Discovery of 3-Arylquinoxaline Derivatives as Potential Anti-Dengue Virus Agents
Abstract
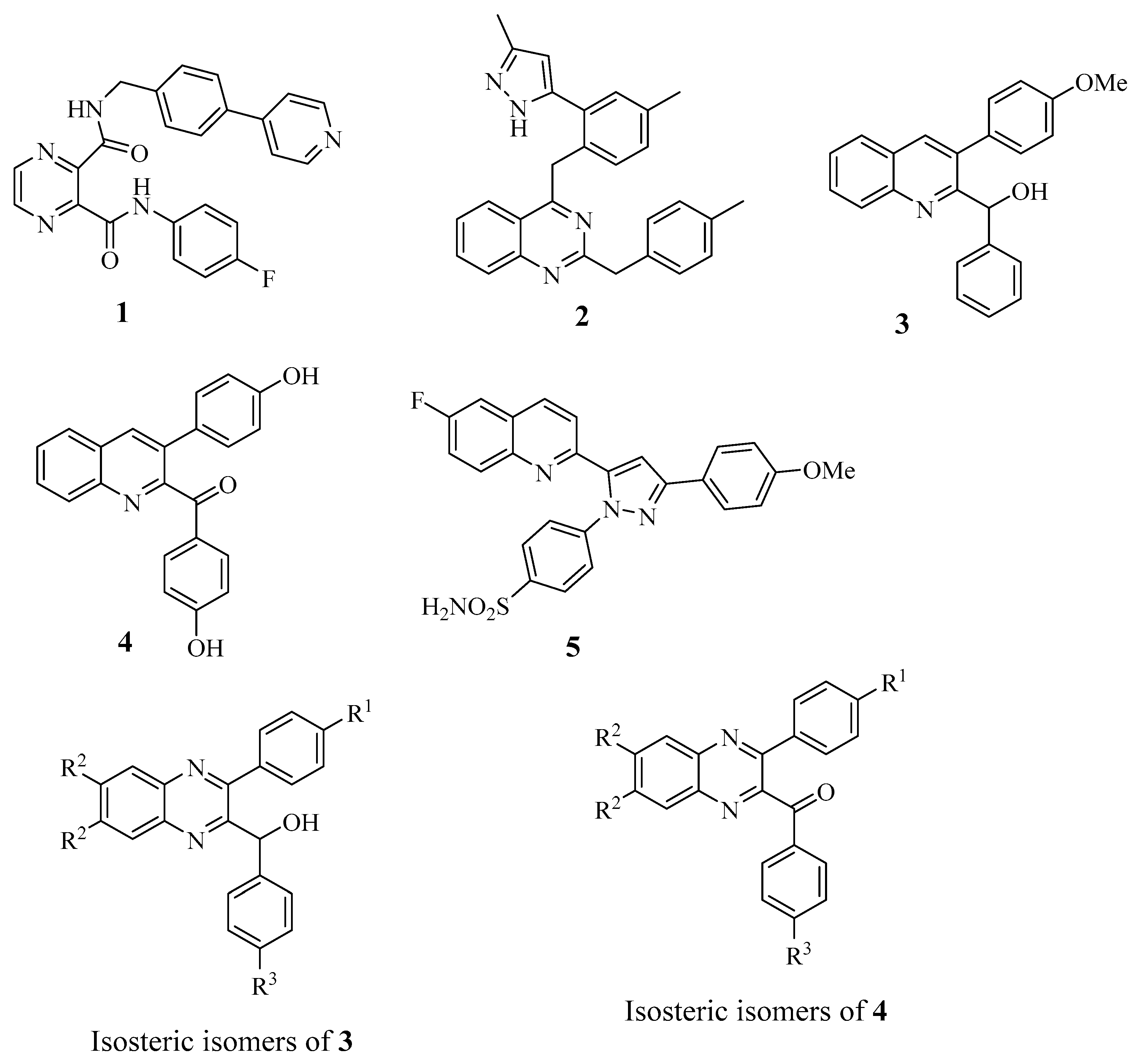
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Anti-DENV Activities and Cytotoxicities
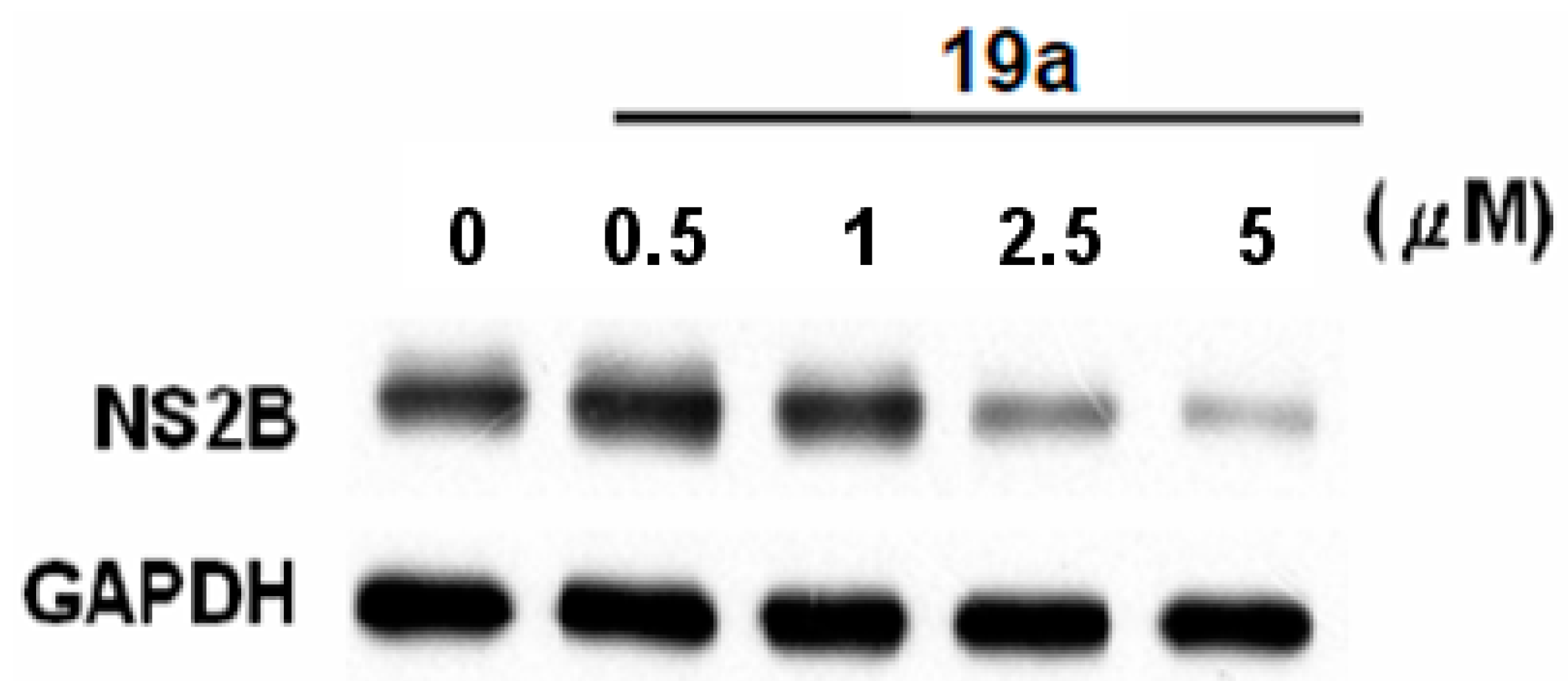
2.2.2. Compound 19a Reduced DENV Replication in Huh-7 cells
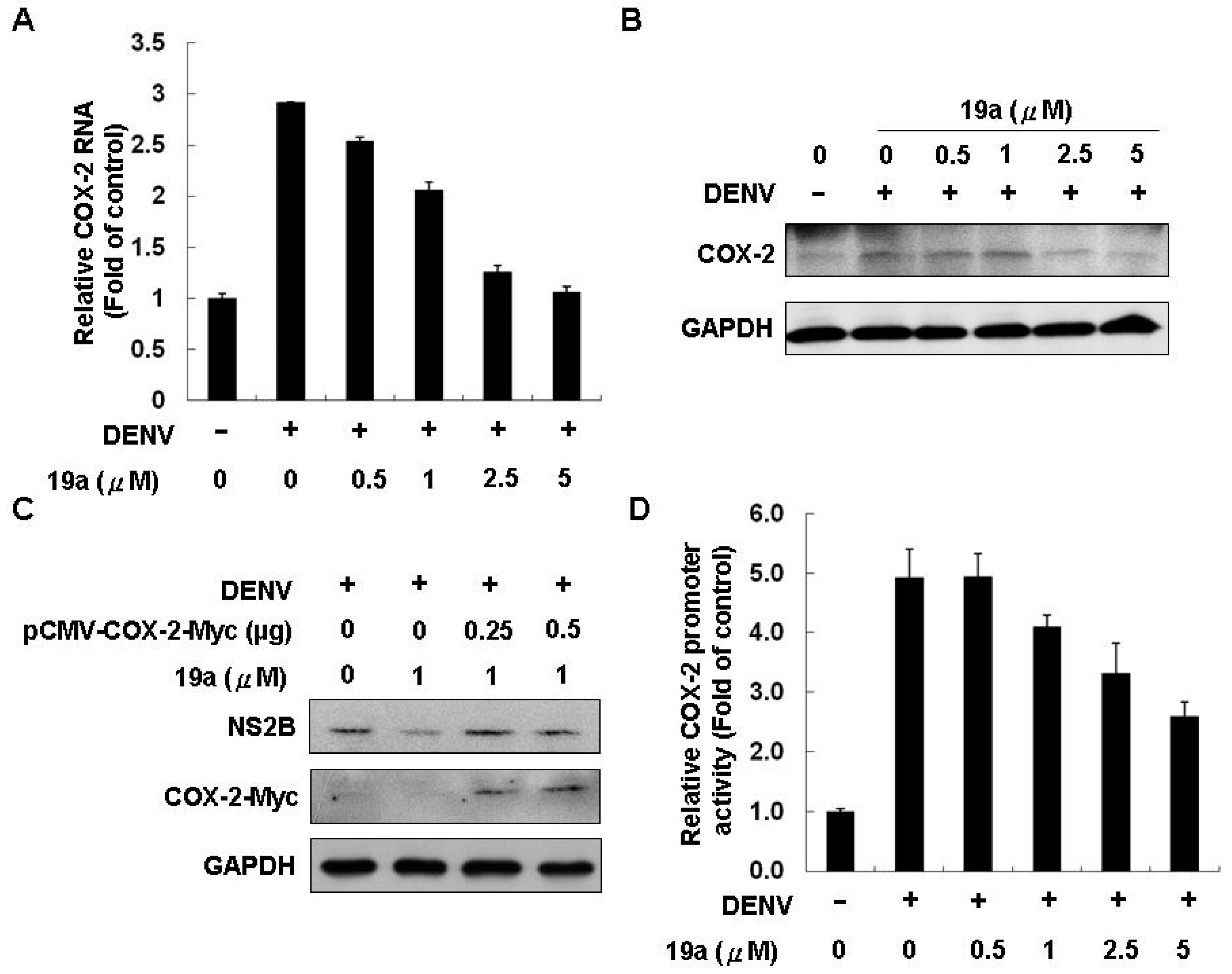
2.2.3. Compound 19a Reduced DENV Replication through Inhibiting COX-2 Expression
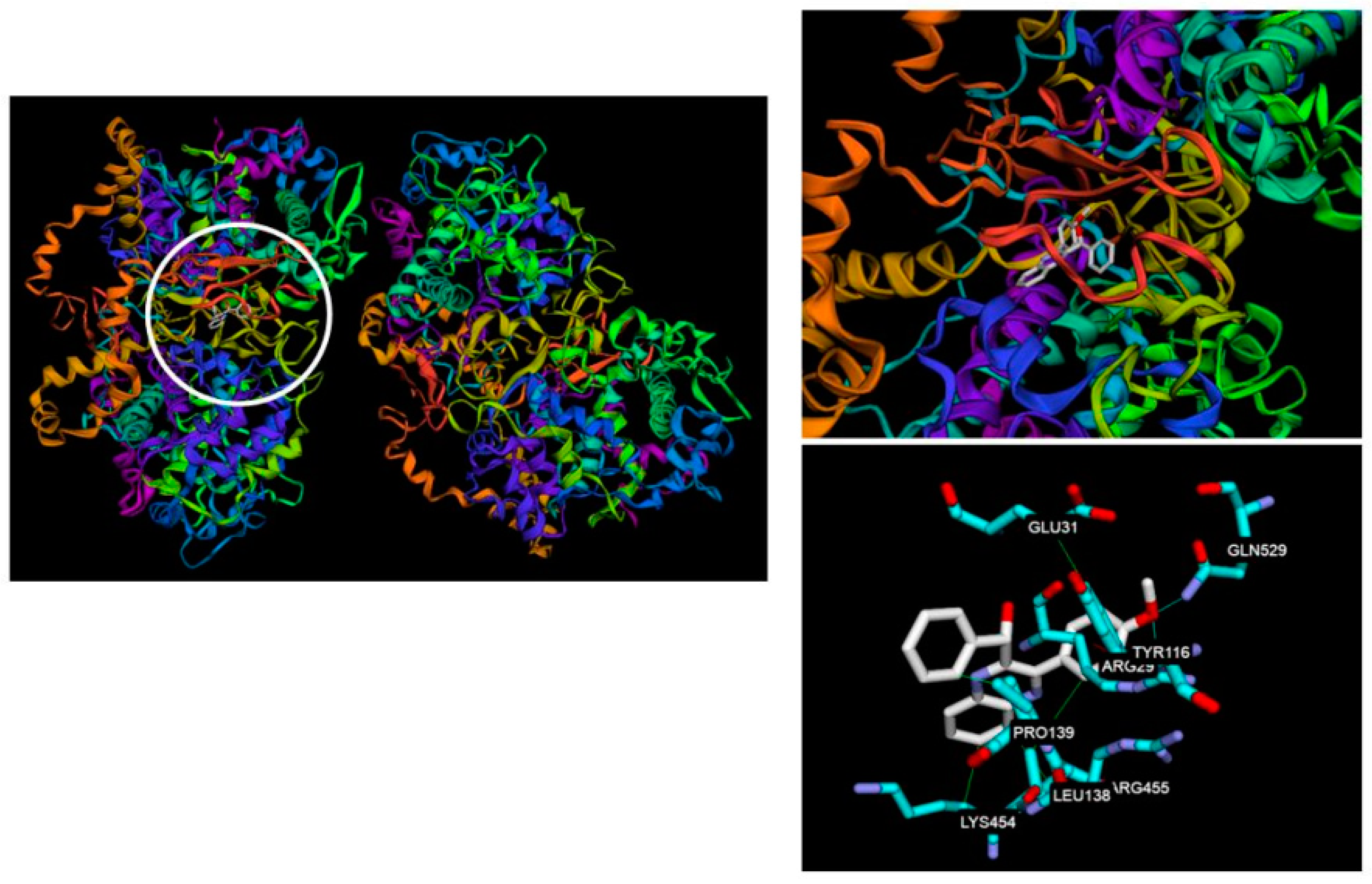
2.3. Molecular Docking
3. Materials and Methods
3.1. Chemistry Section
3.1.1. General Procedure for the Preparation of 2-Methyl-3-phenylquinoxalines 9a–11b
3.1.2. General Procedure for the Preparation of 3-Phenylquinoxaline-2-carbaldehydes 12a–14b
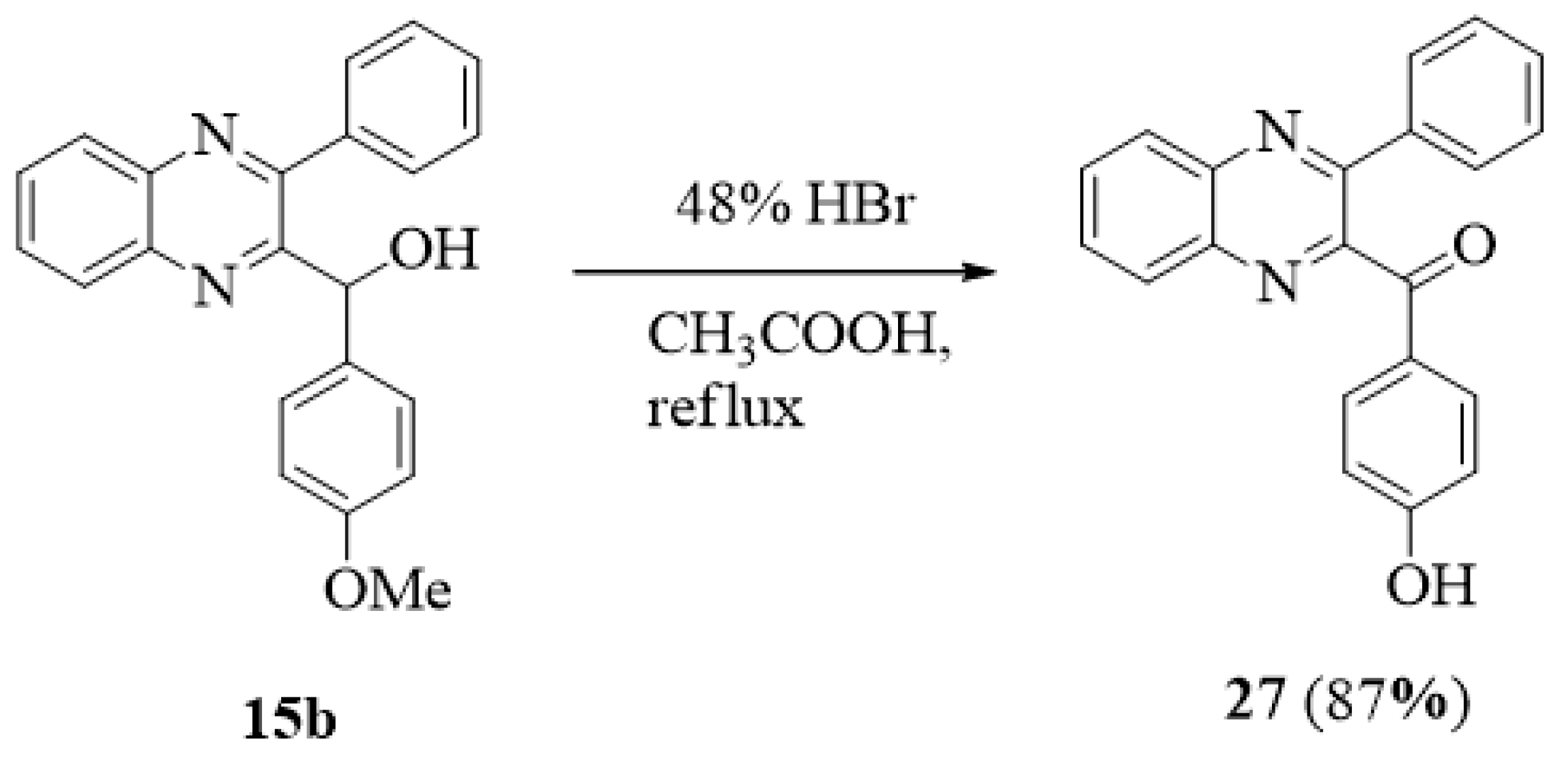
3.1.3. General Procedure for the Preparation of Phenyl(3-phenylquinoxalin-2-yl)methanols 15a–20b
Phenyl(3-phenylquinoxalin-2-yl)methanol (15a)
(4-Methoxyphenyl)(3-phenylquinoxalin-2-yl)methanol (15b)
(6,7-Dichloro-3-phenylquinoxalin-2-yl)(phenyl)methanol (16a)
(6,7-Dichloro-3-phenylquinoxalin-2-yl)(4-methoxyphenyl)methanol (16b)
[3-(4-Fluorophenyl)quinoxalin-2-yl](phenyl)methanol (17a)
[3-(4-Fluorophenyl)quinoxalin-2-yl](4-methoxyphenyl)methanol (17b)
[6,7-Dichloro-3-(4-fluorophenyl)quinoxalin-2-yl](phenyl)methanol (18a)
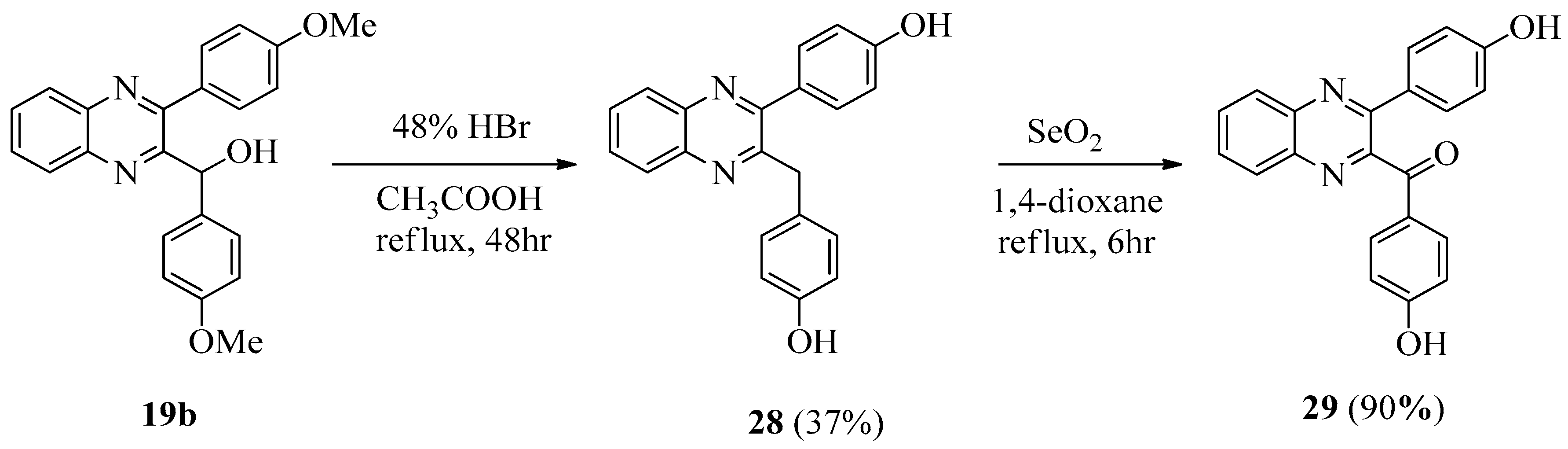
[3-(4-Methoxyphenyl)quinoxalin-2-yl](phenyl)methanol (19a)
(4-Methoxyphenyl)[3-(4-methoxyphenyl)quinoxalin-2-yl]methanol (19b)
