Development of Teleost Intermuscular Bones Undergoing Intramembranous Ossification Based on Histological-Transcriptomic-Proteomic Data
Abstract
1. Introduction
2. Results
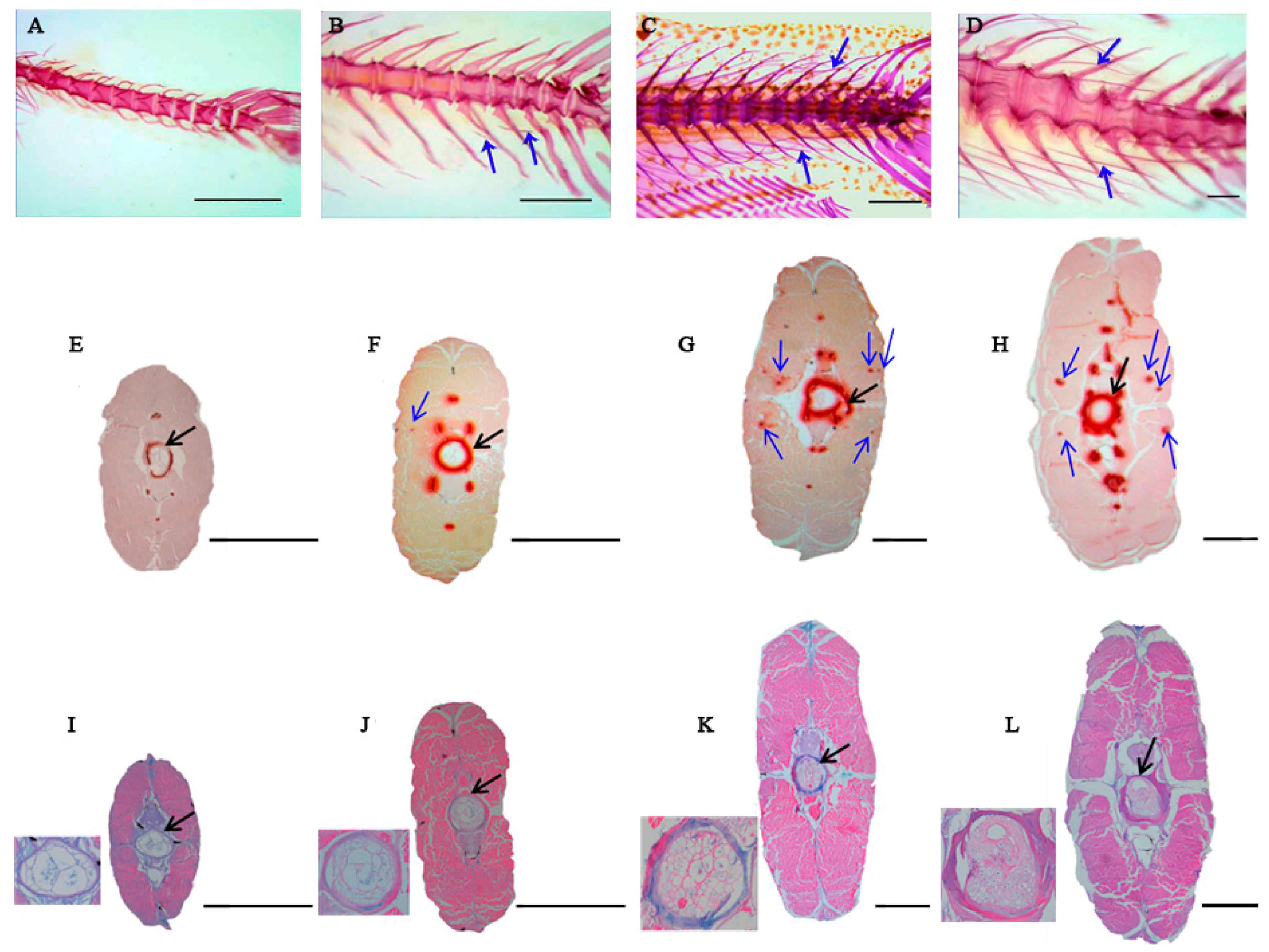
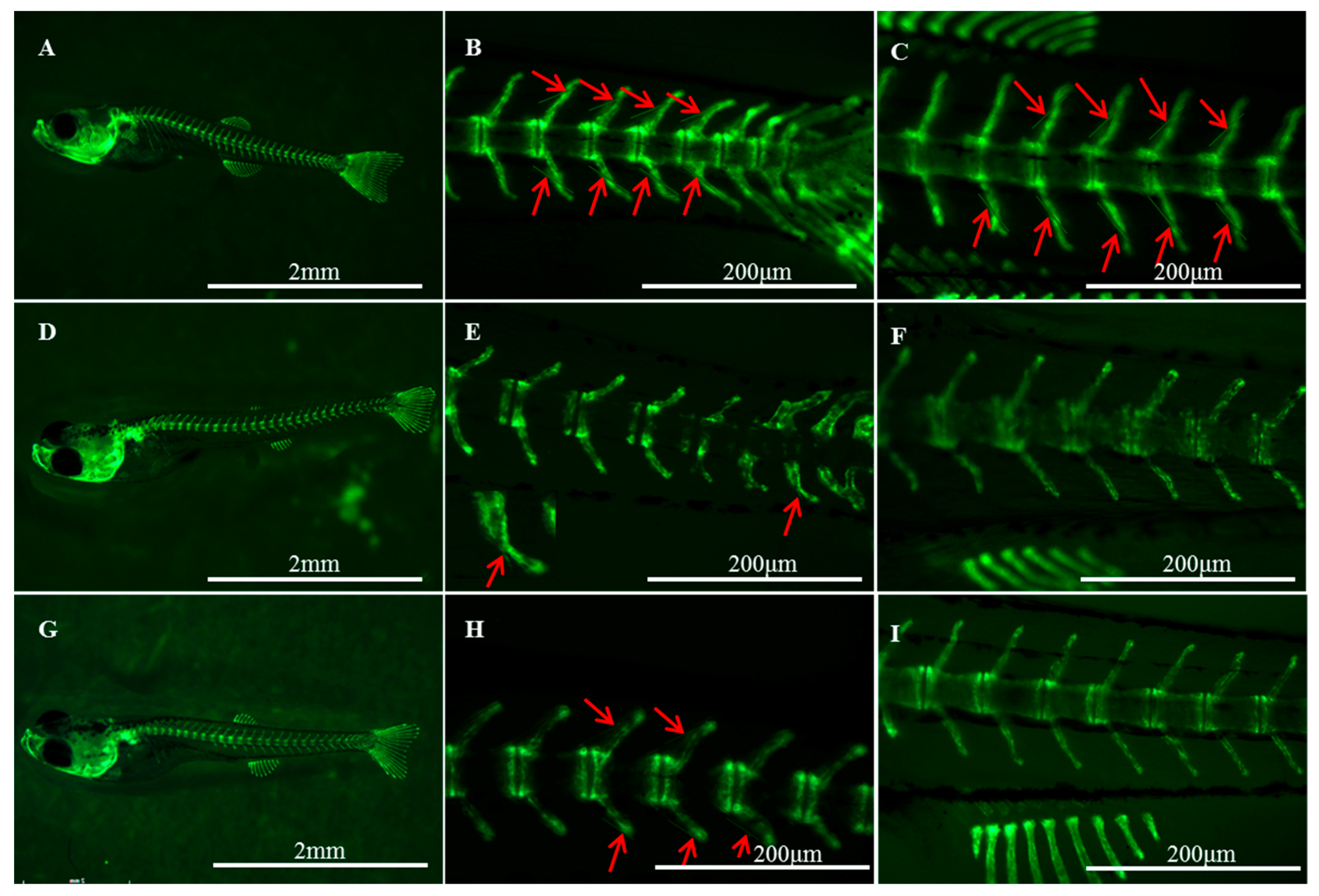
2.1. Histological Structures
2.2. General Proteome Analysis During IB Development
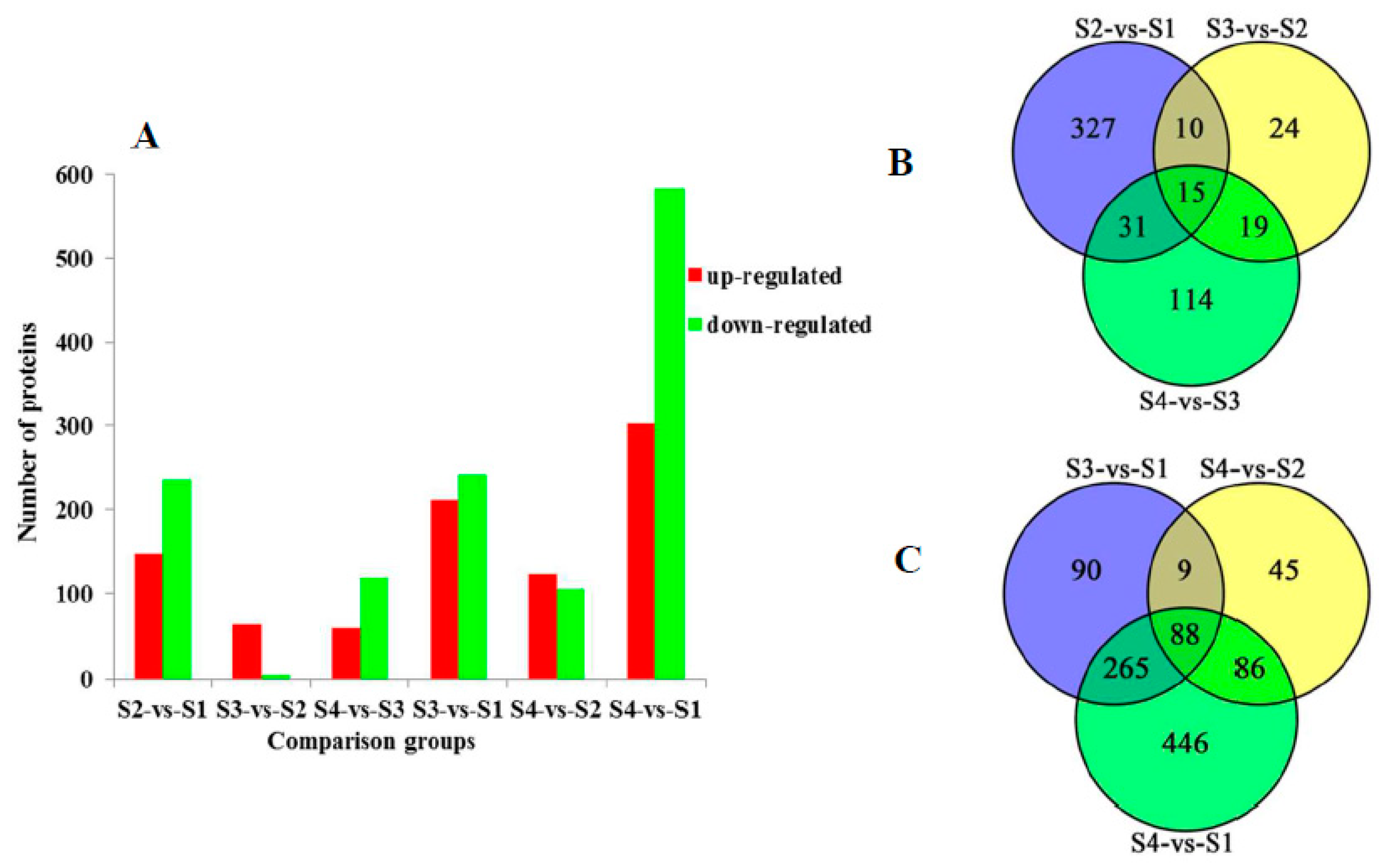
2.3. Comparative Analysis of Differentially Expressed Proteins in Different Comparison Groups
2.4. Correlation Analysis of Protein and mRNA
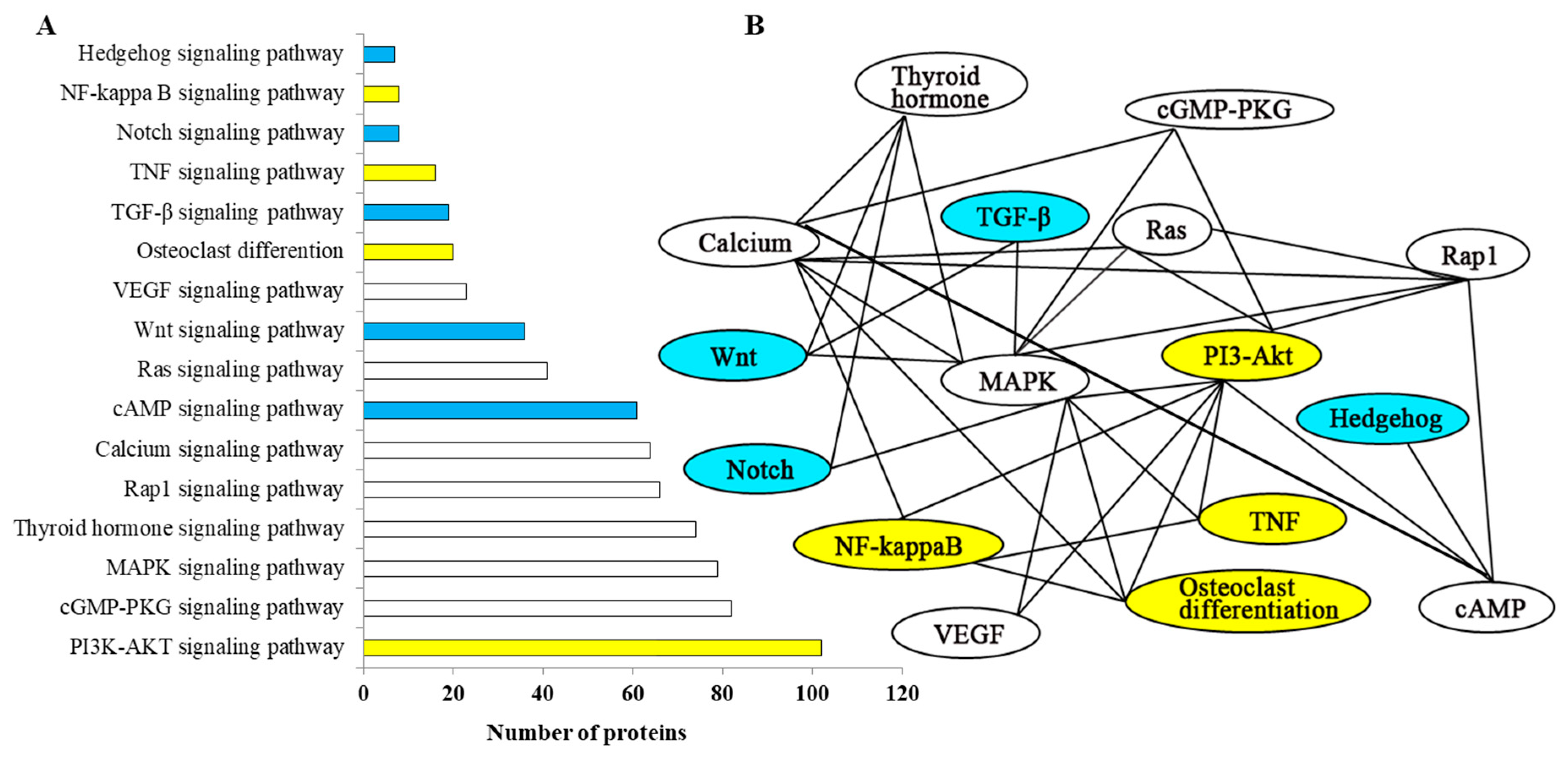
2.5. Network Analysis and Functional Annotation of Altered Bone-Regulated Proteins During IB Development
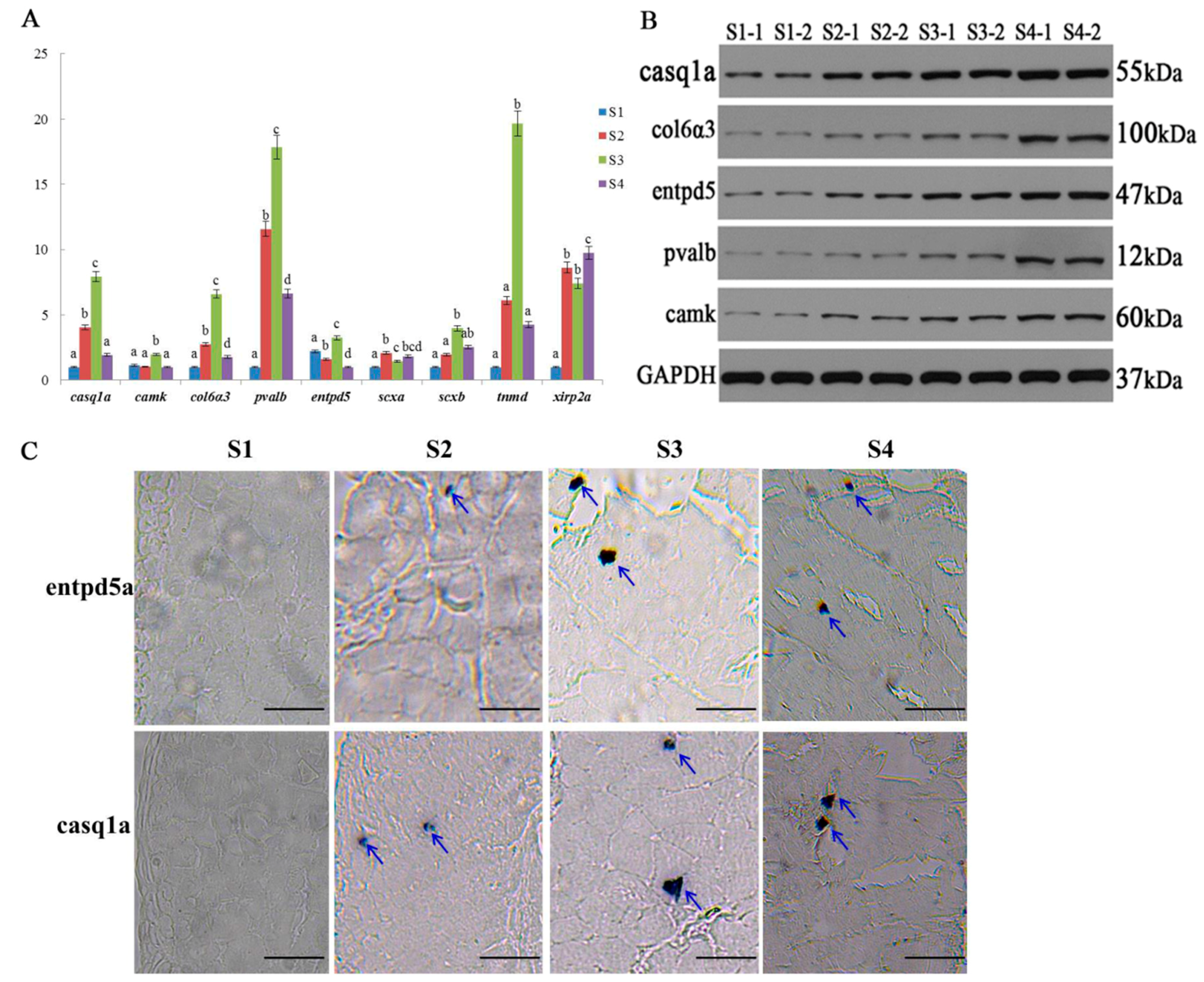
2.6. Expression Validation of Bone-Related Genes
2.7. Pathway Inhibitor Analysis
3. Discussion
4. Methods
4.1. Methods
4.2. Fish Euthanasia
4.3. Histological Analysis
4.4. iTRAQ-Based Proteomics Analysis
4.4.1. Protein Extraction and iTRAQ Labeling
4.4.2. SCX Fractionation and LC-MS/MS Analysis
4.5. Protein Identification and Proteome Analyses
4.6. Enrichment Analysis
4.7. Integrative Analysis of Proteomic and Transcriptomic Data
4.8. Validation of Differentially Expressed Genes
4.9. Pathway Inhibitor Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DMSO | diluted in dimethyl sulfoxide; |
| dph | days post-hatch; |
| GO | gene ontology; |
| HE | hematoxylin-eosin; |
| IB | intermuscular bone; |
| ICAT | isotope-code affinity tags; |
| iTRAQ | Isobaric Tag for Relative Absolute Quantitation; |
| KEGG | Kyoto encyclopedia of genes and genomes; |
| mRNA | messenger RNA; |
| MS-222 | Tricaine methanesulfonate; |
| qRT-PCR | Quantitative real-time polymerase chain reaction; |
| SILAC | stable isotopes labeling by amino acids in cell culture; |
| ARS | alizarin red S; |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry. |
References
- Patterson, C.; Johnson, G.D. The intermuscular bones and ligaments of Teleostean Fishes; Smithsonian Contribution Zool Press: Washington, DC, USA, 1995. [Google Scholar]
- Danos, N.; Ward, A.B. The homology and origins of intermuscular bones in fishes: Phylogenetic or biomechanical determinants? Biol. J. Linn. Soc. Lond. 2012, 106, 607–622. [Google Scholar] [CrossRef]
