Epigenetic Alterations of Heat Shock Proteins (HSPs) in Cancer
Abstract
1. Introduction
2. Epigenetic Alterations of HSPs Expression in Cancer
2.1. HSP60
2.1.1. miRNAs and DNA Methylation of HSP60
2.1.2. Histone and Translational Modifications of HSP60
2.2. HSP70
2.2.1. DNA Methylation of HSP70
2.2.2. miRNAs of HSP70
2.2.3. Histone Modifications of HSP70
2.2.4. Translational Modification of HSP70
2.3. HSP90
2.3.1. miRNAs of HSP90
2.3.2. Translational Modification of HSP90
2.4. Others
2.4.1. HSP27 (HSPB1, HSPB2)
2.4.2. HSP40 Family (DNAJB6, MCJ)
2.4.3. HSP110
3. Clinical Relevance of HSPs in Cancer
HSPs as Cancer Treatment and Therapeutic Targets
4. Conclusions
Funding
Conflicts of Interest
References
- Bakthisaran, R.; Tangirala, R.; Rao Ch, M. Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta 2015, 1854, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.J.; Hageman, J.; Carra, S.; Kampinga, H.H. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry 2008, 47, 7001–7011. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharm. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Ciocca, D.R. Heat shock proteins: stress proteins with Janus-like properties in cancer. Int. J. Hyperth. 2008, 24, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Conway de Macario, E.; Marasa, L.; Zummo, G.; Macario, A.J. Hsp60 expression, new locations, functions and perspectives for cancer diagnosis and therapy. Cancer Biol. 2008, 7, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Ischia, J.; So, A.I. The role of heat shock proteins in bladder cancer. Nat. Rev. Urol 2013, 10, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Han, T.S.; Ban, H.S.; Hur, K.; Cho, H.S. The Epigenetic Regulation of HCC Metastasis. Int. J. Mol. Sci. 2018, 19, 3978. [Google Scholar] [CrossRef] [PubMed]
- Han, T.S.; Voon, D.C.; Oshima, H.; Nakayama, M.; Echizen, K.; Sakai, E.; Yong, Z.W.E.; Murakami, K.; Yu, L.; Minamoto, T.; et al. Interleukin 1 Up-regulates MicroRNA 135b to Promote Inflammation-Associated Gastric Carcinogenesis in Mice. Gastroenterology 2019, 156, 1140–1155. [Google Scholar] [CrossRef]
- Kim, K.; Son, M.Y.; Jung, C.R.; Kim, D.S.; Cho, H.S. EHMT2 is a metastasis regulator in breast cancer. Biochem. Biophys. Res. Commun. 2018, 496, 758–762. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, K.; Ryu, J.W.; Ryu, T.Y.; Lim, J.H.; Oh, J.H.; Min, J.K.; Jung, C.R.; Hamamoto, R.; Son, M.Y.; et al. The novel prognostic marker, EHMT2, is involved in cell proliferation via HSPD1 regulation in breast cancer. Int. J. Oncol. 2019, 54, 65–76. [Google Scholar] [CrossRef]
- Ryu, J.W.; Kim, S.K.; Son, M.Y.; Jeon, S.J.; Oh, J.H.; Lim, J.H.; Cho, S.; Jung, C.R.; Hamamoto, R.; Kim, D.S.; et al. Novel prognostic marker PRMT1 regulates cell growth via downregulation of CDKN1A in HCC. Oncotarget 2017, 8, 115444–115455. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.Y.; Kim, K.; Kim, S.K.; Oh, J.H.; Min, J.K.; Jung, C.R.; Son, M.Y.; Kim, D.S.; Cho, H.S. SETDB1 regulates SMAD7 expression for breast cancer metastasis. Bmb Rep. 2019, 52, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.Y.; Kim, K.; Son, M.Y.; Min, J.K.; Kim, J.; Han, T.S.; Kim, D.S.; Cho, H.S. Downregulation of PRMT1, a histone arginine methyltransferase, by sodium propionate induces cell apoptosis in colon cancer. Oncol. Rep. 2019, 41, 1691–1699. [Google Scholar] [CrossRef]
- Son, M.Y.; Jung, C.R.; Kim, D.S.; Cho, H.S. Comparative in silico profiling of epigenetic modifiers in human tissues. Mol. Biol. Rep. 2018, 45, 309–314. [Google Scholar] [CrossRef]
- Lv, L.H.; Wan, Y.L.; Lin, Y.; Zhang, W.; Yang, M.; Li, G.L.; Lin, H.M.; Shang, C.Z.; Chen, Y.J.; Min, J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J. Biol. Chem. 2012, 287, 15874–15885. [Google Scholar] [CrossRef]
- Tsai, Y.P.; Yang, M.H.; Huang, C.H.; Chang, S.Y.; Chen, P.M.; Liu, C.J.; Teng, S.C.; Wu, K.J. Interaction between HSP60 and beta-catenin promotes metastasis. Carcinogenesis 2009, 30, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Choghaei, E.; Khamisipour, G.; Falahati, M.; Naeimi, B.; Mossahebi-Mohammadi, M.; Tahmasebi, R.; Hasanpour, M.; Shamsian, S.; Hashemi, Z.S. Knockdown of microRNA-29a Changes the Expression of Heat Shock Proteins in Breast Carcinoma MCF-7 Cells. Oncol. Res. 2016, 23, 69–78. [Google Scholar] [CrossRef]
