Compromised Barrier Function in Human Induced Pluripotent Stem-Cell-Derived Retinal Pigment Epithelial Cells from Type 2 Diabetic Patients
Abstract
1. Introduction
2. Results
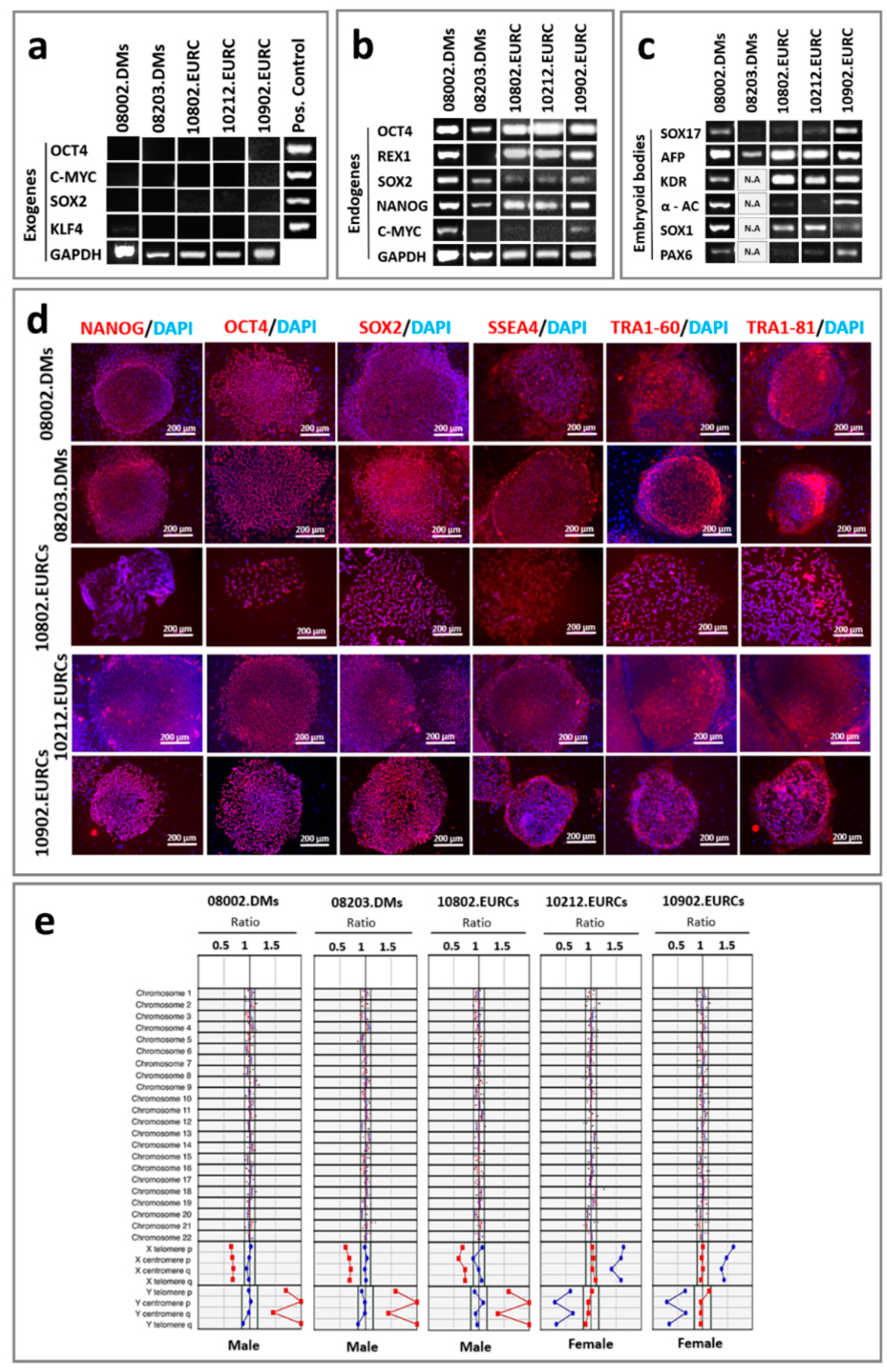
2.1. Pluripotency Assessment
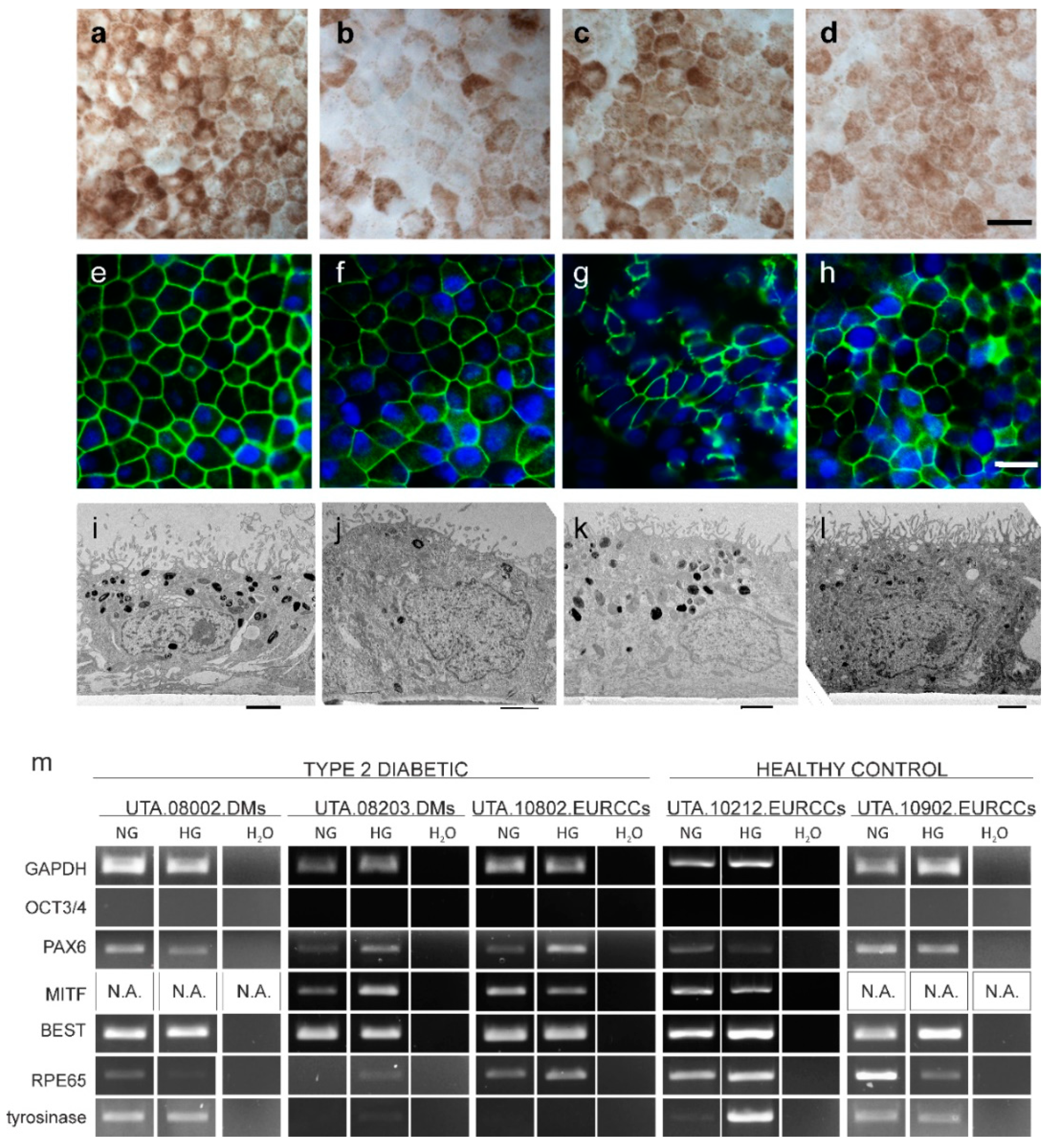
2.2. Maturation Status of Cells Verified with Gene Expression and Morphology
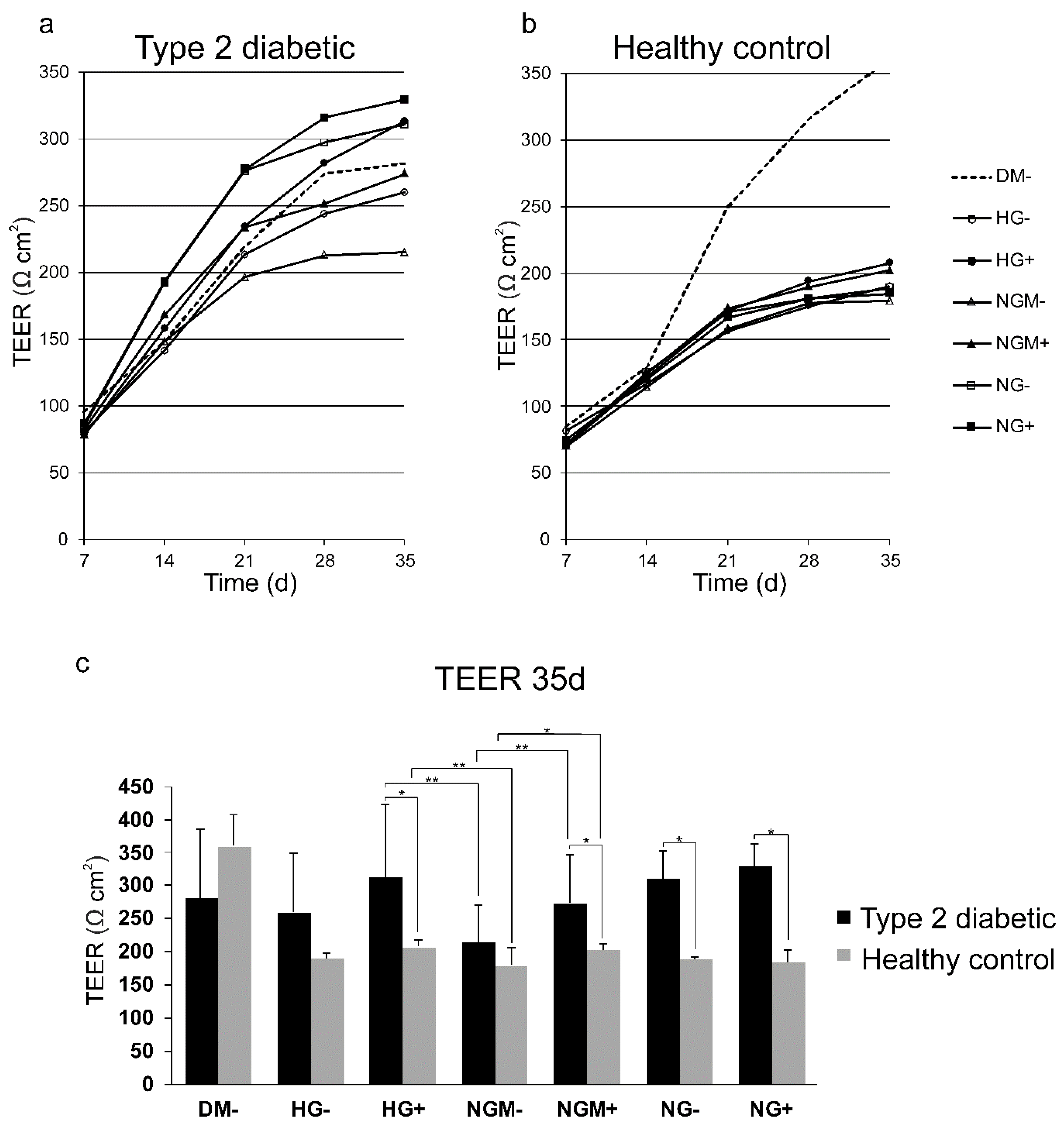
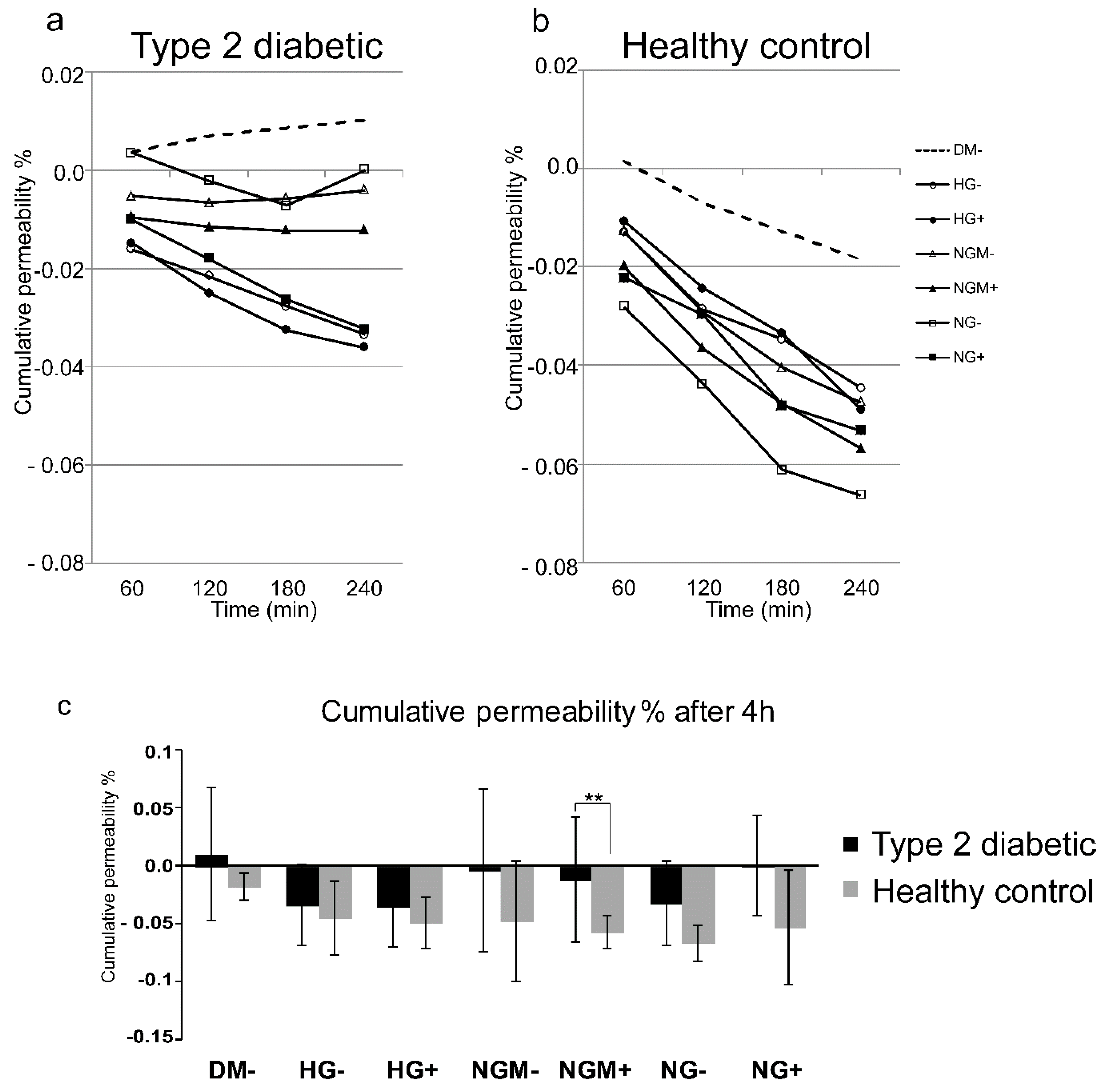
2.3. Barrier Properties in hiPSC-RPE Cells
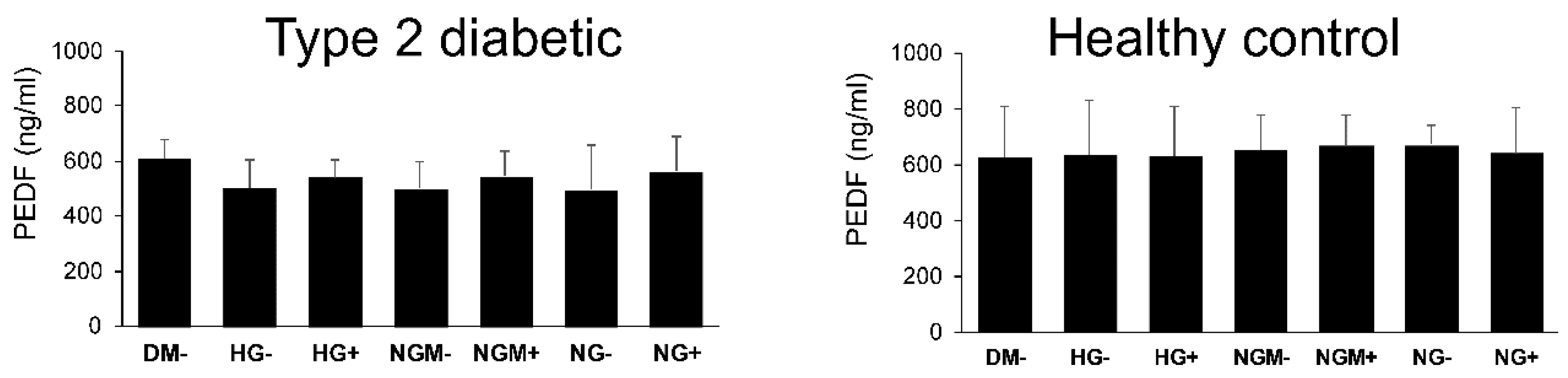
2.4. Effects of Glucose Concentration on the Secretion of the RPE-Specific Growth Factor (PEDF) Secretion
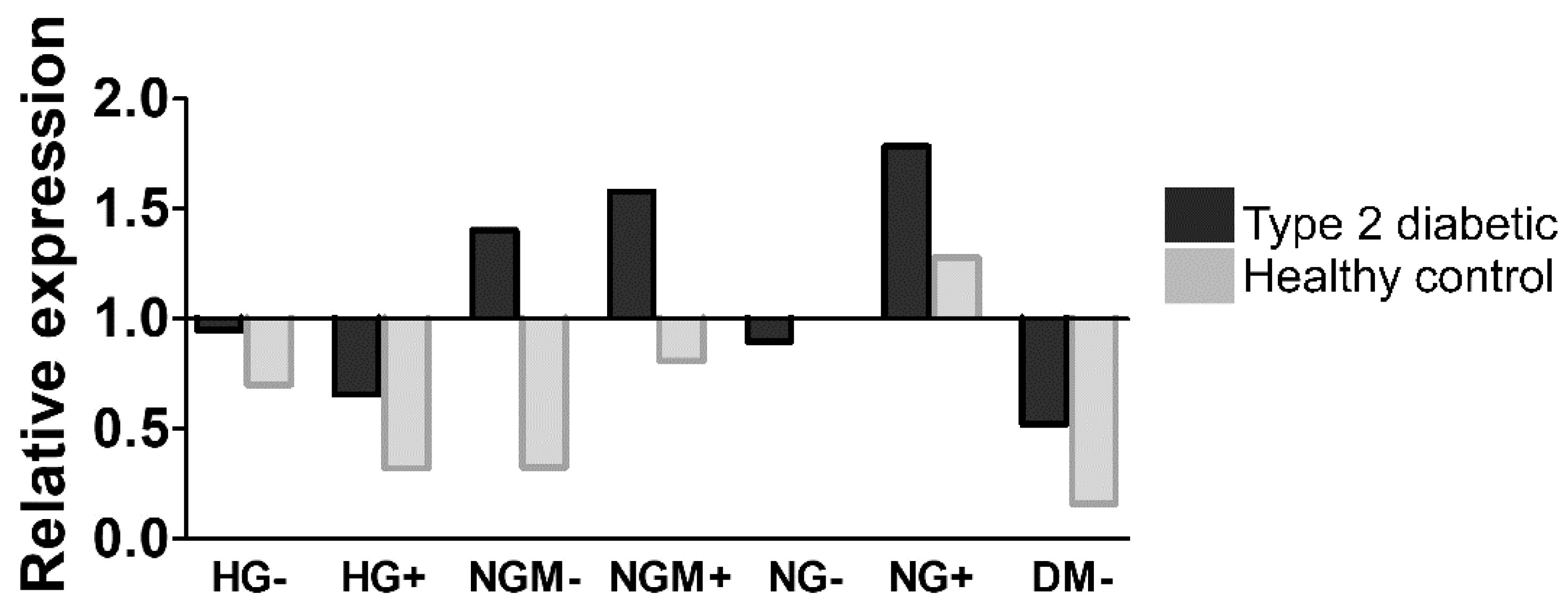
2.5. Effects of Glucose Concertation’s on Glucokinase Gene Expression
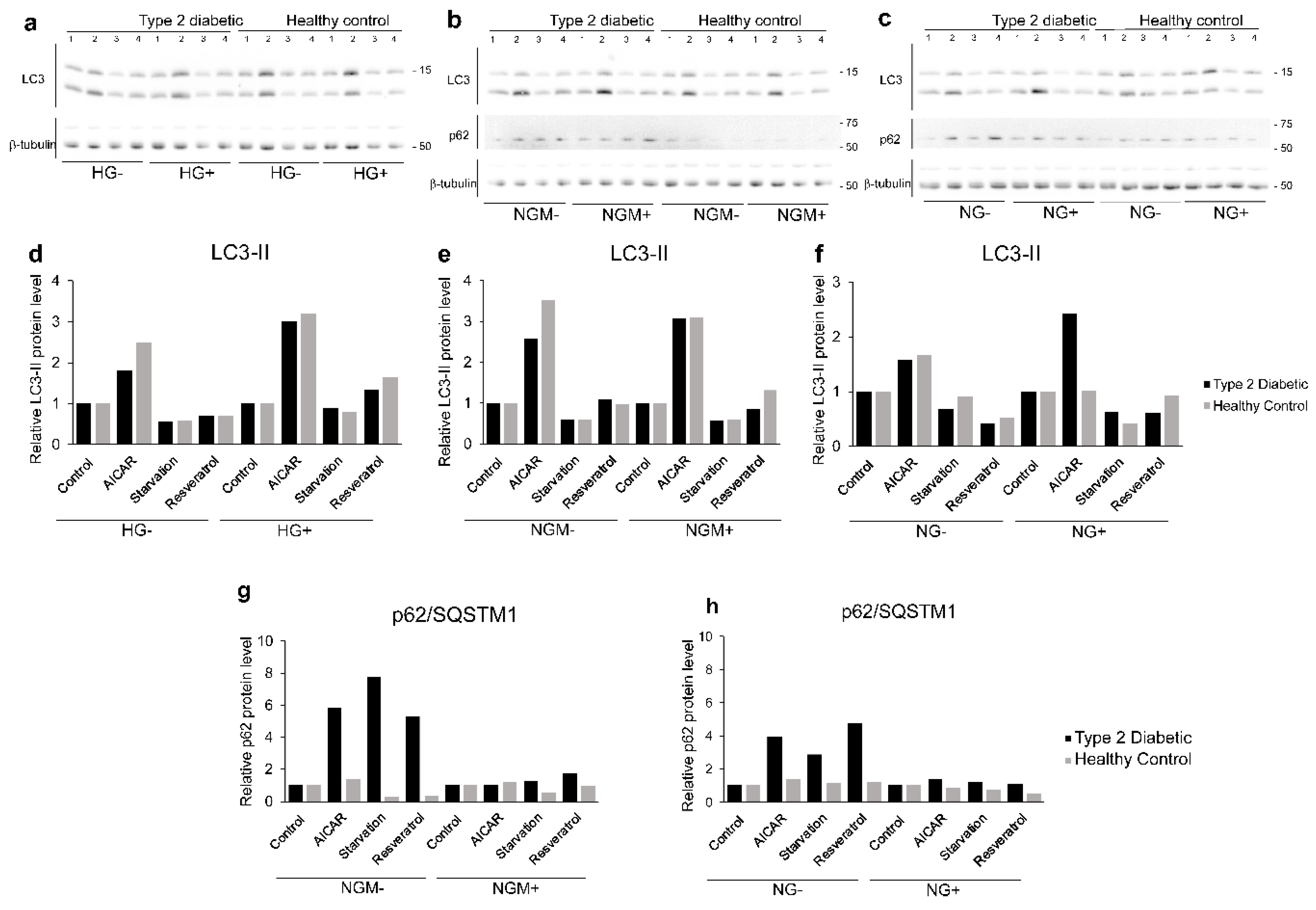
2.6. Effects of Glucose on Autophagy
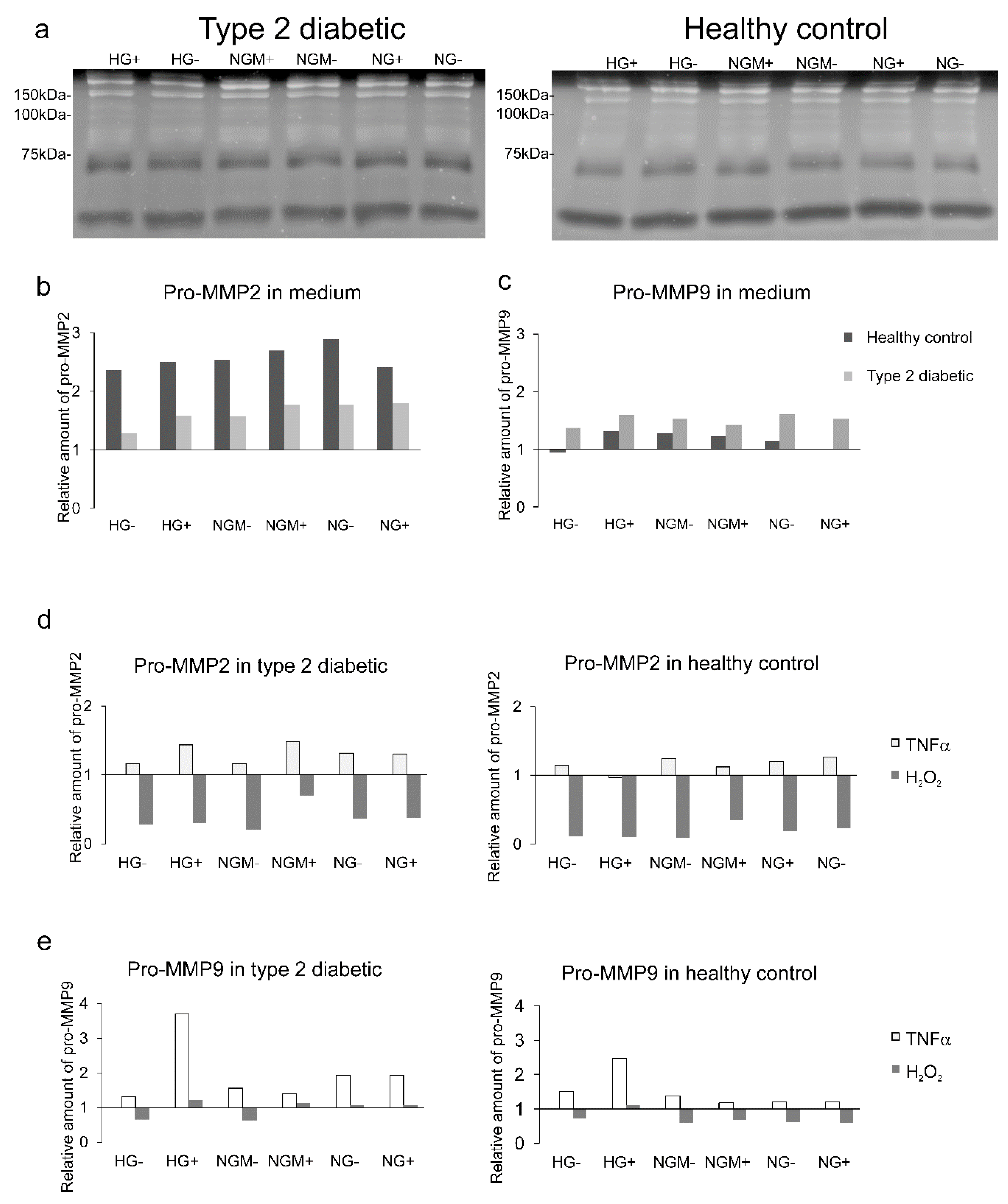
2.7. Effects of Glucose and Insulin Concentration and Stress Inducers on MMP Secretion
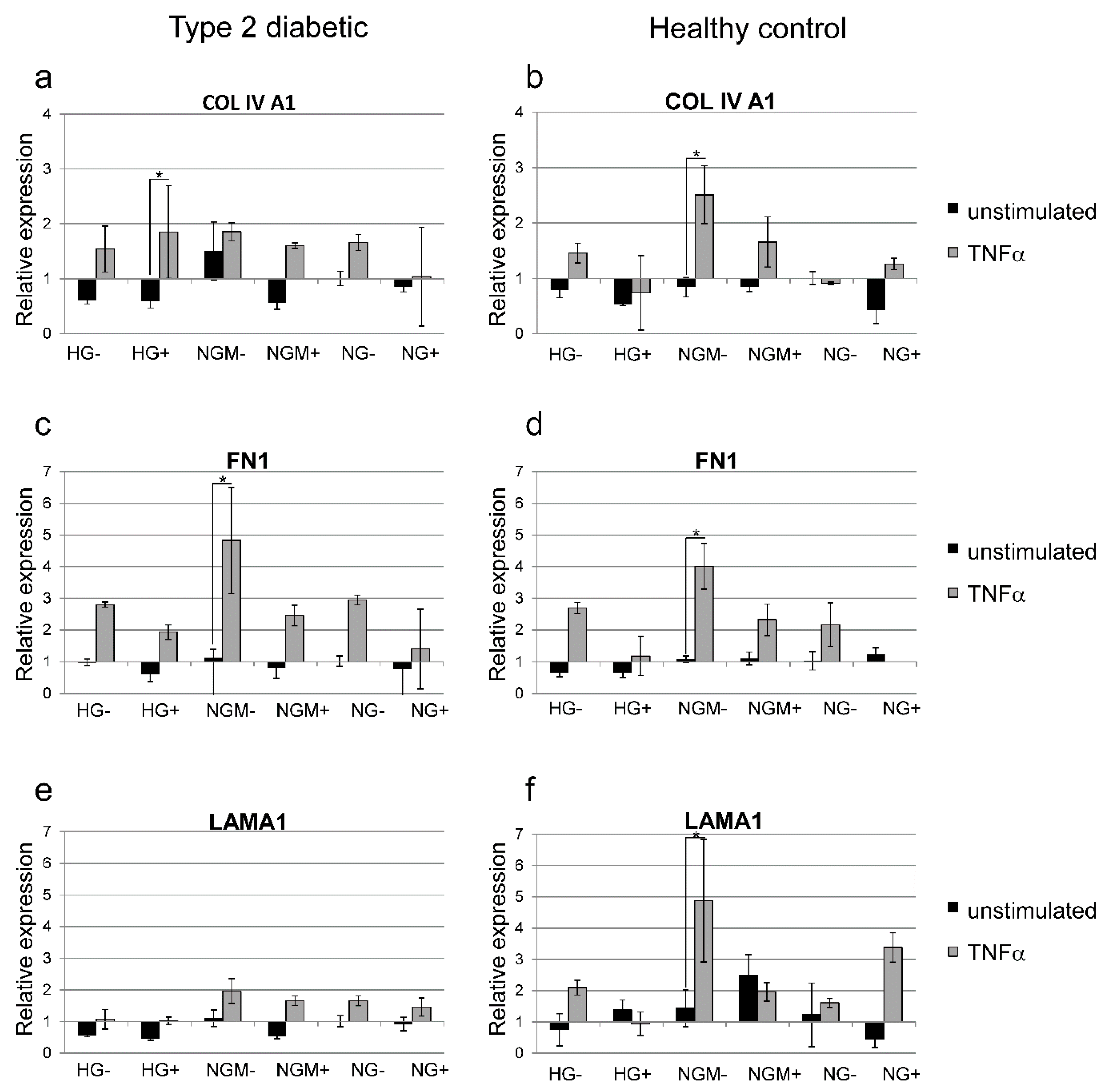
2.8. Effects of Glucose Concentration and Cytokine Stimulation on COL4A1, FN, and LAMA1 Gene Expression
3. Discussion
4. Materials and Methods
4.1. Human Induced Pluripotent Stem Cell Reprogramming
4.2. Human Induced Pluripotent Stem Cell Culture
4.3. Human Induced Pluripotent Stem Cell Characterization
4.4. Differentiation of Human Induced Pluripotent Stem Cells to Retinal Pigment Epithelial Cells
4.5. Cultivation of hiPSC-RPEs for the Experiments
4.6. Experimental Treatments
4.7. Indirect Immunofluorescence Staining
4.8. Preparation of Transmission Electron Microscopy Samples
4.9. RNA Extraction and cDNA Synthesis
4.10. PCR Reaction
4.11. Quantitative RT-PCR
4.12. Trans-Epithelial Electrical Resistance
4.13. Permeability Tests
4.14. Enzyme-Linked Immunosorbent Assay
4.15. Western Blotting
4.16. Zymography
4.17. Statistical Analyses
4.18. Ethical Issues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stitt, A.W.; Curtis, T.M.; Chen, M.; Medina, R.J.; McKay, G.J.; Jenkins, A.; Gardiner, T.A.; Lyons, T.J.; Hammes, H.P.; Simo, R.; et al. The progress in understanding and treatment of diabetic retinopathy. Prog. Retin. Eye Res. 2016, 51, 156–186. [Google Scholar] [CrossRef] [PubMed]