- Nie, C.H.; Hilsdorf, A.W.S.; Wan, S.M.; Gao, Z.X. Understanding the development of intermuscular bones in teleost: Status and future directions for aquaculture. Rev. Aquacult. 2019. [Google Scholar] [CrossRef]
- Gemballa, S.; Britz, R. Homology of intermuscular bones in Acanthomorph fishes. Am. Mus. Novit. 1998, 3241, 1–25. [Google Scholar]
- Bing, Z. On the myoseptal spines of the carp (Cyprinus carpio L.). Acta Zool. Sin. 1962, 14, 175–178. [Google Scholar]
- Bird, N.C.; Mabee, P.M. Developmental morphology of the axial skeleton of the Zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev. Dyn. 2003, 228, 337–357. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.J.; Huang, D.Z.; Li, L.J.; Yuan, X.H.; Miao, W.M.; Chen, Q.Q.; Lu, Z.B.; Zhang, B.L. Preliminary study on intermuscular bones of several cultured cyprinids. J Shanghai Fish Univ. 2006, 15, 425–429. [Google Scholar]
- Ke, Z.H.; Zhang, W.; Jiang, Y.; Bao, B.L. Developmental morphology of the intermuscular bone in Hypophthalmichthys molitrix. Chin. J. Zool. 2008, 43, 88–96. [Google Scholar]
- Wan, S.M.; Yi, S.K.; Zhong, J.; Wang, W.M.; Jiang, E.M.; Chen, B.X.; Gao, Z.X. Developmental and morphological observation of intermuscular bones in Megalobrama amblycephala. Acta Hydrobiol. Sin. 2014, 38, 1144–1152. [Google Scholar]
- Yao, W.J.; Gong, X.L.; Lü, Y.P.; Bao, B.L. The ossificational process of the intermuscular bones in Anguilla japonica. J. Shanghai Ocean Univ. 2015, 23, 810–813. [Google Scholar]
- Hall, B.K. The embryonic development of bone. Am. Sci. 1988, 76, 174–181. [Google Scholar]
- Karsenty, G.; Wagner, E.F. Reaching agenetic and molecular understanding of skeletal development. Dev. Cell 2002, 2, 389–406. [Google Scholar] [CrossRef]
- Schaeffer, B.; Patterson, C. Jurassic Fishes from the Western United States, with Comments on Jurassic Fish Distribution. Am. Mus. Novit. 1984, 2796, 1–86. [Google Scholar]
