FGFR Signaling as a Candidate Therapeutic Target for Cancers Resistant to Carbon Ion Radiotherapy
Abstract
1. Introduction
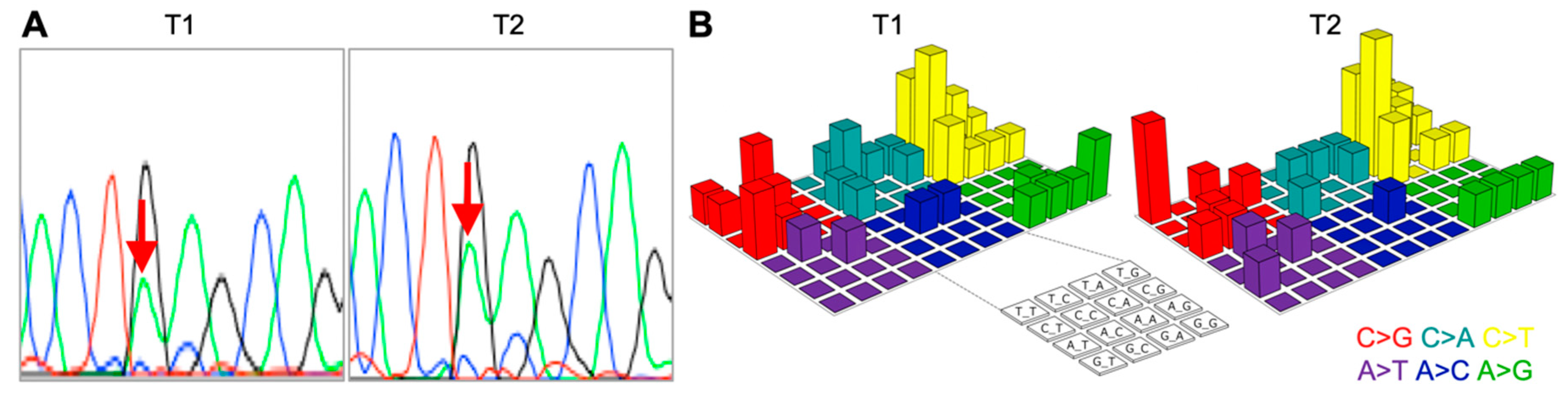
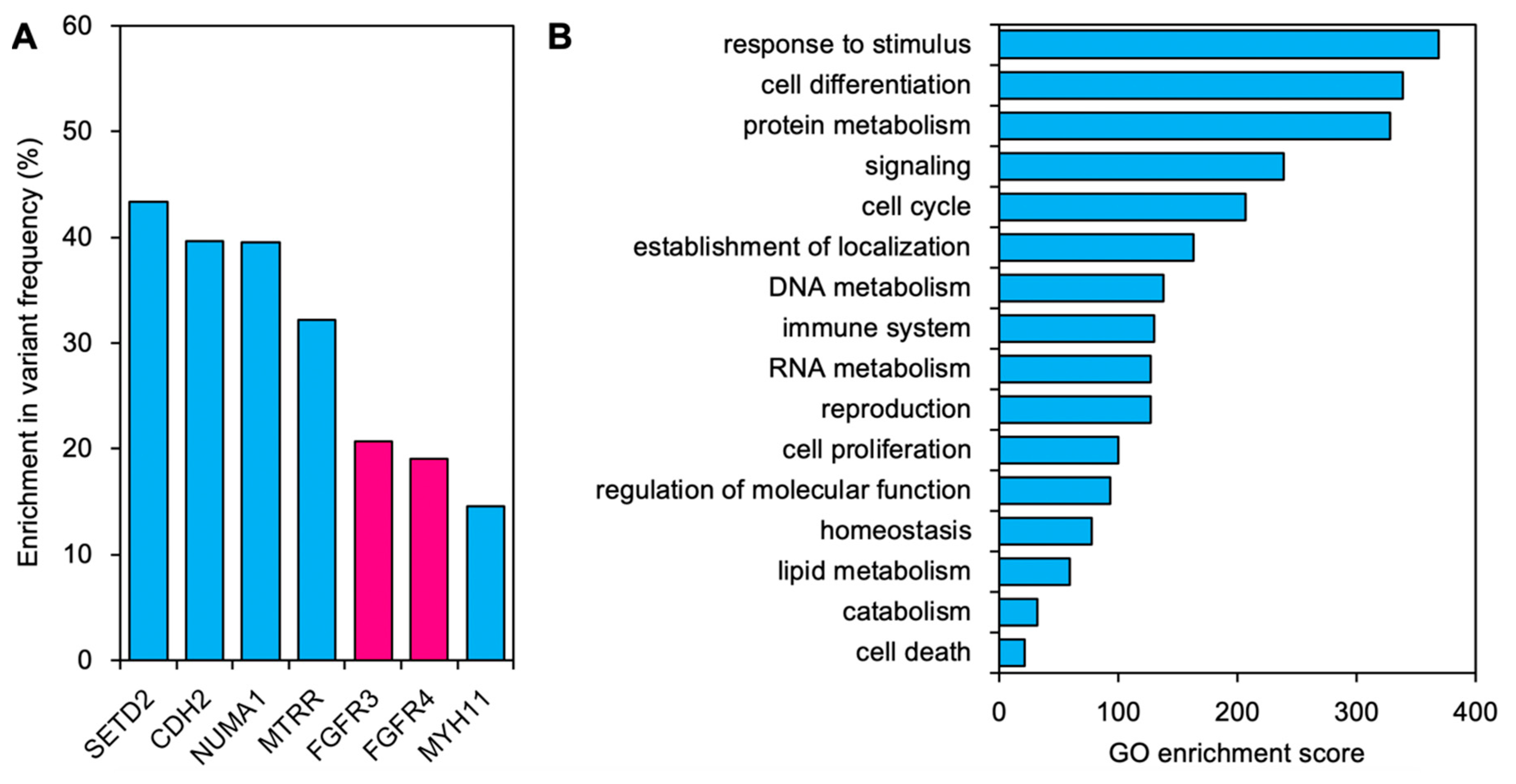
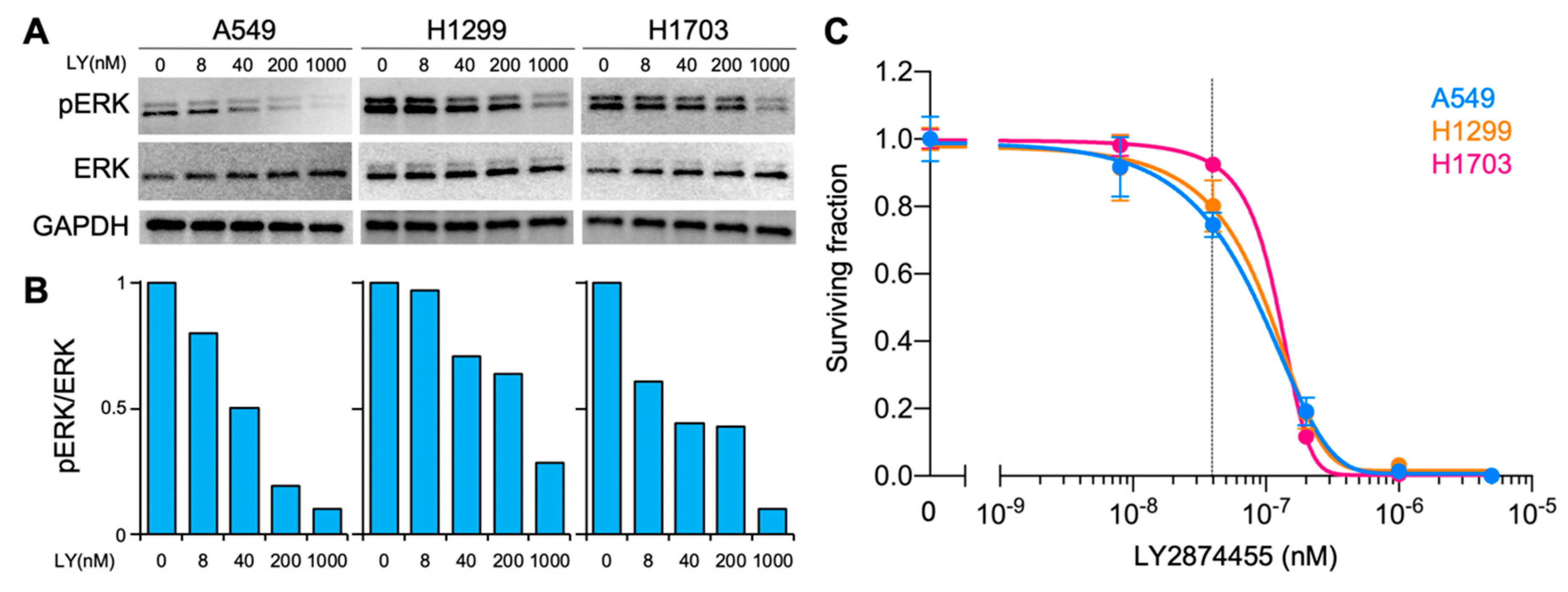
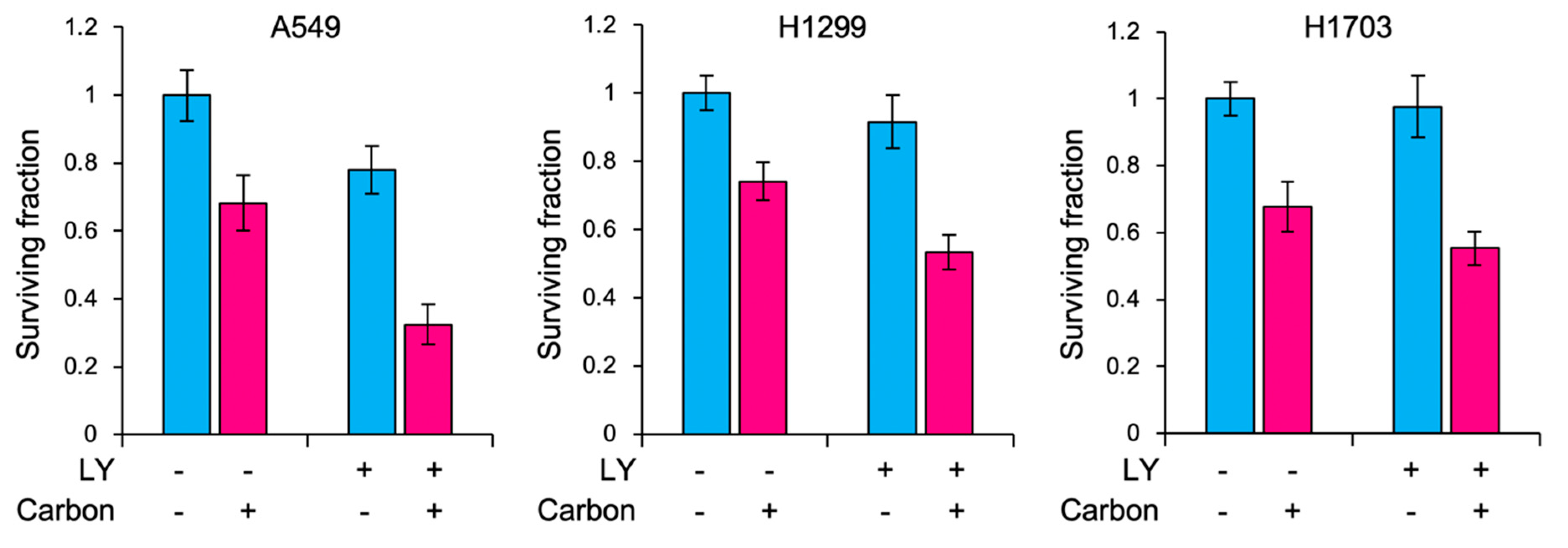
2. Results
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Tissue Sample Collection
4.3. DNA Preparation
4.4. Next-Generation Sequencing
4.5. Identification of Somatic Mutations
4.6. GO Enrichment Analysis
4.7. Cell Culture and Materials
4.8. Carbon Ion Irradiation
4.9. Clonogenic Assays
4.10. Immunoblotting
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CIRT | Carbon ion radiotherapy |
| ERK | Extracellular signal-regulated kinase 1/2 |
| FFPE | Formalin-fixed paraffin-embedded |
| GO | Gene ontology |
| CCP | Ion AmpliSeq Comprehensive Cancer Panel |
| RBE | Relative biological effectiveness |
| SER | Sensitizer enhancement ratio |
| VF | Variant frequency |
References
- Thariat, J.; Hannoun-Levi, J.M.; Sun Myint, A.; Vuong, T.; Gérard, J.P. Past, present, and future of radiotherapy for the benefit of patients. Nat. Rev. Clin. Oncol. 2013, 10, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Orecchia, R.; Loeffler, J.S. Charged-particle therapy in cancer: Clinical uses and future perspectives. Nat. Rev. Clin. Oncol. 2017, 14, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Schlaich, F.; Brons, S.; Haberer, T.; Debus, J.; Combs, S.E.; Weber, K.J. Comparison of the effects of photon versus carbon ion irradiation when combined with chemotherapy in vitro. Radiat. Oncol. 