- Murugeswari, P.; Subramani, M.; Jayadev, C.; Shetty, R.; Das, D. Retinal pigment epithelium-secretome: A diabetic retinopathy perspective. Cytokine 2017, 95, 126–135. [Google Scholar]
- Xia, T.; Rizzolo, L.J. Effects of diabetic retinopathy on the barrier functions of the retinal pigment epithelium. Vision Res. 2017, 139, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Resnikoff, S.; Pascolini, D.; Etya’ale, D.; Kocur, I.; Pararajasegaram, R.; Pokharel, G.P.; Mariotti, S.P. Global data on visual impairment in the year 2002. Bull. World Health Organ 2004, 82, 844–851. [Google Scholar] [PubMed]
- Tarr, J.M.; Kaul, K.; Chopra, M.; Kohner, E.M.; Chibber, R. Pathophysiology of diabetic retinopathy. ISRN Ophthalmol. 2013, 2013, 343560. [Google Scholar] [CrossRef] [PubMed]
- Tarchick, M.J.; Cutler, A.H.; Trobenter, T.D.; Kozlowski, M.R.; Makowski, E.R.; Holoman, N.; Shao, J.; Shen, B.; Anand-Apte, B.; Samuels, I.S. Endogenous insulin signaling in the RPE contributes to the maintenance of rod photoreceptor function in diabetes. Exp. Eye Res. 2019, 180, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Tenconi, P.E.; Bermudez, V.; Oresti, G.M.; Giusto, N.M.; Salvador, G.A.; Mateos, M.V. High glucose-induced phospholipase D activity in retinal pigment epithelium cells: New insights into the molecular mechanisms of diabetic retinopathy. Exp. Eye Res. 2019, 184, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.N.; Li, S.T.; Li, W.; Hua, Y.J.; Wu, Q. The thickness and volume of the choroid, outer retinal layers and retinal pigment epithelium layer changes in patients with diabetic retinopathy. Int. J. Ophthalmol. 2018, 11, 1957–1962. [Google Scholar]
- Xu, H.Z.; Le, Y.Z. Significance of outer blood-retina barrier breakdown in diabetes and ischemia. Investig. Ophthal. Vis. Sci. 2011, 52, 2160–2164. [Google Scholar] [CrossRef]
- Villarroel, M.; Garcia-Ramirez, M.; Corraliza, L.; Hernandez, C.; Simo, R. Effects of high glucose concentration on the barrier function and the expression of tight junction proteins in human retinal pigment epithelial cells. Exp. Eye Res. 2009, 89, 913–920. [Google Scholar] [CrossRef]