- Liu, C.; Cao, Y.; Zhou, S.; Khoso, P.A.; Li, S. Avermectin induced global DNA hypomethylation and over-expression of heat shock proteins in cardiac tissues of pigeon. Pestic. Biochem. Physiol. 2017, 135, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Campanella, C.; D’Anneo, A.; Marino Gammazza, A.; Caruso Bavisotto, C.; Barone, R.; Emanuele, S.; Lo Cascio, F.; Mocciaro, E.; Fais, S.; Conway De Macario, E.; et al. The histone deacetylase inhibitor SAHA induces HSP60 nitration and its extracellular release by exosomal vesicles in human lung-derived carcinoma cells. Oncotarget 2016, 7, 28849–28867. [Google Scholar] [CrossRef] [PubMed]
- Marino Gammazza, A.; Campanella, C.; Barone, R.; Caruso Bavisotto, C.; Gorska, M.; Wozniak, M.; Carini, F.; Cappello, F.; D’Anneo, A.; Lauricella, M.; et al. Doxorubicin anti-tumor mechanisms include Hsp60 post-translational modifications leading to the Hsp60/p53 complex dissociation and instauration of replicative senescence. Cancer Lett. 2017, 385, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Gorska, M.; Marino Gammazza, A.; Zmijewski, M.A.; Campanella, C.; Cappello, F.; Wasiewicz, T.; Kuban-Jankowska, A.; Daca, A.; Sielicka, A.; Popowska, U.; et al. Geldanamycin-induced osteosarcoma cell death is associated with hyperacetylation and loss of mitochondrial pool of heat shock protein 60 (hsp60). PLoS ONE 2013, 8, e71135. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.E. The HSP70 family and cancer. Carcinogenesis 2013, 34, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Han, Y.; Lee, J.E.; Yenari, M.A. The 70-kDa heat shock protein (Hsp70) as a therapeutic target for stroke. Expert Opin. Ther. Targets 2018, 22, 191–199. [Google Scholar] [CrossRef]
- Campanella, C.; Pace, A.; Caruso Bavisotto, C.; Marzullo, P.; Marino Gammazza, A.; Buscemi, S.; Palumbo Piccionello, A. Heat Shock Proteins in Alzheimer’s Disease: Role and Targeting. Int. J. Mol. Sci. 2018, 19, 2603. [Google Scholar] [CrossRef] [PubMed]
- Zeller, C.; Dai, W.; Steele, N.L.; Siddiq, A.; Walley, A.J.; Wilhelm-Benartzi, C.S.M.; Rizzo, S.; van der Zee, A.; Plumb, J.A.; Brown, R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene 2012, 31, 4567. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; White, M.C.; Choi, W.; Guo, C.; Dinney, C.; McConkey, D.J.; Siefker-Radtke, A. Inhibition of inducible heat shock protein-70 (hsp72) enhances bortezomib-induced cell death in human bladder cancer cells. PLoS ONE 2013, 8, e69509. [Google Scholar] [CrossRef]
- Milutinovic, S.; Brown, S.E.; Zhuang, Q.; Szyf, M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J. Biol. Chem. 2004, 279, 27915–27927. [Google Scholar] [CrossRef]
- Jiang, L.; Kwong, D.L.; Li, Y.; Liu, M.; Yuan, Y.F.; Li, Y.; Fu, L.; Guan, X.Y. HBP21, a chaperone of heat shock protein 70, functions as a tumor suppressor in hepatocellular carcinoma. Carcinogenesis 2015, 36, 1111–1120. [Google Scholar] [CrossRef]
- MacKenzie, T.N.; Mujumdar, N.; Banerjee, S.; Sangwan, V.; Sarver, A.; Vickers, S.; Subramanian, S.; Saluja, A.K. Triptolide induces the expression of miR-142–3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol. Cancer Ther. 2013, 12, 1266–1275. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Yuan, Q.; Li, H.; Wang, W.; Xie, G.; Zhu, K.; Li, D. miR-223/Hsp70/JNK/JUN/miR-223 feedback loop modulates the chemoresistance of osteosarcoma to cisplatin. Biochem. Biophys. Res. Commun. 2018, 497, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Wei, M.; Wang, J.; Liu, Y.; Guo, M.; Li, X.; Luo, J.; Zhou, J.; Wang, M.; Guo, D.; et al. Hepatitis B virus-regulated growth of liver cancer cells occurs through the microRNA-340–5p-activating transcription factor 7-heat shock protein A member 1B axis. Cancer Sci. 2019, 110, 1633–1643. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Chen, X.; Lu, P.; Ma, W.; Yue, D.; Song, L.; Fan, Q. miR-202 Promotes Cell Apoptosis in Esophageal Squamous Cell Carcinoma by Targeting HSF2. Oncol. Res. 2017, 25, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Nakagawa, S.Y.; Nakano, T.; Asaumi, J.; Jagetia, G.C.; Kawasaki, S. Effects of KNK437 on heat-induced methylation of histone H3 in human oral squamous cell carcinoma cells. Int. J. Hyperth. Off. J. Eur. Soc. Hyperthermic Oncol. North. Am. Hyperth. Group 2006, 22, 729–735. [Google Scholar] [CrossRef]