2013, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Noda, S.E.; Murata, K.; Yoshimoto, Y.; Okonogi, N.; Ando, K.; Tamaki, T.; Kato, S.; Hirakawa, T.; Kanuma, T.; et al. Phase I Study of Carbon Ion Radiotherapy and Image-Guided Brachytherapy for Locally Advanced Cervical Cancer. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [CrossRef] [PubMed]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef]
- Karakas, B.; Bachman, K.E.; Park, B.H. Mutation of the PIK3CA oncogene in human cancers. Br. J. Cancer 2006, 94, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Ma, P.; Fu, Y.; Cai, M.C.; Yan, Y.; Jing, Y.; Zhang, S.; Chen, M.; Wu, J.; She, Y.; Zhu, L.; et al. Simultaneous evolutionary expansion and constraint of genomic heterogeneity in multifocal lung cancer. Nat. Commun. 2017, 8, 823. [Google Scholar] [CrossRef]
- Burrell, R.A.; Swanton, C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014, 8, 1095–1111. [Google Scholar] [CrossRef]
- Misale, S.; Di Nicolantonio, F.; Sartore-Bianchi, A.; Siena, S.; Bardelli, A. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 2014, 4, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Li, W.Y.; Chen, D.; Henry, J.R.; Li, H.Y.; Chen, Z.; Zia-Ebrahimi, M.; Bloem, L.; Zhai, Y.; Huss, K.; et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol. Cancer Ther. 2011, 10, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.; Bang, Y.J.; Park, Y.S.; Kang, Y.K.; Kim, T.M.; Hamid, O.; Thornton, D.; Tate, S.C.; Raddad, E.; Tie, J. A Phase 1 Study of LY2874455, an Oral Selective pan-FGFR Inhibitor, in Patients with Advanced Cancer. Target Oncol. 