iTRAQ-Based Proteomics Analysis of Autophagy-Mediated Responses against MeJA in Laticifers of Euphorbia kansui L.
Abstract
1. Introduction
2. Results
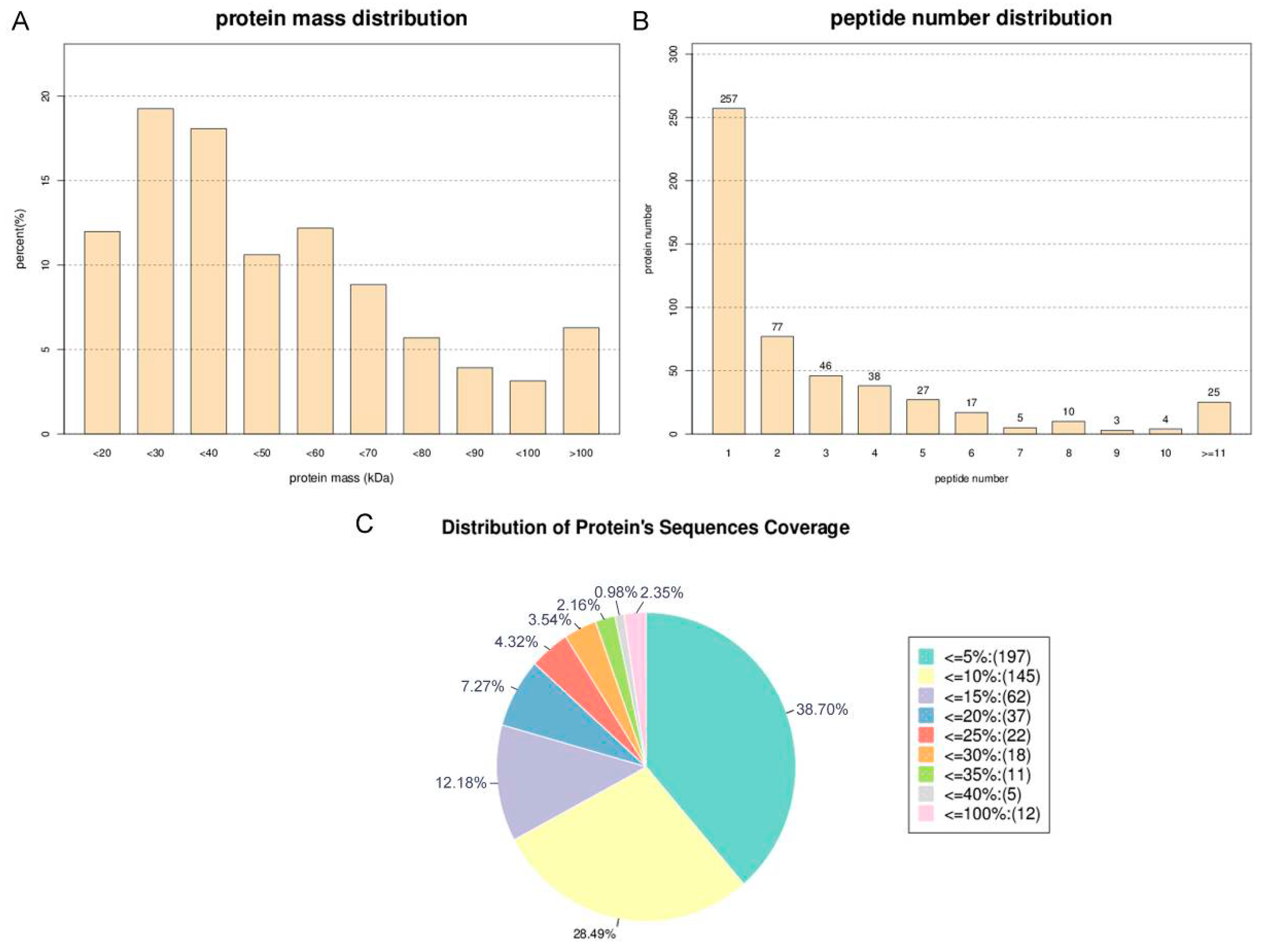
2.1. Primary Data Analysis and Protein Identification