[6,7-Dichloro-3-(4-methoxyphenyl)quinoxalin-2-yl](phenyl)methanol (20a)
[6,7-Dichloro-3-(4-methoxyphenyl)quinoxalin-2-yl](4-methoxyphenyl)methanol (20b)
3.1.4. General procedure for the Preparation of Phenyl(3-phenylquinoxalin-2-yl)methanones (21a–26b)
Phenyl(3-phenylquinoxalin-2-yl)methanone (21a)
(4-Methoxyphenyl)(3-phenylquinoxalin-2-yl)methanone (21b)
(6,7-Dichloro-3-phenylquinoxalin-2-yl)(phenyl)methanone (22a)
(6,7-Dichloro-3-phenylquinoxalin-2-yl)(4-methoxyphenyl)methanone (22b)
[3-(4-Fluorophenyl)quinoxalin-2-yl](phenyl)methanone (23a)
[3-(4-Fluorophenyl)quinoxalin-2-yl](4-methoxyphenyl)methanone (23b)
(6,7-Dichloro-3-(4-fluorophenyl)quinoxalin-2-yl)(phenyl)methanone (24a)
[3-(4-Methoxyphenyl)quinoxalin-2-yl](phenyl)methanone (25a)
(4-Methoxyphenyl)[3-(4-methoxyphenyl)quinoxalin-2-yl]methanone (25b)
[6,7-Dichloro-3-(4-methoxyphenyl)quinoxalin-2-yl](phenyl)methanone (26a)
[6,7-Dichloro-3-(4-methoxyphenyl)quinoxalin-2-yl](4-methoxyphenyl)methanone (26b)
3.1.5. (4-Hydroxyphenyl)(3-phenylquinoxalin-2-yl)methanone (27)
3.1.6. 4-[3-(4-Hydroxybenzyl)quinoxalin-2-yl]phenol (28)
3.1.7. (4-Hydroxyphenyl)(3-(4-hydroxyphenyl)quinoxalin-2-yl)methanone (29)
3.2. Biological Activity
3.2.1. Compounds
3.2.2. Cell
3.2.3. Cytotoxicity Assays
3.2.4. Transfection and Luciferase Activity Assay
3.2.5. Immunoblot Analysis
3.2.6. Quantification of DENV RNA
3.2.7. Molecular Docking Study