- Smith, S.T.; Petruk, S.; Sedkov, Y.; Cho, E.; Tillib, S.; Canaani, E.; Mazo, A. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat. Cell Biol. 2004, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Shimazu, T.; Toyokawa, G.; Daigo, Y.; Maehara, Y.; Hayami, S.; Ito, A.; Masuda, K.; Ikawa, N.; Field, H.I.; et al. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat. Commun. 2012, 3, 1072. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.E.; Moen, A.; Bousset, L.; Egge-Jacobsen, W.; Kernstock, S.; Melki, R.; Falnes, P.O. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 2013, 288, 27752–27763. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Barjau, J.; Sohtome, Y.; Sodeoka, M.; Shinkai, Y. Selenium-based S-adenosylmethionine analog reveals the mammalian seven-beta-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS ONE 2014, 9, e105394. [Google Scholar] [CrossRef]
- Picard, D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol. Life Sci. 2002, 59, 1640–1648. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef]
- Trepel, J.; Mollapour, M.; Giaccone, G.; Neckers, L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 2010, 10, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Wandinger, S.K.; Richter, K.; Buchner, J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008, 283, 18473–18477. [Google Scholar] [CrossRef] [PubMed]
- Whitesell, L.; Lindquist, S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 2005, 5, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cai, M.; Fu, D.; Chen, K.; Sun, M.; Cai, Z.; Cheng, B. Heat shock protein 90B1 plays an oncogenic role and is a target of microRNA-223 in human osteosarcoma. Cell Physiol. Biochem. 2012, 30, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Jiang, F.; Zhou, J.; Wu, D.; Sheng, Z.; Li, M. HSP90: A Novel Target Gene of miRNA-628–3p in A549 Cells. Biomed. Res. Int. 2018, 2018, 4149707. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J.; Saremaslani, P.; Caduff, R. ALG-2: a Ca2+ -binding modulator protein involved in cell proliferation and in cell death. Biochim. Biophys. Acta 2002, 1600, 68–73. [Google Scholar] [CrossRef]
- Mollerup, J.; Krogh, T.N.; Nielsen, P.F.; Berchtold, M.W. Properties of the co-chaperone protein p23 erroneously attributed to ALG-2 (apoptosis-linked gene 2). Febs. Lett. 2003, 555, 478–482. [Google Scholar] [CrossRef]
- Oxelmark, E.; Roth, J.M.; Brooks, P.C.; Braunstein, S.E.; Schneider, R.J.; Garabedian, M.J. The cochaperone p23 differentially regulates estrogen receptor target genes and promotes tumor cell adhesion and invasion. Mol. Cell Biol. 2006, 26, 5205–5213. [Google Scholar] [CrossRef]
- Liu, X.; Zou, L.; Zhu, L.; Zhang, H.; Du, C.; Li, Z.; Gao, C.; Zhao, X.; Bao, S.; Zheng, H. miRNA mediated up-regulation of cochaperone p23 acts as an anti-apoptotic factor in childhood acute lymphoblastic leukemia. Leuk. Res. 2012, 36, 1098–1104. [Google Scholar] [CrossRef]
- Scroggins, B.T.; Neckers, L. Post-translational modification of heat-shock protein 90: impact on chaperone function. Expert Opin. Drug Discov. 2007, 2, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Woodford, M.R.; Truman, A.W.; Dunn, D.M.; Jensen, S.M.; Cotran, R.; Bullard, R.; Abouelleil, M.; Beebe, K.; Wolfgeher, D.; Wierzbicki, S.; et al. Mps1 Mediated Phosphorylation of Hsp90 Confers Renal Cell Carcinoma Sensitivity and Selectivity to Hsp90 Inhibitors. Cell Rep. 2016, 14, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Sims, R.J., 3rd; Gottlieb, P.D.; Tucker, P.W. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 2006, 5, 26. [Google Scholar] [CrossRef]