- Beasley, S.; El-Sherbiny, M.; Megyerdi, S.; El-Shafey, S.; Choksi, K.; Kaddour-Djebbar, I.; Sheibani, N.; Hsu, S.; Al-Shabrawey, M. Caspase-14 expression impairs retinal pigment epithelium barrier function: Potential role in diabetic macular edema. BioMed Res. Int. 2014, 2014, 417986. [Google Scholar] [CrossRef]
- Trudeau, K.; Roy, S.; Guo, W.; Hernandez, C.; Villarroel, M.; Simo, R.; Roy, S. Fenofibric acid reduces fibronectin and collagen type IV overexpression in human retinal pigment epithelial cells grown in conditions mimicking the diabetic milieu: Functional implications in retinal permeability. Investig. Ophthalmol. Vis.Sci. 2011, 52, 6348–6354. [Google Scholar] [CrossRef]
- Qin, D.; Zhang, G.M.; Xu, X.; Wang, L.Y. The PI3K/Akt signaling pathway mediates the high glucose-induced expression of extracellular matrix molecules in human retinal pigment epithelial cells. J. Diabetes Res. 2015, 2015, 920280. [Google Scholar] [CrossRef]
- Coral, K.; Madhavan, J.; Pukhraj, R.; Angayarkanni, N. High glucose induced differential expression of lysyl oxidase and its isoform in ARPE-19 cells. Curr. Eye Res. 2013, 38, 194–203. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chou, H.C.; Lin, S.T.; Chen, Y.W.; Lo, Y.W.; Chan, H.L. Effect of high glucose on secreted proteome in cultured retinal pigmented epithelium cells: Its possible relevance to clinical diabetic retinopathy. J. Proteom. 2012, 77, 111–128. [Google Scholar] [CrossRef]
- Calado, S.M.; Alves, L.S.; Simao, S.; Silva, G.A. GLUT1 activity contributes to the impairment of PEDF secretion by the RPE. Mol. Vis. 2016, 22, 761–770. [Google Scholar]
- Farnoodian, M.; Halbach, C.; Slinger, C.; Pattnaik, B.R.; Sorenson, C.M.; Sheibani, N. High glucose promotes the migration of retinal pigment epithelial cells through increased oxidative stress and PEDF expression. Am. J. Physiol. Cell Physiol. 2016, 311, C418–C436. [Google Scholar] [CrossRef]
- Matteucci, A.; Varano, M.; Mallozzi, C.; Gaddini, L.; Villa, M.; Gabrielli, S.; Formisano, G.; Pricci, F.; Malchiodi-Albedi, F. Primary retinal cultures as a tool for modeling diabetic retinopathy: An overview. BioMed Res. Int. 2015, 2015, 364924. [Google Scholar] [CrossRef]