3.2.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization and World Meteorological Organization. Atlas of health and climate; Section 1: infections.; World Health Organization and World Meteorological Organization: Geneva, Switzerland, 2012; p. 21. [Google Scholar]
- Halstead, S.B. Pathogenesis of dengue: Challenges to molecular biology. Science 1988, 239, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Kurane, I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Kalayanarooj, S. Clinical manifestations and management of dengue/DHF/DSS. Trop. Med. Health. 2011, 39, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Low, J.G.; Ooi, E.E.; Vasudevan, S.G. Current status of dengue therapeutics research and development. J. Infect. Dis. 2017, 215, S96–S102. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Wu, Y.H.; Tseng, C.K.; Lin, C.K.; Chen, W.C.; Hsu, Y.C.; Lee, J.C. Green tea phenolic epicatechins inhibit hepatitis C virus replication via cycloxygenase-2 and attenuate virus-induced inflammation. PLoS ONE 2013, 8, e54466. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Cong, J.P.; Yu, D.; Bresnahan, W.A.; Shenk, T.E. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc. Natl. Acad. Sci. USA 2002, 99, 3932–3937. [Google Scholar] [CrossRef] [PubMed]
- Schröer, J.; Shenk, T. Inhibition of cyclooxygenase activity blocks cell-to-cell spread of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2008, 105, 19468–19473. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Siddiqui, A. Hepatitis C virus stimulates the expression of cyclooxygenase-2 via oxidative stress: Role of prostaglandin E2 in RNA replication. J. Virol. 2005, 79, 9725–9734. [Google Scholar] [CrossRef]