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Gygi, S.P.; Rist, B.; Gerber, S.A.; Turecek, F.; Gelb, M.H.; Aebersold, R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999, 17, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Bouchal, P.; Dvořáková, M.; Roumeliotis, T.; Bortlíček, Z.; Ihnatová, I.; Procházková, I.; Ho, J.T.; Maryáš, J.; Imrichová, H.; Budinská, E.; et al. Combined Proteomics and Transcriptomics Identifies Carboxypeptidase B1 and Nuclear Factor κB (NF-κB) Associated Proteins as Putative Biomarkers of Metastasis in Low Grade Breast Cancer. Mol. Cell Proteom. 2015, 4, 1814–1830. [Google Scholar] [CrossRef] [PubMed]
- Petersen, H.O.; Höger, S.K.; Mario, L.; Lengfeld, T.; Kuhn, A.; Warnken, U.; Nishimiya-Fujisawa, C.; Schnölzer, M.; Krüger, M.; Özbek, S.; et al. A Comprehensive Transcriptomic and Proteomic Analysis of Hydra Head Regeneration. Mol. Biol. Evol. 2015, 32, 1928–1947. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteomics 2015, 122, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Tse, W.K.; Sun, J.; Zhang, H.; Lai, K.P.; Gu, J.; Qiu, J.W.; Wong, C.K. iTRAQ-based quantitative proteomic analysis reveals acute hypo-osmotic responsive proteins in the gills of the Japanese eel (Anguilla japonica). J. Proteom. 2014, 105, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.M.; Yi, S.K.; Zhong, J.; Nie, C.H.; Guan, N.N.; Zhang, W.Z.; Gao, Z.X. Dynamic mRNA and miRNA expression analysis in response to intermuscular bone development of blunt snout bream (Megalobrama amblycephala). Sci. Rep. 2016, 6, 31050. [Google Scholar] [CrossRef] [PubMed]
- Muers, M. Gene expression: Transcriptome to proteome and back to genome. Nat. Rev. Genet. 2011, 12, 518. [Google Scholar] [CrossRef] [PubMed]
- Eames, B.F.; Yan, Y.L.; Swartz, M.E.; Levic, D.S.; Knapik, E.W.; Postlethwait, J.H.; Kimmel, C.B. Mutations in fam20b and xylosyltransferase1 reveal that cartilage matrix controls timing of endochondral ossification through inhibition of chondrocyte maturation. PLoS Genet. 2011, 7, e1002246. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.B.; Kimmel, C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef]
- Witten, P.E.; Huysseune, A. A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol. Rev. Camb. Philos. Soc. 2009, 84, 315–346. [Google Scholar] [CrossRef] [PubMed]