2017, 12, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Amornwichet, N.; Oike, T.; Shibata, A.; Nirodi, C.S.; Ogiwara, H.; Makino, H.; Kimura, Y.; Hirota, Y.; Isono, M.; Yoshida, Y.; et al. The EGFR mutation status affects the relative biological effectiveness of carbon-ion beams in non-small cell lung carcinoma cells. Sci. Rep. 2015, 5, 11305. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, D.; Oike, T.; Shibata, A.; Niimi, A.; Kubota, Y.; Sakai, M.; Amornwhichet, N.; Yoshimoto, Y.; Hagiwara, Y.; Kimura, Y.; et al. Mitotic catastrophe is a putative mechanism underlying the weak correlation between sensitivity to carbon ions and cisplatin. Sci. Rep. 2017, 7, 40588. [Google Scholar] [CrossRef] [PubMed]
- Nuryadi, E.; Sasaki, Y.; Hagiwara, Y.; Permata, T.B.M.; Sato, H.; Komatsu, S.; Yoshimoto, Y.; Murata, K.; Ando, K.; Kubo, N.; et al. Mutational analysis of uterine cervical cancer that survived multiple rounds of radiotherapy. Oncotarget 2018, 9, 32642–32652. [Google Scholar] [CrossRef]
- McMahon, C.M.; Ferng, T.; Canaani, J.; Wang, E.S.; Morrissette, J.J.D.; Eastburn, D.J.; Pellegrino, M.; Durruthy-Durruthy, R.; Watt, C.D.; Asthana, S.; et al. Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov. 2019, 9, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Savage, S.; Schultz, A.R.; Bottomly, D.; White, L.; Segerdell, E.; Wilmot, B.; McWeeney, S.K.; Eide, C.A.; Nechiporuk, T.; et al. Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nat. Commun. 2019, 10, 244. [Google Scholar] [CrossRef]