- Jhaveri, R.; Kundu, P.; Shapiro, A.M.; Venkatesan, A.; Dasgupta, A. Effect of hepatitis C virus core protein on cellular gene expression: Specific inhibition of cyclooxygenase 2. J. Infect. Dis. 2005, 191, 1498–1506. [Google Scholar] [CrossRef]
- Lin, C.K.; Tseng, C.K.; Wu, Y.H.; Liaw, C.C.; Lin, C.Y.; Huang, C.H.; Chen, Y.H.; Lee, J.C. Cyclooxygenase-2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents. Sci. Rep. 2017, 7, 44701. [Google Scholar] [CrossRef]
- Saudi, M.; Zmurko, J.; Kaptein, S.; Rozenski, J.; Neyts, J.; Van Aerschot, A. Synthesis and evaluation of imidazole-4,5- and pyrazine-2,3-dicarboxamides targeting dengue and yellow fever virus. Eur. J. Med. Chem. 2014, 87, 529–539. [Google Scholar] [CrossRef]
- Venkatesham, A.; Saudi, M.; Kaptein, S.; Neyts, J.; Rozenski, J.; Froeyen, M.; Van Aerschot, A. Aminopurine and aminoquinazoline scaffolds for development of potential dengue virus inhibitors. Eur. J. Med. Chem. 2017, 126, 101–109. [Google Scholar] [CrossRef]
- Tseng, C.H.; Lin, C.K.; Chen, Y.L.; Hsu, C.Y.; Wu, H.N.; Tseng, C.K.; Lee, J.C. Synthesis, antiproliferative and anti-dengue virus evaluations of 2-aroyl-3-arylquinoline derivatives. Eur. J. Med. Chem. 2014, 79, 66–76. [Google Scholar] [CrossRef]