- Kessels, M.Y.; Huitema, L.F.; Boeren, S.; Kranenbarg, S.; Schulte-Merker, S.; van Leeuwen, J.L.; de Vries, S.C. Proteomics analysis of the zebrafish skeletal extracellular matrix. PLoS ONE 2014, 9, e90568. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.H.; Wan, S.M.; Tomljanovic, T.; Treer, T.; Hsiao, C.D.; Wang, W.M.; Gao, Z.X. Comparative proteomics analysis of teleost intermuscular bones and ribs provides insight into their development. BMC Genom. 2017, 18, 147. [Google Scholar] [CrossRef] [PubMed]
- Amend, S.R.; Uluckan, O.; Hurchla, M.; Leib, D.; Novack, D.V.; Silva, M.; Frazier, W.; Weilbaecher, K.N. Thrombospondin-1 regulates bone homeostasis through effects on bone matrix integrity and nitric oxide signaling in osteoclasts. J. Bone Miner. Res. 2015, 30, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Beard, N.A.; Laver, D.R.; Dulhunty, A.F. Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog. Biophys. Mol. Biol. 2004, 85, 33–69. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.A.; Thorne, M.A.; Stueber, K.; Darias, M.; Reinhardt, R.; Clark, M.S.; Gisbert, E.; Power, D.M. Comparative analysis of a teleost skeleton transcriptome provides insight into its regulation. Gen. Comp. Endocrinol. 2013, 191, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Talmage, R.V.; Matthews, J.L.; Mobley, H.T.; Lester, G.E. Calcium homeostasis and bone surface proteins, a postulated vital process for plasma calcium control. J. Musculoskelet Neuronal Interact 2003, 3, 194–200. [Google Scholar] [PubMed]
- Vandenberge, J.C.; Storer, R.W. Intratendinous Ossification in Birds-a Review. J. Morphol. 1995, 226, 47–77. [Google Scholar]
- Qu, J.; Thoreson, A.R.; Chen, Q.; An, K.N.; Amadio, P.C.; Zhao, C. Tendon gradient mineralization for tendon to bone interface integration. J. Orthop. Res. 2013, 31, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Ishida, H.; Vogel, H.J. Structural Characterization of the Interaction of Human Lactoferrin with Calmodulin. PLoS ONE 2012, 7, e51026. [Google Scholar] [CrossRef] [PubMed]
- Doroudi, M.; Plaisance, M.C.; Boyan, B.D.; Schwartz, Z. Membrane actions of 1α, 25(OH)2D3 are mediated by Ca(2+)/calmodulin-dependent protein kinase II in bone and cartilage cells. J. Steroid Biochem. Mol. Biol. 2015, 145, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Smits, P.; Li, P.; Mandel, J.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B.; Lefebvre, V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 2001, 1, 277–290. [Google Scholar] [CrossRef]