- Abu-Farha, M.; Lambert, J.P.; Al-Madhoun, A.S.; Elisma, F.; Skerjanc, I.S.; Figeys, D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell Proteom. 2008, 7, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Hayami, S.; Toyokawa, G.; Maejima, K.; Yamane, Y.; Suzuki, T.; Dohmae, N.; Kogure, M.; Kang, D.; Neal, D.E.; et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia 2012, 14, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Toyokawa, G.; Nakakido, M.; Ueda, K.; Nakamura, Y. SMYD2-dependent HSP90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett 2014, 351, 126–133. [Google Scholar] [CrossRef]
- Passarino, G.; Cavalleri, G.L.; Stecconi, R.; Franceschi, C.; Altomare, K.; Dato, S.; Greco, V.; Luca Cavalli Sforza, L.; Underhill, P.A.; de Benedictis, G. Molecular variation of human HSP90alpha and HSP90beta genes in Caucasians. Hum. Mutat. 2003, 21, 554–555. [Google Scholar] [CrossRef] [PubMed]
- Eustace, B.K.; Sakurai, T.; Stewart, J.K.; Yimlamai, D.; Unger, C.; Zehetmeier, C.; Lain, B.; Torella, C.; Henning, S.W.; Beste, G.; et al. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat. Cell Biol. 2004, 6, 507–514. [Google Scholar] [CrossRef]
- Yang, Y.; Rao, R.; Shen, J.; Tang, Y.; Fiskus, W.; Nechtman, J.; Atadja, P.; Bhalla, K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008, 68, 4833–4842. [Google Scholar] [CrossRef]
- Rogalla, T.; Ehrnsperger, M.; Preville, X.; Kotlyarov, A.; Lutsch, G.; Ducasse, C.; Paul, C.; Wieske, M.; Arrigo, A.P.; Buchner, J.; et al. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J. Biol. Chem. 1999, 274, 18947–18956. [Google Scholar] [CrossRef]
- Vahid, S.; Thaper, D.; Gibson, K.F.; Bishop, J.L.; Zoubeidi, A. Molecular chaperone Hsp27 regulates the Hippo tumor suppressor pathway in cancer. Sci. Rep. 2016, 6, 31842. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bao, Y.; Yang, G.K.; Wan, J.; Du, L.J.; Ma, Z.H. MiR-214 sensitizes human colon cancer cells to 5-FU by targeting Hsp27. Cell Mol. Biol. Lett. 2019, 24, 22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ju, H.; Zhang, L.; Lu, H.; Jie, K. microRNA-577 suppresses tumor growth and enhances chemosensitivity in colorectal cancer. J. Biochem. Mol. Toxicol. 2017, 31. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yin, Y.; Wang, F.; Wang, Y.; Zhang, L.; Tang, Y.; Sun, S. miR-17–5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology 2010, 51, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Chiang, W.F.; Huang, H.H.; Huang, S.K.; Chiang, H.C. Promoter hypermethylation of the gene encoding heat shock protein B1 in oral squamous carcinoma cells. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 115, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Yamashita, K.; Sidransky, D.; Kim, M.S. Promoter methylation of heat shock protein B2 in human esophageal squamous cell carcinoma. Int. J. Oncol. 2011, 38, 1129–1135. [Google Scholar] [PubMed]
- Mitra, A.; Rostas, J.W.; Dyess, D.L.; Shevde, L.A.; Samant, R.S. Micro-RNA-632 downregulates DNAJB6 in breast cancer. Lab. Invest. 2012, 92, 1310–1317. [Google Scholar] [CrossRef]
- Lindsey, J.C.; Lusher, M.E.; Strathdee, G.; Brown, R.; Gilbertson, R.J.; Bailey, S.; Ellison, D.W.; Clifford, S.C. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int. J. Cancer 2006, 118, 346–352. [Google Scholar] [CrossRef]
- Strathdee, G.; Vass, J.K.; Oien, K.A.; Siddiqui, N.; Curto-Garcia, J.; Brown, R. Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol. Oncol. 2005, 97, 898–903. [Google Scholar] [CrossRef]
- Zappasodi, R.; Ruggiero, G.; Guarnotta, C.; Tortoreto, M.; Tringali, C.; Cavane, A.; Cabras, A.D.; Castagnoli, L.; Venerando, B.; Zaffaroni, N.; et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood 2015, 125, 1768–1771. [Google Scholar] [CrossRef]