- Borooah, S.; Phillips, M.J.; Bilican, B.; Wright, A.F.; Wilmut, I.; Chandran, S.; Gamm, D.; Dhillon, B. Using human induced pluripotent stem cells to treat retinal disease. Prog. Retin. Eye Res. 2013, 37, 163–181. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Xu, H.; Kauppinen, A. Mechanistical retinal drug targets and challenges. Adv. Drug Deliv. Rev. 2018, 126, 177–184. [Google Scholar] [CrossRef]
- Johansson, I.; Monsen, V.T.; Pettersen, K.; Mildenberger, J.; Misund, K.; Kaarniranta, K.; Schonberg, S.; Bjorkoy, G. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy 2015, 11, 1636–1651. [Google Scholar] [CrossRef]
- Felszeghy, S.; Viiri, J.; Paterno, J.J.; Hyttinen, J.M.T.; Koskela, A.; Chen, M.; Leinonen, H.; Tanila, H.; Kivinen, N.; Koistinen, A.; et al. Loss of NRF-2 and PGC-1alpha genes leads to retinal pigment epithelium damage resembling dry age-related macular degeneration. Redox Biol. 2019, 20, 1–12. [Google Scholar] [CrossRef]
- Uchiki, T.; Weikel, K.A.; Jiao, W.; Shang, F.; Caceres, A.; Pawlak, D.; Handa, J.T.; Brownlee, M.; Nagaraj, R.; Taylor, A. Glycation-altered proteolysis as a pathobiologic mechanism that links dietary glycemic index, aging, and age-related disease (in nondiabetics). Aging Cell 2012, 11, 1–13. [Google Scholar] [CrossRef]
- Kikuchi, C.; Bienengraeber, M.; Canfield, S.; Koopmeiner, A.; Schafer, R.; Bosnjak, Z.J.; Bai, X. Comparison of Cardiomyocyte Differentiation Potential Between Type 1 Diabetic Donor- and Nondiabetic Donor-Derived Induced Pluripotent Stem Cells. Cell Transplant. 2015, 24, 2491–2504. [Google Scholar] [CrossRef]
- Shaer, A.; Azarpira, N.; Vahdati, A.; Karimi, M.H.; Shariati, M. Differentiation of human-induced pluripotent stem cells into insulin-producing clusters. Exp. Clin. Transplant. 2015, 13, 68–75. [Google Scholar]
- Sternisha, S.M.; Miller, B.G. Molecular and cellular regulation of human glucokinase. Arch. Biochem. Biophys. 2019, 663, 199–213. [Google Scholar] [CrossRef]
- Klaassen, I.; Van Noorden, C.J.; Schlingemann, R.O. Molecular basis of the inner blood-retinal barrier and its breakdown in diabetic macular edema and other pathological conditions. Prog. Retin. Eye Res. 2013, 34, 19–48. [Google Scholar] [CrossRef]
- Maugeri, G.; D’Amico, A.G.; Rasa, D.M.; La Cognata, V.; Saccone, S.; Federico, C.; Cavallaro, S.; D’Agata, V. Nicotine promotes blood retinal barrier damage in a model of human diabetic macular edema. Toxicol. In Vitro 2017, 44, 182–189. [Google Scholar] [CrossRef]