- Núñez, C.; Esteve-Núñez, A.; Giometti, C.; Tollaksen, S.; Khare, T.; Lin, W.; Lovley, D.R.; Methé, B.A. DNA microarray and proteomic analyses of the RpoS regulon in Geobacter sulfurreducens. J. Bacteriol. 2006, 188, 2792–2800. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anderson, L.; Seilhamer, J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997, 18, 533–537. [Google Scholar] [CrossRef]
- Hegde, P.S.; White, I.R.; Debouck, C. Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 2003, 14, 647–651. [Google Scholar] [CrossRef]
- Nie, L.; Wu, G.; Culley, D.E.; Scholten, J.C.; Zhang, W. Integrative analysis of transcriptomic and proteomic data: Challenges, solutions and applications. Crit. Rev. Biotechnol. 2007, 27, 63–75. [Google Scholar] [CrossRef]
- Karve, T.M.; Cheema, A.K. Small changes huge impact: The role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids 2011, 2, 207691. [Google Scholar] [CrossRef]
- Huitema, L.F.; Apschner, A.; Logister, I.; Spoorendonk, K.M.; Bussmann, J.; Hammond, C.L.; Schulte-Merker, S. Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc. Natl. Acad. Sci. USA 2012, 109, 21372–21377. [Google Scholar] [CrossRef]
- Irwin, W.A.; Bergamin, N.; Sabatelli, P.; Reggiani, C.; Megighian, A.; Merlini, L.; Braghetta, P.; Columbaro, M.; Volpin, D.; Bressan, G.M.; et al. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 2003, 35, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, V.G.; Rosa, J.; Laizé, V.; Gavaia, P.J.; Cancela, M.L. Identification of a new cartilage-specific S100-like protein up-regulated during endo/perichondral mineralization in gilthead seabream. Gene Expr. Patterns 2011, 11, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Moss, S.E.; Morgan, R.O. The annexins. Genome Biol. 2004, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, T. Annexins-their role in cartilage mineralization. Front Biosci. 