- Yu, N.; Kakunda, M.; Pham, V.; Lill, J.R.; Du, P.; Wongchenko, M.; Yan, Y.; Firestein, R.; Huang, X. HSP105 recruits protein phosphatase 2A to dephosphorylate beta-catenin. Mol. Cell Biol. 2015, 35, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Kariya, A.; Furusawa, Y.; Yunoki, T.; Kondo, T.; Tabuchi, Y. A microRNA-27a mimic sensitizes human oral squamous cell carcinoma HSC-4 cells to hyperthermia through downregulation of Hsp110 and Hsp90. Int. J. Mol. Med. 2014, 34, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Burns, T.F. Targeting Heat Shock Proteins in Cancer: A Promising Therapeutic Approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Daniels, C.K.; Cao, S. Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol. Ther. 2012, 136, 354–374. [Google Scholar] [CrossRef] [PubMed]
- Rerole, A.L.; Jego, G.; Garrido, C. Hsp70: Anti-apoptotic and tumorigenic protein. Methods Mol. Biol. 2011, 787, 205–230. [Google Scholar] [PubMed]
- Chiosis, G.; Vilenchik, M.; Kim, J.; Solit, D. Hsp90: The vulnerable chaperone. Drug Discov. Today 2004, 9, 881–888. [Google Scholar] [CrossRef]
- Neckers, L. Using natural product inhibitors to validate Hsp90 as a molecular target in cancer. Curr. Top. Med. Chem. 2006, 6, 1163–1171. [Google Scholar] [CrossRef]
- Workman, P. Combinatorial attack on multistep oncogenesis by inhibiting the Hsp90 molecular chaperone. Cancer Lett. 2004, 206, 149–157. [Google Scholar] [CrossRef]
- Zhang, H.; Burrows, F. Targeting multiple signal transduction pathways through inhibition of Hsp90. J. Mol. Med. (Berl.) 2004, 82, 488–499. [Google Scholar] [CrossRef]
- Conroy, S.E.; Latchman, D.S. Do heat shock proteins have a role in breast cancer? Br. J. Cancer 1996, 74, 717–721. [Google Scholar] [CrossRef]
- Roman, E.; Lunde, M.L.; Miron, T.; Warnakulasauriya, S.; Johannessen, A.C.; Vasstrand, E.N.; Ibrahim, S.O. Analysis of protein expression profile of oral squamous cell carcinoma by MALDI-TOF-MS. Anticancer. Res. 2013, 33, 837–845. [Google Scholar]
- Thomas, X.; Campos, L.; Mounier, C.; Cornillon, J.; Flandrin, P.; Le, Q.H.; Piselli, S.; Guyotat, D. Expression of heat-shock proteins is associated with major adverse prognostic factors in acute myeloid leukemia. Leuk Res. 2005, 29, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhi, J.; Peng, X.; Zhong, X.; Xu, A. Clinical significance of HSP27 expression in colorectal cancer. Mol Med. Rep. 2010, 3, 953–958. [Google Scholar] [PubMed]
- Li, Y.; Zhang, T.; Schwartz, S.J.; Sun, D. New developments in Hsp90 inhibitors as anti-cancer therapeutics: mechanisms, clinical perspective and more potential. Drug Resist. Updates. Rev. Comment. Antimicrob. Anticancer Chemother. 2009, 12, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Burrows, F.; Zhang, H.; Kamal, A. Hsp90 activation and cell cycle regulation. Cell Cycle (Georget. Tex.) 2004, 3, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Gibert, B.; Hadchity, E.; Czekalla, A.; Aloy, M.T.; Colas, P.; Rodriguez-Lafrasse, C.; Arrigo, A.P.; Diaz-Latoud, C. Inhibition of heat shock protein 27 (HspB1) tumorigenic functions by peptide aptamers. Oncogene 2011, 30, 3672–3681. [Google Scholar] [CrossRef] [PubMed]
- de La Motte Rouge, T.; Galluzzi, L.; Olaussen, K.A.; Zermati, Y.; Tasdemir, E.; Robert, T.; Ripoche, H.; Lazar, V.; Dessen, P.; Harper, F.; et al. A novel epidermal growth factor receptor inhibitor promotes apoptosis in non-small cell lung cancer cells resistant to erlotinib. Cancer Res. 2007, 67, 6253–6262. [Google Scholar] [CrossRef] [PubMed]
- Fendrich, V.; Wichmann, S.; Wiese, D.; Waldmann, J.; Lauth, M.; Rexin, P.; L.-Lopez, C.; Schlitt, H.J.; Bartsch, D.K.; Lang, S.A. Inhibition of heat shock protein 90 with AUY922 represses tumor growth in a transgenic mouse model of islet cell neoplasms. Neuroendocrinology 2014, 100, 300–309. [Google Scholar] [CrossRef]
- Shan, Z.X.; Lin, Q.X.; Deng, C.Y.; Zhu, J.N.; Mai, L.P.; Liu, J.L.; Fu, Y.H.; Liu, X.Y.; Li, Y.X.; Zhang, Y.Y.; et al. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. Febs. Lett. 2010, 584, 3592–3600. [Google Scholar] [CrossRef] [PubMed]
- Nagumo, Y.; Kakeya, H.; Shoji, M.; Hayashi, Y.; Dohmae, N.; Osada, H. Epolactaene binds human Hsp60 Cys442 resulting in the inhibition of chaperone activity. Biochem. J. 2005, 387, 835–840. [Google Scholar] [CrossRef]