- Lee, J.C.; Tseng, C.K.; Lin, C.K.; Tseng, C.H. Discovery of novel diarylpyrazolylquinoline derivatives as potent anti-dengue virus agents. Eur. J. Med. Chem. 2017, 141, 282–292. [Google Scholar] [CrossRef]
- Radwan, A.A.; Abdel-Mageed, W.M. In silico studies of quinoxaline-2-carboxamide 1,4-di-n-oxide derivatives as antimycobacterial agents. Molecules 2014, 19, 2247–2260. [Google Scholar] [CrossRef]
- Tariq, S.; Somakala, K.; Amir, M. Quinoxaline: An insight into the recent pharmacological advances. Eur. J. Med. Chem. 2018, 143, 542–557. [Google Scholar] [CrossRef]
- Pereira, J.A.; Pessoa, A.M.; Cordeiro, M.N.; Fernandes, R.; Prudêncio, C.; Noronha, J.P.; Vieira, M. Quinoxaline, its derivatives and applications: A State of the Art review. Eur. J. Med. Chem. 2015, 97, 664–672. [Google Scholar] [CrossRef]
- González, M.; Cerecetto, H. Quinoxaline derivatives: A patent review (2006--present). Expert Opin. Ther. Pat. 2012, 22, 1289–1302. [Google Scholar] [CrossRef]
- Shi, L.; Hu, W.; Wu, J.; Zhou, H.; Zhou, H.; Li, X. Quinoxalinone as a privileged platform in drug development. Mini. Rev. Med. Chem. 2018, 18, 392–413. [Google Scholar] [CrossRef]
- Jampilek, J. Recent advances in design of potential quinoxaline anti-infectives. Curr. Med. Chem. 2014, 21, 4347–4373. [Google Scholar] [CrossRef]
- Tseng, C.H.; Chen, Y.R.; Tzeng, C.C.; Liu, W.; Chou, C.K.; Chiu, C.C.; Chen, Y.L. Discovery of indeno[1,2-b]quinoxaline derivatives as potential anticancer agents. Eur. J. Med. Chem. 2016, 108, 258–273. [Google Scholar] [CrossRef]