- Wang, S.; Du, S.; Wu, Q.; Hu, J.; Li, T. Decorin Prevents Retinal Pigment Epithelial Barrier Breakdown Under Diabetic Conditions by Suppressing p38 MAPK Activation. Investig. Ophthalmol. Vis.Sci. 2015, 56, 2971–2979. [Google Scholar] [CrossRef]
- Maugeri, G.; D’Amico, A.G.; Rasa, D.M.; La Cognata, V.; Saccone, S.; Federico, C.; Cavallaro, S.; D’Agata, V. Caffeine Prevents Blood Retinal Barrier Damage in a Model, In Vitro, of Diabetic Macular Edema. J. Cell. Biochem. 2017, 118, 2371–2379. [Google Scholar] [CrossRef]
- Maugeri, G.; D’Amico, A.G.; Gagliano, C.; Saccone, S.; Federico, C.; Cavallaro, S.; D’Agata, V. VIP Family Members Prevent Outer Blood Retinal Barrier Damage in a Model of Diabetic Macular Edema. J. Cell. Physiol. 2017, 232, 1079–1085. [Google Scholar] [CrossRef]
- Adamis, A.P.; Shima, D.T.; Yeo, K.T.; Yeo, T.K.; Brown, L.F.; Berse, B.; Damore, P.A.; Folkman, J. Synthesis and Secretion of Vascular-Permeability Factor Vascular Endothelial Growth-Factor by Human Retinal-Pigment Epithelial-Cells. Biochem. Biophys. Res. Commun. 1993, 193, 631–638. [Google Scholar] [CrossRef]
- Vaajasaari, H.; Ilmarinen, T.; Juuti-Uusitalo, K.; Rajala, K.; Onnela, N.; Narkilahti, S.; Suuronen, R.; Hyttinen, J.; Uusitalo, H.; Skottman, H. Toward the defined and xeno-free differentiation of functional human pluripotent stem cell-derived retinal pigment epithelial cells. Mol. Vis. 2011, 17, 558–575. [Google Scholar]
- Rosa, M.D.; Distefano, G.; Gagliano, C.; Rusciano, D.; Malaguarnera, L. Autophagy in Diabetic Retinopathy. Curr. Neuropharmacol. 2016, 14, 810–825. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, Z.; Wang, X.; Li, R.; Hou, W.; Bi, W.; Zhang, X. Inhibition of autophagy induces IL-1beta release from ARPE-19 cells via ROS mediated NLRP3 inflammasome activation under high glucose stress. Biochem. Biophys. Res. Commun. 2015, 463, 1071–1076. [Google Scholar] [CrossRef]
- Zhang, Y.; Xi, X.; Mei, Y.; Zhao, X.; Zhou, L.; Ma, M.; Liu, S.; Zha, X.; Yang, Y. High-glucose induces retinal pigment epithelium mitochondrial pathways of apoptosis and inhibits mitophagy by regulating ROS/PINK1/Parkin signal pathway. Biomed. Pharmacother. 2019, 111, 1315–1325. [Google Scholar] [CrossRef]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef]