- Chapman, E.; Farr, G.W.; Furtak, K.; Horwich, A.L. A small molecule inhibitor selective for a variant ATP-binding site of the chaperonin GroEL. Bioorganic Med. Chem. Lett. 2009, 19, 811–813. [Google Scholar] [CrossRef] [PubMed]
- Massey, A.J.; Williamson, D.S.; Browne, H.; Murray, J.B.; Dokurno, P.; Shaw, T.; Macias, A.T.; Daniels, Z.; Geoffroy, S.; Dopson, M.; et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 2010, 66, 535–545. [Google Scholar] [CrossRef]
- Brierley-Hobson, S. Binding of (-)-epigallocatechin-3-gallate to the Hsp70 ATPase domain may promote apoptosis in colorectal cancer. Biosci. Horiz. Int. J. Stud. Res. 2008, 1, 9–18. [Google Scholar] [CrossRef]
- Balaburski, G.M.; Leu, J.I.; Beeharry, N.; Hayik, S.; Andrake, M.D.; Zhang, G.; Herlyn, M.; Villanueva, J.; Dunbrack, R.L., Jr.; Yen, T.; et al. A modified HSP70 inhibitor shows broad activity as an anticancer agent. Mol. Cancer Res. Mcr. 2013, 11, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Sugihara, T.; Yoshida, A.; Nomura, H.; Reddel, R.R.; Simpson, R.; Maruta, H.; Kaul, S.C. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000, 60, 6818–6821. [Google Scholar]
- Supko, J.G.; Hickman, R.L.; Grever, M.R.; Malspeis, L. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother. Pharmacol. 1995, 36, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Eiseman, J.L.; Lan, J.; Lagattuta, T.F.; Hamburger, D.R.; Joseph, E.; Covey, J.M.; Egorin, M.J. Pharmacokinetics and pharmacodynamics of 17-demethoxy 17-[[(2-dimethylamino)ethyl]amino]geldanamycin (17DMAG, NSC 707545) in C.B-17 SCID mice bearing MDA-MB-231 human breast cancer xenografts. Cancer Chemother. Pharmacol. 2005, 55, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.R.; Schoepfer, J.; Radimerski, T.; Massey, A.; Guy, C.T.; Brueggen, J.; Quadt, C.; Buckler, A.; Cozens, R.; Drysdale, M.J.; et al. NVP-AUY922: A small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 2008, 10, R33. [Google Scholar] [CrossRef]
- Graham, B.; Curry, J.; Smyth, T.; Fazal, L.; Feltell, R.; Harada, I.; Coyle, J.; Williams, B.; Reule, M.; Angove, H.; et al. The heat shock protein 90 inhibitor, AT13387, displays a long duration of action in vitro and in vivo in non-small cell lung cancer. Cancer Sci. 2012, 103, 522–527. [Google Scholar] [CrossRef]
- Sessa, C.; Shapiro, G.I.; Bhalla, K.N.; Britten, C.; Jacks, K.S.; Mita, M.; Papadimitrakopoulou, V.; Pluard, T.; Samuel, T.A.; Akimov, M.; et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 3671–3680. [Google Scholar] [CrossRef]
- Wang, Y.; Trepel, J.B.; Neckers, L.M.; Giaccone, G. STA-9090, a small-molecule Hsp90 inhibitor for the potential treatment of cancer. Curr. Opin. Investig. Drugs 2010, 11, 1466–1476. [Google Scholar]
- Lundgren, K.; Zhang, H.; Brekken, J.; Huser, N.; Powell, R.E.; Timple, N.; Busch, D.J.; Neely, L.; Sensintaffar, J.L.; Yang, Y.C.; et al. BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol. Cancer Ther. 2009, 8, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Thao, L.; Sensintaffar, J.; Zhang, L.; Boehm, M.F.; Fritz, L.C.; Burrows, F.J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003, 425, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Lamoureux, F.; Thomas, C.; Yin, M.J.; Fazli, L.; Zoubeidi, A.; Gleave, M.E. Suppression of heat shock protein 27 using OGX-427 induces endoplasmic reticulum stress and potentiates heat shock protein 90 inhibitors to delay castrate-resistant prostate cancer. Eur. Urol. 2014, 66, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Musiani, D.; Konda, J.D.; Pavan, S.; Torchiaro, E.; Sassi, F.; Noghero, A.; Erriquez, J.; Perera, T.; Olivero, M.; Di Renzo, M.F. Heat-shock protein 27 (HSP27, HSPB1) is up-regulated by MET kinase inhibitors and confers resistance to MET-targeted therapy. Faseb. J. 2014, 28, 4055–4067. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.H.; Zhao, C.Y.; Zhang, J.; Chen, Y.; Gao, L.; Ni, C.Y.; Zhu, M.H. Role of heat shock protein 27 in gemcitabine-resistant human pancreatic cancer: comparative proteomic analyses. Mol. Med. Rep. 2012, 6, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, R.; Ishii, Y.; Ochiai, H.; Matsunaga, A.; Endo, T.; Hasegawa, H.; Kitagawa, Y. Suppression of heat shock protein 27 expression promotes 5-fluorouracil sensitivity in colon cancer cells in a xenograft model. Oncol. Rep. 2012, 28, 1269–1274. [Google Scholar] [CrossRef]
