CrossRef]
- Billerey, C.; Boussaha, M.; Esquerre, D.; Rebours, E.; Djari, A.; Meersseman, C.; Klopp, C.; Gautheret, D.; Rocha, D. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 2014, 15, 499. [Google Scholar] [CrossRef]
- Włodarski, K. Lactoferrin a promising bone-growth promoting milk-derived glycoprotein. Chir. Narz. Ruchu. Ortop. Pol. 2009, 74, 257–259. [Google Scholar]
- Montesi, M.; Panseri, S.; Iafisco, M.; Adamiano, A.; Tampieri, A. Effect of hydroxyapatite nanocrystals functionalized with lactoferrin in osteogenic differentiation of mesenchymal stem cells. J. Biomed. Mater. Res. Part A 2015, 103, 224–234. [Google Scholar] [CrossRef]
- Park, S.Y.; Jeong, A.J.; Kim, G.Y.; Jo, A.; Lee, J.E.; Leem, S.H.; Yoon, J.H.; Ye, S.K.; Chung, J.W. Lactoferrin Protects Human Mesenchymal Stem Cells from Oxidative Stress-Induced Senescence and Apoptosis. J. Microbiol. Biotechnol. 2017, 27, 1877–1884. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.E.; Yun, Y.P.; Shim, K.S.; Park, K.; Choi, S.W.; Suh, D.H. Effect of lactoferrin-impregnated porous poly(lactide-co-glycolide) (PLGA) microspheres on osteogenic differentiation of rabbit adipose-derived stem cells (rADSCs). Colloids Surf. B-Biointerfaces 2014, 122, 457–464. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Xu, Y.; Kong, Z.; Xie, Y.; Tabys, D.; Ma, M.; Cao, X.; Ren, H.; Liu, N. Effect of lactoferrin and its digests on differentiation activities of bone mesenchymal stem cells. J. Funct. Foods 2019, 57, 202–210. [Google Scholar] [CrossRef]
- Zou, L.; Zou, X.; Chen, L.; Li, H.; Mygind, T.; Kassem, M.; Bunger, C. Multilineage differentiation of porcine bone marrow stromal cells associated with specific gene expression pattern. J. Orthop. Res. 2008, 26, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Mizumachi, K. Effect of lactoferrin-embedded collagen membrane on osteogenic differentiation of human osteoblast-like cells. J. Biosci. Bioeng. 2009, 107, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Hong, T.; Wang, T.; Wang, X.; Cao, L.; Xu, X.; Zheng, M. Gene expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation. J. Cell. Physiol. 2019, 234, 7070–7077. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, Y.; Du, F.; Chen, M.; Dong, X.; Chen, Y.; Huang, W. IL-17A Inhibits osteogenic Differentiation of bone mesenchymal stem cells via wnt signaling pathway. Med. Sci. Monit. 2017, 23, 4095. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.Y.; Tang, S.A.; Ye, G.W.; Wang, P.; Li, J.T.; Liu, W.J.; Li, M.; Wang, S.; Wu, X.H.; Cen, S.Z.; et al. Interleukin-6/interleukin-6 receptor complex promotes osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Stem Cell Res. Ther. 2018, 9, 13. [Google Scholar] [CrossRef]
- Li, G.; Liu, J.Y.; Zhao, M.Z.; Wang, Y.Y.; Yang, K.; Liu, C.; Xiao, Y.; Wen, X.J.; Liu, L.C. SOST, an LNGFR target, inhibits the osteogenic differentiation of rat ectomesenchymal stem cells. Cell Prolif. 2018, 51, e12412. [Google Scholar] [CrossRef]
- Kim, S.E.; Lee, D.W.; Yun, Y.P.; Shim, K.S.; Jeon, D.I.; Rhee, J.K.; Kim, H.J.; Park, K. Heparin-immobilized hydroxyapatite nanoparticles as a lactoferrin delivery system for improving osteogenic differentiation of adipose-derived stem cells. Biomed. Mater. 2016, 11, 025004. [Google Scholar] [CrossRef]