- Kano, S.; Shibuya, S.; Yuasa, Y. Synthesis of quinoxaline derivatives through condensation of 1,2-diaminobenzenes with β-keto sulfoxides. J. Heterocycl. Chem. 1980, 17, 1559–1561. [Google Scholar] [CrossRef]
- Lou, S.J.; Xu, D.Q.; Shen, D.F.; Wang, Y.F.; Liu, Y.K.; Xu, Z.Y. Highly efficient vinylaromatics generation via iron-catalyzed sp3 C-H bond functionalization CDC reaction: A novel approach to preparing substituted benzo[α]phenazines. Chem. Commun. 2012, 48, 11993–11995. [Google Scholar] [CrossRef]
- Roehm, N.W.; Rodgers, G.H.; Hatfield, S.M.; Glasebrook, A.L. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazoliumsalt XTT. J. Immunol. Methods 1991, 142, 257–265. [Google Scholar] [CrossRef]
- Sánchez-Linares, I.; Pérez-Sánchez, H.; Cecilia, J.M.; García, J.M. High-throughput parallel blind virtual screening using BINDSURF. BMC Bioinform. 2012, 13, S13. [Google Scholar] [CrossRef]
- Rego, N.; Koes, D. 3Dmol.js: Molecular visualization with WebGL. Bioinformatics 2015, 31, 1322–1324. [Google Scholar] [CrossRef]
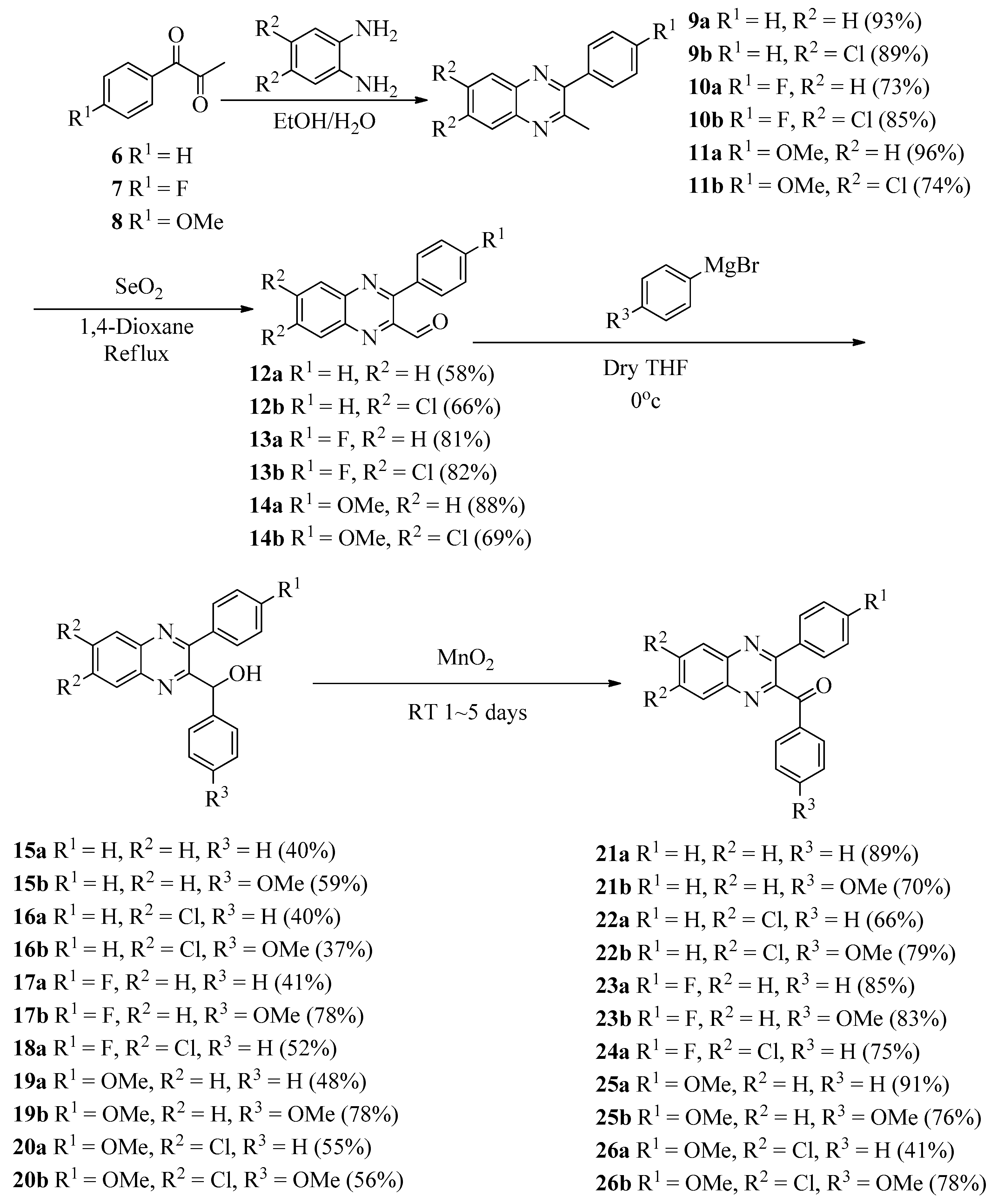

| Compounds | DENV2 | Huh-7 Cell | ||
|---|---|---|---|---|
| a % Inhibition at 1 μM | % Inhibition at 10 μM | % viability at 20 μM | % viability at 200 μM | |
| 3 | 12.37 ± 2.67 | 50.14 ± 3.47 | 90.28 ± 2.62 | 21.87 ± 3.55 |
| 4 | 11.21 ± 2.31 | 41.24 ± 2.88 | 96.62 ± 8.78 | 41.38 ± 5.71 |
| 15a | 2.13 ± 1.12 | 5.31 ± 1.73 | 104.71 ± 5.66 | 59.94 ± 8.33 |