- Chen, R.; Dai, R.Y.; Duan, C.Y.; Liu, Y.P.; Chen, S.K.; Yan, D.M.; Chen, C.N.; Wei, M.; Li, H. Unfolded protein response suppresses cisplatin-induced apoptosis via autophagy regulation in human hepatocellular carcinoma cells. Folia Biol. (Praha) 2011, 57, 87–95. [Google Scholar]
- Chen, S.F.; Nieh, S.; Jao, S.W.; Liu, C.L.; Wu, C.H.; Chang, Y.C.; Yang, C.Y.; Lin, Y.S. Quercetin suppresses drug-resistant spheres via the p38 MAPK-Hsp27 apoptotic pathway in oral cancer cells. Plos One 2012, 7, e49275. [Google Scholar] [CrossRef]
- Heinrich, J.C.; Tuukkanen, A.; Schroeder, M.; Fahrig, T.; Fahrig, R. RP101 (brivudine) binds to heat shock protein HSP27 (HSPB1) and enhances survival in animals and pancreatic cancer patients. J. Cancer Res. Clin. Oncol. 2011, 137, 1349–1361. [Google Scholar] [CrossRef]
- Sharma, A.; Upadhyay, A.K.; Bhat, M.K. Inhibition of Hsp27 and Hsp40 potentiates 5-fluorouracil and carboplatin mediated cell killing in hepatoma cells. Cancer Biol. 2009, 8, 2106–2113. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan-Sunol, C.; Gabai, V.L.; Sherman, M.Y. Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Res. 2007, 67, 11779–11788. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Kitahara, M.; Nagata, K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer Res. 2000, 60, 2942–2948. [Google Scholar] [PubMed]
- Wong, C.S.; Wong, V.W.; Chan, C.M.; Ma, B.B.; Hui, E.P.; Wong, M.C.; Lam, M.Y.; Au, T.C.; Chan, W.H.; Cheuk, W.; et al. Identification of 5-fluorouracil response proteins in colorectal carcinoma cell line SW480 by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Oncol. Rep. 2008, 20, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Su, T.R.; Lin, J.J.; Chiu, C.C.; Chen, J.Y.; Su, J.H.; Cheng, Z.J.; Hwang, W.I.; Huang, H.H.; Wu, Y.J. Proteomic investigation of anti-tumor activities exerted by sinularin against A2058 melanoma cells. Electrophoresis 2012, 33, 1139–1152. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Lee, S.; Kim, D.E.; Kim, M.; Kwon, B.M.; Han, D.C. Fisetin, a dietary flavonoid, induces apoptosis of cancer cells by inhibiting HSF1 activity through blocking its binding to the hsp70 promoter. Carcinogenesis 2015, 36, 696–706. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, Y.; Kwon, B.M.; Han, D.C. The natural compound cantharidin induces cancer cell death through inhibition of heat shock protein 70 (HSP70) and Bcl-2-associated athanogene domain 3 (BAG3) expression by blocking heat shock factor 1 (HSF1) binding to promoters. J. Biol. Chem. 2013, 288, 28713–28726. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, L.; Dong, X.; Zhao, H.; Li, S.; Han, D.; Liu, X. Apoptin selectively induces the apoptosis of tumor cells by suppressing the transcription of HSP70. Tumour. Biol. 2013, 34, 577–585. [Google Scholar] [CrossRef]
- Nomura, M.; Nomura, N.; Newcomb, E.W.; Lukyanov, Y.; Tamasdan, C.; Zagzag, D. Geldanamycin induces mitotic catastrophe and subsequent apoptosis in human glioma cells. J. Cell Physiol. 2004, 201, 374–384. [Google Scholar] [CrossRef]
- Georgakis, G.V.; Li, Y.; Younes, A. The heat shock protein 90 inhibitor 17-AAG induces cell cycle arrest and apoptosis in mantle cell lymphoma cell lines by depleting cyclin D1, Akt, Bid and activating caspase 9. Br. J. Haematol. 2006, 135, 68–71. [Google Scholar] [CrossRef]
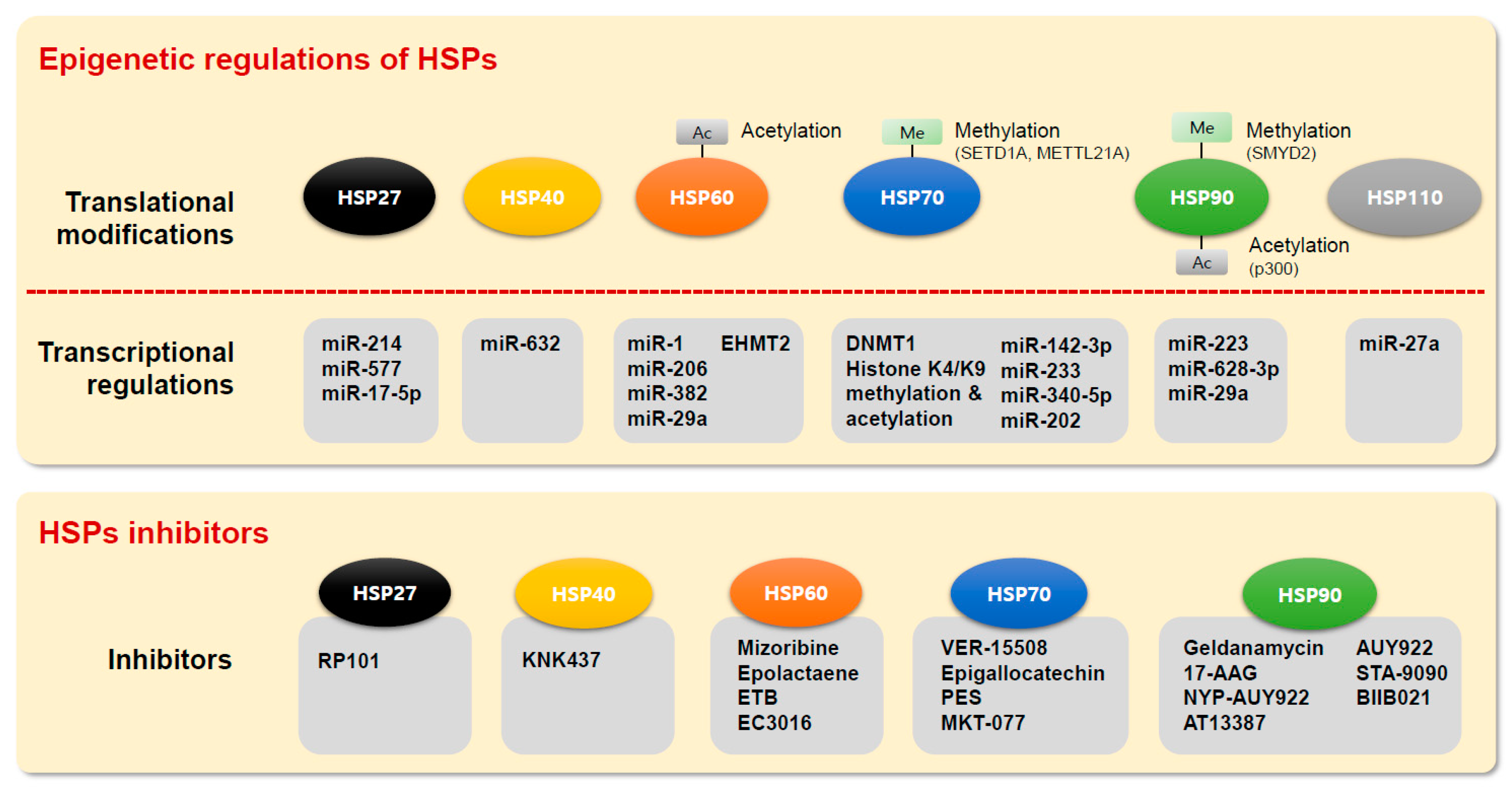
| Name | Function |
|---|---|
| miR-1, miR-206 | High-glucose-mediated apoptosis via HSP60 regulation |
| miR-382 | Attenuation of renal interstitial fibrosis by the regulation of HSP60 |
| miR-29a | Cell apoptosis by the regulation of HSP60 in MCF-7 breast cancer cells |
| EHMT2 | Cell apoptosis by the regulation of HSP60 in MCF-7 breast cancer cells |