- Giebel, S.J.; Menicucci, G.; McGuire, P.G.; Das, A. Matrix metalloproteinases in early diabetic retinopathy and their role in alteration of the blood-retinal barrier. Lab. Investig. 2005, 85, 597–607. [Google Scholar] [CrossRef]
- Huang, H.; Gandhi, J.K.; Zhong, X.; Wei, Y.; Gong, J.; Duh, E.J.; Vinores, S.A. TNFalpha is required for late BRB breakdown in diabetic retinopathy, and its inhibition prevents leukostasis and protects vessels and neurons from apoptosis. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1336–1344. [Google Scholar] [CrossRef]
- Abu el Asrar, A.M.; Maimone, D.; Morse, P.H.; Gregory, S.; Reder, A.T. Cytokines in the vitreous of patients with proliferative diabetic retinopathy. Am. J. Ophthalmol. 1992, 114, 731–736. [Google Scholar] [CrossRef]
- Joussen, A.M.; Poulaki, V.; Dohmen, S.; Koizumi, K.; Kirchhof, B.; Adamis, A.P. Non-steroid alanti-inflammatory drugs prevent early diabetic retinopathy: Aspirinand COX-2 inhibition prevent blood-retinal barrier breakdown and leukocyte adhesion via TNF-asuppression. Investig. Ophthalmol. Vis. Sci. 2002, 43, 2969. [Google Scholar]
- Ohnuki, M.; Takahashi, K.; Yamanaka, S. Generation and characterization of human induced pluripotent stem cells. Curr. Protocols Stem Cell Biol. 2009, 9, 4A-2. [Google Scholar]
- Takahashi, K.; Okita, K.; Nakagawa, M.; Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protocols 2007, 2, 3081–3089. [Google Scholar] [CrossRef]
- Manzini, S.; Viiri, L.E.; Marttila, S.; Aalto-Setala, K. A Comparative View on Easy to Deploy non-Integrating Methods for Patient-Specific iPSC Production. Stem Cell Rev. 2015, 11, 900–908. [Google Scholar] [CrossRef]
- Kiamehr, M.; Viiri, L.E.; Vihervaara, T.; Koistinen, K.M.; Hilvo, M.; Ekroos, K.; Kakela, R.; Aalto-Setala, K. Lipidomic profiling of patient-specific iPSC-derived hepatocyte-like cells. Dis. Model. Mech. 2017, 10, 1141–1153. [Google Scholar] [CrossRef]
- Lund, R.J.; Nikula, T.; Rahkonen, N.; Narva, E.; Baker, D.; Harrison, N.; Andrews, P.; Otonkoski, T.; Lahesmaa, R. High-throughput karyotyping of human pluripotent stem cells. Stem Cell Res. 2012, 9, 192–195. [Google Scholar] [CrossRef]