| 15b | 24.34 ± 2.51 | 48.67 ± 4.71 | 103.29 ± 1.79 | 61.42 ± 2.67 |
| 16a | 1.97 ± 0.72 | 6.42 ± 2.97 | 108.06 ± 8.91 | 20.62 ± 5.37 |
| 16b | 2.41 ± 1.48 | 9.46 ± 2.88 | 104.05 ± 4.18 | 72.96 ± 9.05 |
| 17a | 2.58 ± 0.85 | 4.78 ± 1.93 | 96.35 ± 3.52 | 73.26 ± 6.57 |
| 17b | 19.17 ± 3.5 | 22.89 ± 4.54 | 102.33 ± 1.98 | 100.73 ± 1.27 |
| 18a | 3.41 ± 1.11 | 6.74 ± 1.57 | 102.04 ± 3.96 | 21.50 ± 1.60 |
| 19a | 36.74 ± 3.71 | 82.71 ± 2.88 | 99.66 ± 1.65 | 85.26 ± 5.63 |
| 19b | 35.47 ± 4.82 | 47.69 ± 5.41 | 96.43 ± 1.24 | 21.34 ± 1.55 |
| 20a | 16.31 ± 3.42 | 58.71 ± 2.86 | 99.42 ± 3.31 | 27.12 ± 1.24 |
| 20b | 2.75 ± 1.69 | 4.39 ± 1.52 | 95.47 ± 3.96 | 34.21 ± 4.97 |
| 21a | 2.76 ± 1.55 | 7.98 ± 2.14 | 107.34 ± 2.30 | 79.45 ± 7.07 |
| 21b | 42.78 ± 5.13 | 51.85 ± 3.52 | 106.46 ± 3.12 | 95.13 ± 4.32 |
| 22a | 12.74 ± 3.91 | 19.74 ± 4.17 | 108.76 ± 1.36 | 103.05 ± 2.67 |
| 22b | 5.19 ± 2.78 | 24.68 ± 1.89 | 103.09 ± 8.30 | 92.02 ± 3.08 |
| 23a | 34.17 ± 5.12 | 40.78 ± 2.58 | 98.61 ± 5.68 | 87.59 ± 5.75 |
| 23b | 16.44 ± 3.11 | 33.58 ± 4.21 | 104.26 ± 1.85 | 103.32 ± 1.03 |
| 24a | 2.54 ± 1.21 | 23.47 ± 3.71 | 106.06 ± 2.12 | 91.94 ± 2.83 |
| 25a | 40.44 ± 2.22 | 47.53 ± 3.51 | 101.13 ± 2.39 | 100.30 ± 4.59 |
| 25b | 41.51 ± 3.25 | 49.74 ± 4.38 | 106.59 ± 3.03 | 99.42 ± 2.14 |
| 26a | 1.95 ± 0.82 | 14.47 ± 2.55 | 99.39 ± 6.50 | 35.29 ± 4.95 |
| 26b | 1.17 ± 0.41 | 19.28 ± 3.85 | 104.35 ± 4.22 | 99.11 ± 2.40 |