| HSP60 acetylation | Inhibition of the interaction between p53 and HSP60 in lung cancer |
| HSP60 acetylation | Induction of cell apoptosis via the hyperacetylated HSP60 mitochondrial protein in osteosarcoma |
| Name | Function |
|---|---|
| HSPA1A promoter methylation | Decreased expression of HSPA1A by promoter methylation in bladder cancer cells |
| DNA methyltransferase 1 (DNMT1)/HSPA2 | DNA methylation-independent regulation of HSPA2 expression through interaction with the Sp1 protein in lung cancer cells |
| HSP21 promoter methylation | Reduced expression of the HSP70 cochaperone HSP21 by promoter methylation in hepatocellular carcinoma cells |
| Histone H3 methylation | Heat-mediated HSP70 expression through histone H3 methylation in oral squamous carcinoma cells |
| Histone H3 methylation | Recruitment of TAC1 to the HSP70 promoter and methylation and acetylation of histone H3 |
| HSP70 methylation | Enhancement of Aurora kinase B activity and cancer cell proliferation by methylated HSP70 |
| HSP70 methylation | Reduction in the affinity for soluble α-Syn by methylated HSPA8 |
| HSP70 methylation | Methylation of HSP70, glucose-regulated protein 75 (GRP75), GRP78 and HSP7C by the METTL21A methyltransferase |
| miR-142-3p | Regulation of pancreatic cancer cell proliferation by the suppression of HSP70 expression |
| miR-233 | Negative regulation of HSP70 expression and modulation of chemoresistance in osteosarcoma |
| miR-340-5p | Regulation of proliferation by the miR-340–5p/ATF7/HSPA1B axis in HCC |
| miR-202 | Regulation of HSF2-mediated HSP70 expression in esophageal squamous cell carcinoma |
| Name | Function |
|---|---|
| miR-223 | Inhibition of cell growth and promotion of apoptosis by suppressing HSP90B1 in osteosarcoma |
| miR-628-3p | Tumor suppression by negative regulation of HSP90 in A549 lung cancer cells |
| miR-29a | Anti-miR-29a induces sensitivity to the anticancer drug Taxol by inhibiting HSPs, including HSP90 |
| HSP90 methylation | SMYD2-mediated HSP90AB1 methylation promotes cancer cell proliferation |
| HSP90α acetylation | Extracellular hyperacetylated HSP90α increases breast cancer cell invasion |
| HSPs | Name | Function |
|---|---|---|
| HSP27 | miR-214 | Inhibition of cell clone formation and cell growth and enhancement of cell apoptosis in colon cancer cells |
| miR-577 | Inhibition of cell growth and xenograft tumor growth and enhancement of 5-FU sensitivity in colorectal cancer cells | |
| miR-17-5p | Enhancement of cell migration in hepatocellular carcinoma cells through the p38 MAPK-HSP27 pathway | |
| HSP27 promoter methylation | Reduced expression level of HSP27 gene by promoter hypermethylation in oral squamous carcinoma cells and esophageal squamous cell carcinoma | |
| HSP40 | miR-632 | Exogenous miR-632 expression increases the invasive ability by targeting DNAJB6 in breast cancer |
| HSP40 promoter methylation | Epigenetic inactivation of MCJ gene by promoter hypermethylation is associated with disease pathogenesis in malignant pediatric brain tumor and epithelial ovarian cancer | |
| HSP110 | miR-27a | Increased hyperthermia-induced cell death in oral squamous cell carcinoma |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ban, H.S.; Han, T.-S.; Hur, K.; Cho, H.-S. Epigenetic Alterations of Heat Shock Proteins (HSPs) in Cancer. Int. J. Mol. Sci. 2019, 20, 4758. https://doi.org/10.3390/ijms20194758
Ban HS, Han T-S, Hur K, Cho H-S. Epigenetic Alterations of Heat Shock Proteins (HSPs) in Cancer. International Journal of Molecular Sciences. 2019; 20(19):4758. https://doi.org/10.3390/ijms20194758
Chicago/Turabian StyleBan, Hyun Seung, Tae-Su Han, Keun Hur, and Hyun-Soo Cho. 2019. "Epigenetic Alterations of Heat Shock Proteins (HSPs) in Cancer" International Journal of Molecular Sciences 20, no. 19: 4758. https://doi.org/10.3390/ijms20194758
APA StyleBan, H. S., Han, T.-S., Hur, K., & Cho, H.-S. (2019). Epigenetic Alterations of Heat Shock Proteins (HSPs) in Cancer. International Journal of Molecular Sciences, 20(19), 4758. https://doi.org/10.3390/ijms20194758