- Viiri, J.; Amadio, M.; Marchesi, N.; Hyttinen, J.M.; Kivinen, N.; Sironen, R.; Rilla, K.; Akhtar, S.; Provenzani, A.; D’Agostino, V.G.; et al. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS ONE 2013, 8, e69563. [Google Scholar] [CrossRef]
- Kiamehr, M.; Alexanova, A.; Viiri, L.E.; Heiskanen, L.; Vihervaara, T.; Kauhanen, D.; Ekroos, K.; Laaksonen, R.; Kakela, R.; Aalto-Setala, K. hiPSC-derived hepatocytes closely mimic the lipid profile of primary hepatocytes: A future personalised cell model for studying the lipid metabolism of the liver. J. Cell. Physiol. 2019, 234, 3744–3761. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Skottman, H.; Muranen, J.; Lahdekorpi, H.; Pajula, E.; Makela, K.; Koivusalo, L.; Koistinen, A.; Uusitalo, H.; Kaarniranta, K.; Juuti-Uusitalo, K. Contacting co-culture of human retinal microvascular endothelial cells alters barrier function of human embryonic stem cell derived retinal pigment epithelial cells. Exp. Cell Res. 2017, 359, 101–111. [Google Scholar] [CrossRef]
- Juuti-Uusitalo, K.; Nieminen, M.; Treumer, F.; Ampuja, M.; Kallioniemi, A.; Klettner, A.; Skottman, H. Effects of Cytokine Activation and Oxidative Stress on the Function of the Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 6265–6274. [Google Scholar] [CrossRef]

| Gene | Forward | Reverse | bp | Tm |
|---|---|---|---|---|
| GAPDH | GTTCGACAGTCAGCCGCATC | GGAATTTGCCATGGGTGGA | 229 | 55 |
| OCT 3/4 | CGTGAAGCTGGAGAAGGAGAAGCTG | AAGGGCCGCAGCTTACACATGTTC | 245 | 55 |
| PAX6 | AACAGACACAGCCCTCACAAACA | CGGGAACTTGAACTGGAACTGAC | 274 | 60 |
| BEST | GAATTTGCAGGTGTCCCTGT | ATCAGGAGGACGAGGAGGAT | 214 | 60 |
| RPE65 | TCC CCA ATA CAA CTG CCA CT | CAC CACC ACA CTC AGA ACT A | 316 | 52 |
| Tyrosinase | TGC CAA CGA TCC TAT CTT CC | GAC ACA GCA AGC TCA CAA GC | 316 | 52 |
| Protein | Primary Antibody Staining | Secondary Antibody Staining | Buffer Used for Dilution and Washing | ||
|---|---|---|---|---|---|
| Antibody | Cat, Producer, Dilution | Antibody | Producer, Dilution, Time | ||
| p62 | Mouse monoclonal p62 antibody | sc-28359, Santa Cruz Biotechnology Inc, CA, USA, 1:1000 | Horseradish peroxidase-conjugated anti-mouse IgG | GE Healthcare, Little Chalfont, Buckinghamshire, UK, 1:10,000, 2 h | 0.5% BSA in 0.3% Tween-20/PBS |