| 27 | 44.85 ± 3.68 | 48.69 ± 3.48 | 101.58 ± 2.84 | 18.43 ± 2.33 |
| 29 | 2.36 ± 1.11 | 4.25 ± 1.39 | 103.62 ± 2.91 | 51.91 ± 1.62 |
| Ribavirin b | 10.14 ± 1.98 | 32.53 ± 2.30 | 71.37 ± 1.31 | 14.36 ± 2.14 |
| Compounds | EC50 a | CC50 b | SI c |
|---|---|---|---|
| 3 | 12.57 ± 2.16 | 139.11 ± 2.32 | 11.07 |
| 4 | 13.16 ± 1.23 | 163.67 ± 2.32 | 12.44 |
| 19a | 1.29 ± 0.74 | >200 | >155.04 |
| 20a | 5.68 ± 0.81 | >200 | >35.21 |
| 21b | 3.25 ± 0.89 | >200 | >61.54 |
| Ribavirin | 12.61 ± 1.17 | 56.31 ± 2.32 | 4.47 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, C.-H.; Han, C.-R.; Tang, K.-W. Discovery of 3-Arylquinoxaline Derivatives as Potential Anti-Dengue Virus Agents. Int. J. Mol. Sci. 2019, 20, 4786. https://doi.org/10.3390/ijms20194786
Tseng C-H, Han C-R, Tang K-W. Discovery of 3-Arylquinoxaline Derivatives as Potential Anti-Dengue Virus Agents. International Journal of Molecular Sciences. 2019; 20(19):4786. https://doi.org/10.3390/ijms20194786
Chicago/Turabian StyleTseng, Chih-Hua, Cheng-Ruei Han, and Kai-Wei Tang. 2019. "Discovery of 3-Arylquinoxaline Derivatives as Potential Anti-Dengue Virus Agents" International Journal of Molecular Sciences 20, no. 19: 4786. https://doi.org/10.3390/ijms20194786
APA StyleTseng, C.-H., Han, C.-R., & Tang, K.-W. (2019). Discovery of 3-Arylquinoxaline Derivatives as Potential Anti-Dengue Virus Agents. International Journal of Molecular Sciences, 20(19), 4786. https://doi.org/10.3390/ijms20194786