- Deng, L.D.; Hong, H.; Zhang, X.Q.; Chen, D.R.; Chen, Z.Y.; Ling, J.Q.; Wu, L.P. Down-regulated lncRNA MEG3 promotes osteogenic differentiation of human dental follicle stem cells by epigenetically regulating Wnt pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2061–2067. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Liu, Z.G.; Yu, B.; Zhang, X.R. MicroRNA-874 targeting SUFU involves in osteoblast proliferation and differentiation in osteoporosis rats through the hedgehog signaling pathway. Biochem. Biophys. Res. Commun. 2018, 506, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhang, W.W.; Huang, E.Y.; Wang, J.; Liao, J.Y.; Li, R.D.; Yu, X.Y.; Zhao, C.; Zeng, Z.Y.; Shu, Y.; et al. BMP9-induced osteoblastic differentiation requires functional Notch signaling in mesenchymal stem cells. Lab. Investig. 2019, 99, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Xue, D.T.; Chen, E.M.; Zhang, W.; Gao, X.; Yu, J.W.; Feng, Y.D.; Pan, Z.J. HMGB1 promotes the secretion of multiple cytokines and potentiates the osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Exp. Ther. Med. 2016, 12, 3941–3947. [Google Scholar] [CrossRef] [PubMed]
- Viti, F.; Landini, M.; Mezzelani, A.; Petecchia, L.; Milanesi, L.; Scaglione, S. Osteogenic differentiation of msc through calcium signaling activation: Transcriptomics and functional analysis. PLoS ONE 2016, 11, e0148173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, H.Y.; Li, Y.X.; Ren, F.Z.; Guo, H.Y. Lactoferrin-induced growth factors and cytokines expression profile in pre-osteoblast MC3T3-E1 cell and LRP1 stable knockdown MC3T3-E1 cell. J. Funct. Foods 2017, 37, 147–156. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Zhao, Q.; Liu, Y.; Zhang, L.; Li, D.X.; Zhu, Z.L.; Gan, X.Q.; Yu, H.Y. Vibration loading promotes osteogenic differentiation of bone marrow-derived mesenchymal stem cells via p38 MAPK signaling pathway. J. Biomech. 2018, 71, 67–75. [Google Scholar] [CrossRef]
- He, Y.L.; de Castro, L.F.; Shin, M.H.; Dubois, W.; Yang, H.H.; Jiang, S.L.; Mishra, P.J.; Ren, L.; Gou, H.F.; Lal, A.; et al. p53 Loss Increases the Osteogenic Differentiation of Bone Marrow Stromal Cells. Stem Cells 2015, 33, 1304–1319. [Google Scholar] [CrossRef]
- Artigas, N.; Gamez, B.; Cubillos-Rojas, M.; Sanchez-de Diego, C.; Valer, J.A.; Pons, G.; Rosa, J.L.; Ventura, F. p53 inhibits SP7/Osterix activity in the transcriptional program of osteoblast differentiation. Cell Death Differ. 2017, 24, 2022–2031. [Google Scholar] [CrossRef]
- Molchadsky, A.; Shats, I.; Goldfinger, N.; Pevsner-Fischer, M.; Olson, M.; Rinon, A.; Tzahor, E.; Lozano, G.; Zipori, D.; Sarig, R.; et al. p53 Plays a Role in Mesenchymal Differentiation Programs, in a Cell Fate Dependent Manner. PLoS ONE 2008, 3, e3707. [Google Scholar] [CrossRef]
- Kim, M.O.; Jung, H.; Kim, S.C.; Park, J.K.; Seo, Y.K. Electromagnetic fields and nanomagnetic particles increase the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Int. J. Mol. Med. 2015, 35, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.Y.; Li, J.T.; Wang, P.; Li, Y.X.; Wu, X.H.; Wang, S.; Su, H.J.; Deng, W.; Liu, Z.H.; Cen, S.Z.; et al. Differential Expression Profiles of Long Noncoding RNA and mRNA of Osteogenically Differentiated Mesenchymal Stem Cells in Ankylosing Spondylitis. J. Rheumatol. 2016, 43, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.R.; Liu, N.; Wang, W.B. High glucose prevents osteogenic differentiation of mesenchymal stem cells via lncRNA AK028326/CXCL13 pathway. Biomed. Pharmacother. 2016, 84, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jia, L.F.; Zhang, S.; Zheng, Y.F.; Zhou, Y.S. DEPTOR regulates osteogenic differentiation via inhibiting MEG3-mediated activation of BMP4 signaling and is involved in osteoporosis. Stem Cell Res. Ther. 2018, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Q.; Zhang, Z.H.; Chen, Z.Q.; Zhang, D.D. Osteogenic growth peptide promotes osteogenic differentiation of mesenchymal stem cells mediated by LncRNA AK141205-induced upregulation of CXCL13. Biochem. Biophys. Res. Commun. 2015, 466, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.L.; Jia, B.; Sun, X.; Hu, W.T.; Chu, H.X.; Xu, S.M.; Zhao, J.J. The Critical Role of Long Noncoding RNA in Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. Biomed. Res. Int. 2017, 2017, 11. [Google Scholar] [CrossRef]