2005, 10, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Kobayashi, M.; Nakamura, M.; Suzuki, N.; Yashima, S.; Iwamuro, S.; Ikegame, M.; Yamamoto, T.; Hattori, A. Two osteoclastic markers expressed in multinucleate osteoclasts of goldfish scales. Biochem. Biophys. Res. Commun. 2007, 362, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Sahni, M.; Ambrosetti, D.C.; Mansukhani, A.; Gertner, R.; Levy, D.; Basilico, C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999, 13, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Windhausen, T.; Squifflet, S.; Renn, J.; Muller, M. BMP Signaling Regulates Bone Morphogenesis in Zebrafish through Promoting Osteoblast Function as Assessed by Their Nitric Oxide Production. Molecules 2015, 20, 7586–7601. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Moon, J.; Dodgem, M.E.; Pan, X.; Zhang, L.; Hanson, J.M.; Tuladhar, R.; Ma, Z.; Shi, H.; Williams, N.S.; et al. The development of highly potent inhibitors for porcupine. J. Med. Chem. 2013, 56, 2700–2704. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Wu, J.; Jing, J.; Huang, P.P.; Li, Z.; Mei, J.; Gui, J.F. Loss of stat3 function leads to spine malformation and immune disorder in zebrafish. Sci. Bull. 2017, 62, 185–196. [Google Scholar] [CrossRef]
- Luzio, A.; Monteiro, S.M.; Rocha, E.; Fontaínhas-Fernandes, A.A.; Coimbra, A.M. Development and recovery of histopathological alterations in the gonads of zebrafish (Danio rerio) after single and combined exposure to endocrine disruptors (17α-ethinylestradiol and fadrozole). Aquat. Toxicol. 2016, 175, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, F.; Takeuchi, I.; Agata, H.; Kagami, H.; Shiono, H.; Kiyota, Y.; Honda, H.; Kato, R. Morphology-based prediction of osteogenic differentiation potential of human mesenchymal stem cells. PLoS ONE 2013, 8, e55082. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Sun, J.; Zhang, Y.; He, F.; Xu, Y.; Matsumura, K.; He, L.S.; Qiu, J.W.; Qi, S.H.; Qian, P.Y. iTRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J. Proteome Res. 2013, 12, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Dineshram, R.; Sharma, R.; Chandramouli, K.; Yalamanchili, H.K.; Chu, I.; Thiyagarajan, V. Comparative and quantitative proteomics reveal the adaptive strategies of oyster larvae to ocean acidification. Proteomics 2015, 15, 4120–4134. [Google Scholar] [CrossRef] [PubMed]