- Ishigami, T.; Uzawa, K.; Higo, M.; Nomura, H.; Saito, K.; Kato, Y.; Nakashima, D.; Shiiba, M.; Bukawa, H.; Yokoe, H.; et al. Genes and molecular pathways related to radioresistance of oral squamous cell carcinoma cells. Int. J. Cancer 2007, 120, 2262–2270. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.M.; Raghav, D.; Singh, H.; Raghava, G.P. CCDB: A curated database of genes involved in cervix cancer. Nucleic Acids Res. 2011, 39, D975–D979. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.; Butler, T.; Rhodes, K.; Quist, M.; Neff, T.L.; Moore, S.; Tomlins, S.A.; Reinig, E.; Beadling, C.; Andersen, M.; et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J. Mol. Diagn. 2015, 17, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishiguro, K.; Igarashi, T.; Aoki, Y.; Hayashi, T.; Ishida, T.; Sasaki, Y.; Tokino, T.; Shinomura, Y. Molecular diagnostics of a single drug-resistant multiple myeloma case using targeted next-generation sequencing. Onco. Targets Ther. 2015, 8, 2805–2815. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Singh, R.R.; Patel, K.P.; Routbort, M.J.; Reddy, N.G.; Barkoh, B.A.; Handal, B.; Kanagal-Shamanna, R.; Greaves, W.O.; Medeiros, L.J.; Aldape, K.D.; et al. Clinical validation of a next- generation sequencing screen for mutational hotspots in 46 cancer-related genes. J. Mol. Diagn. 2013, 15, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.broadinstitute.org/igv (accessed on 5 August 2019).