- Nardocci, G.; Carrasco, M.E.; Acevedo, E.; Hodar, C.; Meneses, C.; Montecino, M. Identification of a novel long noncoding RNA that promotes osteoblast differentiation. J. Cell. Biochem. 2018, 119, 7657–7666. [Google Scholar] [CrossRef]
- Tye, C.E.; Gordon, J.A.R.; Martin-Buley, L.A.; Stein, J.L.; Lian, J.B.; Stein, G.S. Could lncRNAs be the Missing Links in Control of Mesenchymal Stem Cell Differentiation? J. Cell. Physiol. 2015, 230, 526–534. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Dong, R.; Diao, S.; Du, J.; Fan, Z.P.; Wang, F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 2017, 8, 30. [Google Scholar] [CrossRef]
- Li, H.L.; Fan, J.F.; Fan, L.Y.; Li, T.P.; Yang, Y.L.; Xu, H.Y.; Deng, L.C.; Li, J.; Li, T.; Weng, X.S.; et al. MiRNA-10b Reciprocally Stimulates Osteogenesis and Inhibits Adipogenesis Partly through the TGF-beta/SMAD2 Signaling Pathway. Aging Dis. 2018, 9, 1058–1073. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, V.; Rattan, V.; Jha, V.; Bhattacharyya, S. Secretome proteins regulate comparative osteogenic and adipogenic potential in bone marrow and dental stem cells. Biochimie 2018, 155, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Miao, J.; Meng, X.; Chen, N.; Wang, Y.L. Expression of long non-coding RNAs in human bone marrow mesenchymal stem cells co-cultured with human amnion-derived mesenchymal stem cells. Mol. Med. Rep. 2017, 16, 6683–6689. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.T.; Zhang, C.H.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Bateman, A.; Finn, R.D. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 2007, 8, 298. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
| Sample | Total Reads | Total Mapped Reads | Unique Mapped Reads | Reads Mapped in Paired | Detected Gene Number | Detected SNP Number | Detected InDel Number |
|---|---|---|---|---|---|---|---|
| CON 1 | 152,017,088 | 94.17% | 75.18% | 92.02% | 9793 | 50,655 | 8142 |
| CON 2 | 151,919,042 | 93.78% | 74.16% | 91.80% | 9485 | 46,504 | 7442 |
| CON 3 | 152,842,644 | 96.66% | 78.92% | 94.02% | 9777 | 50,140 | 8047 |
| LF 1 | 151,975,658 | 96.84% | 77.27% | 94.59% | 9890 | 51,318 | 8190 |
| LF 2 | 152,415,436 | 95.30% | 73.01% | 92.95% | 9171 | 43,676 | 7003 |
| LF 3 | 151,898,670 | 91.47% | 65.73% | 88.37% | 9219 | 42,587 | 6739 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; An, J.-J.; Tabys, D.; Xie, Y.-D.; Zhao, T.-Y.; Ren, H.-W.; Liu, N. Effect of Lactoferrin on the Expression Profiles of Long Non-coding RNA during Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 4834. https://doi.org/10.3390/ijms20194834
Xu Y, An J-J, Tabys D, Xie Y-D, Zhao T-Y, Ren H-W, Liu N. Effect of Lactoferrin on the Expression Profiles of Long Non-coding RNA during Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2019; 20(19):4834. https://doi.org/10.3390/ijms20194834
Chicago/Turabian StyleXu, Yan, Jing-Jing An, Dina Tabys, Yin-Dan Xie, Tian-Yu Zhao, Hao-Wei Ren, and Ning Liu. 2019. "Effect of Lactoferrin on the Expression Profiles of Long Non-coding RNA during Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells" International Journal of Molecular Sciences 20, no. 19: 4834. https://doi.org/10.3390/ijms20194834
APA StyleXu, Y., An, J.-J., Tabys, D., Xie, Y.-D., Zhao, T.-Y., Ren, H.-W., & Liu, N. (2019). Effect of Lactoferrin on the Expression Profiles of Long Non-coding RNA during Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. International Journal of Molecular Sciences, 20(19), 4834. https://doi.org/10.3390/ijms20194834