- Larionov, A.; Krause, A.; Miller, W. A standard curve based method for relative real time PCR data processing. BMC Bioinf. 2005, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.F.; Houweling, A.C.; de Boer, P.A.; Christoffels, V.M. Sensitive nonradioactive detection of mRNA in tissue sections: Novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 2001, 49, 1–8. [Google Scholar] [CrossRef]
- Kague, E.; Roy, P.; Asselin, G.; Hu, G.; Simonet, J.; Stanley, A.; Albertson, C.; Fisher, S. Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev. Biol. 2016, 413, 160–172. [Google Scholar] [CrossRef]
- Kadowaki, T.; Wilder, E.; Klingensmith, J.; Zachary, K.; Perrimon, N. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev. 1996, 10, 3116–3128. [Google Scholar] [CrossRef]
- Komekado, H.; Yamamoto, H.; Chiba, T.; Kikuchi, A. Glycosylation and palmitoylation of Wnt-3a are coupled to produce an active form of Wnt-3a. Genes Cells 2007, 12, 521–534. [Google Scholar] [CrossRef]
- Nie, C.H.; Chen, Z.X.; Dai, C.J.; Wan, S.H.; Gao, Z.X. Ossification patterns of intermusclar bones in different fish species. Acta Hydrobiol. Sin. 2018, 42, 1–7. [Google Scholar]


| NCBI nr Description | Abbreviation | S2-vs-S1 | S3-vs-S2 | S4-vs-S3 | |||
|---|---|---|---|---|---|---|---|
| Protein | mRNA | Protein | mRNA | Protein | mRNA | ||
| Calsequestrin 1a precursor | casq1a | 0.69 | 0.22 | 0.06 | −0.13 | −0.15 | 0.47 |
| Calcium/calmodulin-dependent protein kinase type II subunit beta-like | camk | 0.93 | 0.93 | −0.17 | 0.77 | 0.30 | −1.28 |
| Parvalbumin | pvalb | 0.68 | 1.27 | 1.32 | 0.73 | 0.11 | 1.66 |
| Calreticulin precursor | calrl | −0.38 | −1.71 | −0.03 | −4.08 | −0.12 | 1.03 |
| Annexin A1 | anxa1 | −0.12 | 0.09 | 0.21 | −0.05 | −0.43 | 0.43 |
| Annexin A2a | anxa2a | −0.49 | 0.17 | 0.11 | −0.13 | −0.15 | 0.75 |
| Annexin A5 | anxa5 | −0.40 | −0.22 | 0.54 | −0.88 | 0.08 | −0.04 |
| Ectonucleoside triphosphate diphosphohydrolase 5-like | entpd5 | 0.18 | 0.67 | 0.19 | −0.18 | −0.04 | −0.23 |
| Beta-catenin-like protein 1 | β-catenin | −1.09 | −0.49 | −0.07 | −0.42 | 0.11 | −0.29 |