| LC3 | Rabbit polyclonal LC3 antibody | 3868, Cell Signaling, Danvers, MA, USA, 1:1000 | Horseradish peroxidase-conjugated anti-rabbit IgG | Novex™, ThermoFisher Scientific, 1:10,000, 2 h | 5% BSA in 0.1% Tween-20/TBS (tris-buffered saline) overnight at 4 °C |
| α-tubulin | Mouse monoclonal alpha-tubulin antibody | T5168, Sigma-Aldrich, 1:8000 | Horseradish peroxidase-conjugated anti-rabbit IgG | GE Healthcare, Little Chalfont, Buckinghamshire, UK, 1:10,000, 1 h | 1% milk powder in 0.05 % Tween-20/PBS for 1hrou at RT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiamehr, M.; Klettner, A.; Richert, E.; Koskela, A.; Koistinen, A.; Skottman, H.; Kaarniranta, K.; Aalto-Setälä, K.; Juuti-Uusitalo, K. Compromised Barrier Function in Human Induced Pluripotent Stem-Cell-Derived Retinal Pigment Epithelial Cells from Type 2 Diabetic Patients. Int. J. Mol. Sci. 2019, 20, 3773. https://doi.org/10.3390/ijms20153773
Kiamehr M, Klettner A, Richert E, Koskela A, Koistinen A, Skottman H, Kaarniranta K, Aalto-Setälä K, Juuti-Uusitalo K. Compromised Barrier Function in Human Induced Pluripotent Stem-Cell-Derived Retinal Pigment Epithelial Cells from Type 2 Diabetic Patients. International Journal of Molecular Sciences. 2019; 20(15):3773. https://doi.org/10.3390/ijms20153773
Chicago/Turabian StyleKiamehr, Mostafa, Alexa Klettner, Elisabeth Richert, Ali Koskela, Arto Koistinen, Heli Skottman, Kai Kaarniranta, Katriina Aalto-Setälä, and Kati Juuti-Uusitalo. 2019. "Compromised Barrier Function in Human Induced Pluripotent Stem-Cell-Derived Retinal Pigment Epithelial Cells from Type 2 Diabetic Patients" International Journal of Molecular Sciences 20, no. 15: 3773. https://doi.org/10.3390/ijms20153773
APA StyleKiamehr, M., Klettner, A., Richert, E., Koskela, A., Koistinen, A., Skottman, H., Kaarniranta, K., Aalto-Setälä, K., & Juuti-Uusitalo, K. (2019). Compromised Barrier Function in Human Induced Pluripotent Stem-Cell-Derived Retinal Pigment Epithelial Cells from Type 2 Diabetic Patients. International Journal of Molecular Sciences, 20(15), 3773. https://doi.org/10.3390/ijms20153773