- Anakura, M.; Nachankar, A.; Kobayashi, D.; Amornwichet, N.; Hirota, Y.; Shibata, A.; Oike, T.; Nakano, T. Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses. Int. J. Mol. Sci. 2019, 20, 15. [Google Scholar] [CrossRef]
- Shibata, A.; Moiani, D.; Arvai, A.S.; Perry, J.; Harding, S.M.; Genois, M.M.; Maity, R.; van Rossum-Fikkert, S.; Kertokalio, A.; Romoli, F.; et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 2014, 53, 7–18. [Google Scholar] [CrossRef]
| Event | Treatment | Months | Sample |
|---|---|---|---|
| Diagnosis | 0 | T1 | |
| CIRT | 1 | ||
| Local recurrence | 13 | T2 | |
| Surgery | 15 | Normal | |
| Deceased | 25 |
| Cell line | LY | SF at 3Gy (RBE) | p-Values | SER |
|---|---|---|---|---|
| A549 | - | 0.68 ± 0.06 | 0.00050 | 1.66 ± 0.17 |
| + | 0.41 ± 0.05 | |||
| H1299 | - | 0.74 ± 0.06 | 0.00049 | 1.27 ± 0.09 |
| + | 0.58 ± 0.03 | |||
| H1703 | - | 0.67 ± 0.05 | 0.026 | 1.20 ± 0.18 |
| + | 0.56 ± 0.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darwis, N.D.M.; Nachankar, A.; Sasaki, Y.; Matsui, T.; Noda, S.-e.; Murata, K.; Tamaki, T.; Ando, K.; Okonogi, N.; Shiba, S.; et al. FGFR Signaling as a Candidate Therapeutic Target for Cancers Resistant to Carbon Ion Radiotherapy. Int. J. Mol. Sci. 2019, 20, 4563. https://doi.org/10.3390/ijms20184563
Darwis NDM, Nachankar A, Sasaki Y, Matsui T, Noda S-e, Murata K, Tamaki T, Ando K, Okonogi N, Shiba S, et al. FGFR Signaling as a Candidate Therapeutic Target for Cancers Resistant to Carbon Ion Radiotherapy. International Journal of Molecular Sciences. 2019; 20(18):4563. https://doi.org/10.3390/ijms20184563
Chicago/Turabian StyleDarwis, Narisa Dewi Maulany, Ankita Nachankar, Yasushi Sasaki, Toshiaki Matsui, Shin-ei Noda, Kazutoshi Murata, Tomoaki Tamaki, Ken Ando, Noriyuki Okonogi, Shintaro Shiba, and et al. 2019. "FGFR Signaling as a Candidate Therapeutic Target for Cancers Resistant to Carbon Ion Radiotherapy" International Journal of Molecular Sciences 20, no. 18: 4563. https://doi.org/10.3390/ijms20184563
APA StyleDarwis, N. D. M., Nachankar, A., Sasaki, Y., Matsui, T., Noda, S.-e., Murata, K., Tamaki, T., Ando, K., Okonogi, N., Shiba, S., Irie, D., Kaminuma, T., Kumazawa, T., Anakura, M., Yamashita, S., Hirakawa, T., Kakoti, S., Hirota, Y., Tokino, T., ... Nakano, T. (2019). FGFR Signaling as a Candidate Therapeutic Target for Cancers Resistant to Carbon Ion Radiotherapy. International Journal of Molecular Sciences, 20(18), 4563. https://doi.org/10.3390/ijms20184563