| Transforming growth factor, beta-induced | TGF-β | −0.71 | −0.02 | −0.04 | −1.25 | −0.09 | 0.47 |
| Tenascin C | tnc | −0.40 | −1.08 | −0.09 | −1.48 | 0.06 | 0.25 |
| Signal transducer and activator of transcription 1-alpha/beta | stat1 | −0.97 | 0.24 | −0.03 | 0.72 | 0.28 | −1.12 |
| Collagen alpha-1(IX) chain-like, partial | col9α1 | −3.32 | −9.38 | 0.73 | 0.00 | −0.56 | 0.00 |
| Laminin subunit beta-2 precursor | lamb2 | −0.07 | −0.42 | 0.01 | −0.002 | −0.64 | −0.31 |
| nidogen-1-like | nid1 | 0.11 | −0.58 | 0.04 | −0.67 | 0.15 | −0.45 |
| Collagen alpha-3(VI) chain-like | col6α3 | 0.57 | 1.67 | 0.56 | 0.06 | −0.18 | −1.12 |
| Matrix metalloproteinase-14 precursor | mmp14b | 0.44 | −0.04 | −0.17 | −1.12 | 0.11 | −0.40 |
| Decorin variant 1 | dcn | 0.04 | 0.32 | 0.37 | −1.60 | −0.06 | 0.63 |
| Cathepsin K | ctsk | −0.94 | −0.22 | 0.82 | −0.53 | −0.79 | −0.62 |
| Prostaglandin E synthase 2-like | PGE2 | −0.22 | −0.40 | 0.12 | −0.13 | −0.20 | −0.38 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, C.-H.; Wan, S.-M.; Liu, Y.-L.; Liu, H.; Wang, W.-M.; Gao, Z.-X. Development of Teleost Intermuscular Bones Undergoing Intramembranous Ossification Based on Histological-Transcriptomic-Proteomic Data. Int. J. Mol. Sci. 2019, 20, 4698. https://doi.org/10.3390/ijms20194698
Nie C-H, Wan S-M, Liu Y-L, Liu H, Wang W-M, Gao Z-X. Development of Teleost Intermuscular Bones Undergoing Intramembranous Ossification Based on Histological-Transcriptomic-Proteomic Data. International Journal of Molecular Sciences. 2019; 20(19):4698. https://doi.org/10.3390/ijms20194698
Chicago/Turabian StyleNie, Chun-Hong, Shi-Ming Wan, Yu-Long Liu, Han Liu, Wei-Min Wang, and Ze-Xia Gao. 2019. "Development of Teleost Intermuscular Bones Undergoing Intramembranous Ossification Based on Histological-Transcriptomic-Proteomic Data" International Journal of Molecular Sciences 20, no. 19: 4698. https://doi.org/10.3390/ijms20194698
APA StyleNie, C.-H., Wan, S.-M., Liu, Y.-L., Liu, H., Wang, W.-M., & Gao, Z.-X. (2019). Development of Teleost Intermuscular Bones Undergoing Intramembranous Ossification Based on Histological-Transcriptomic-Proteomic Data. International Journal of Molecular Sciences, 20(19), 4698. https://doi.org/10.3390/ijms20194698