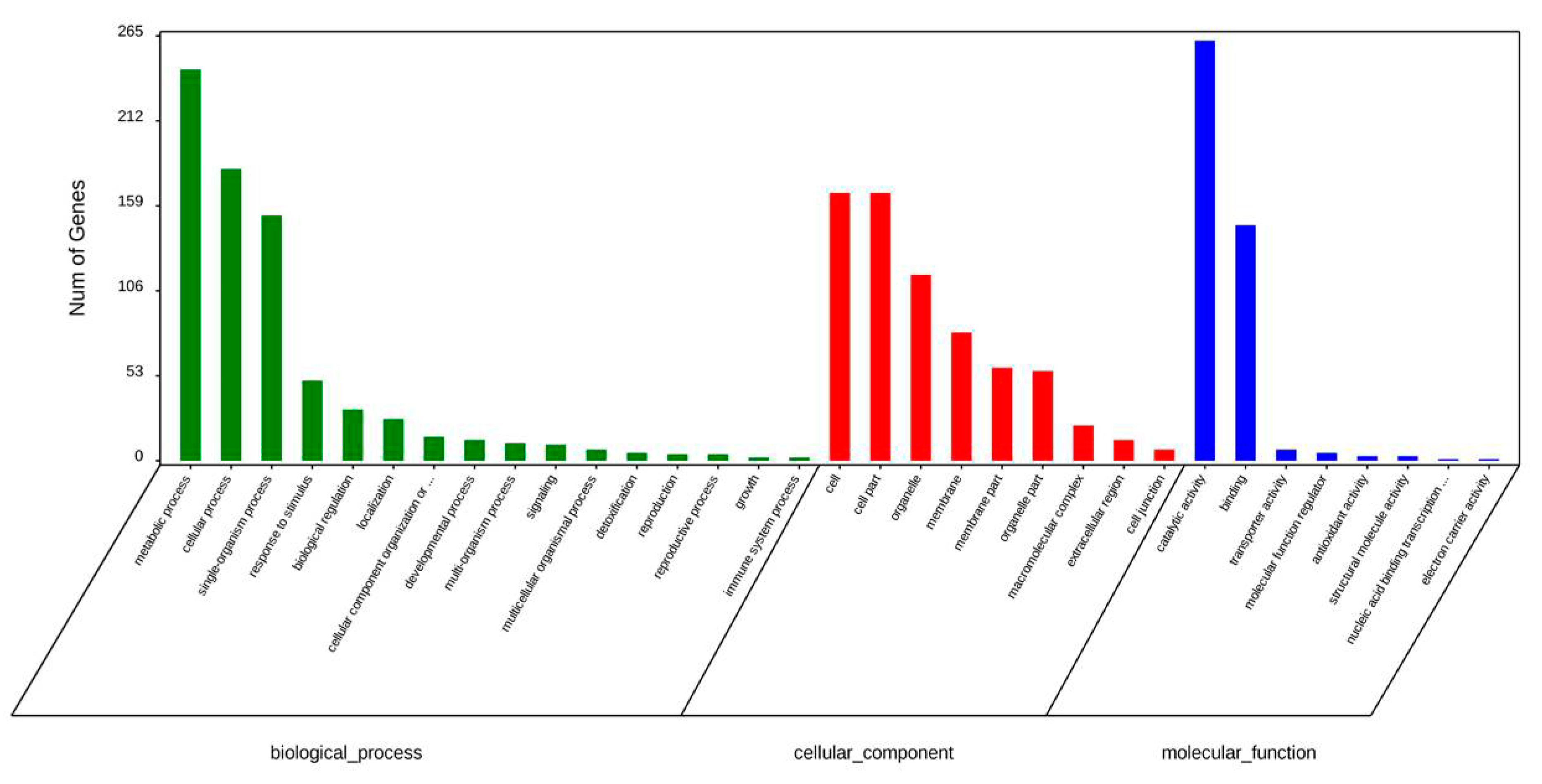
2.2. Gene Ontology Analysis
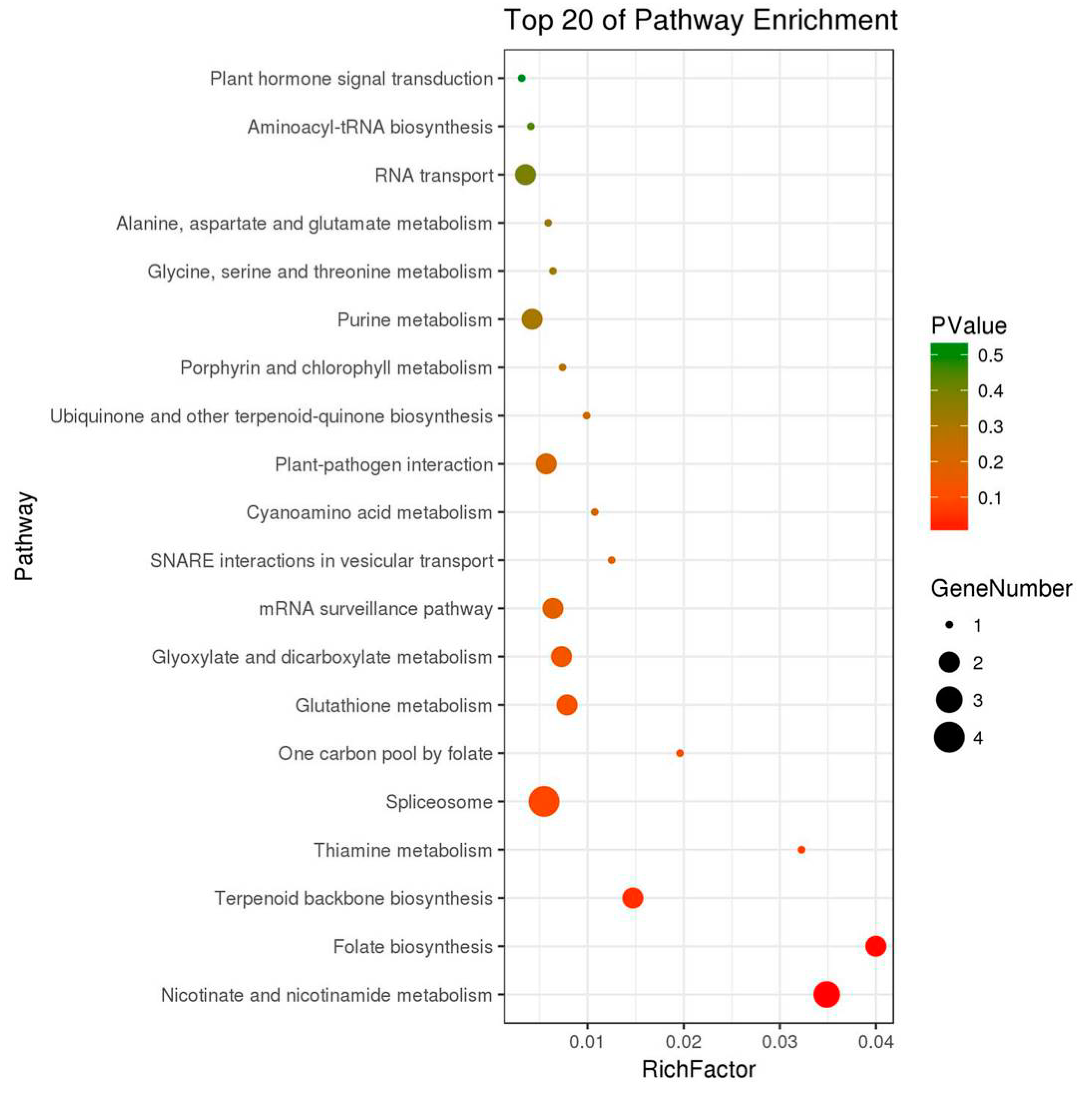
2.3. KEGG Enriched Pathways
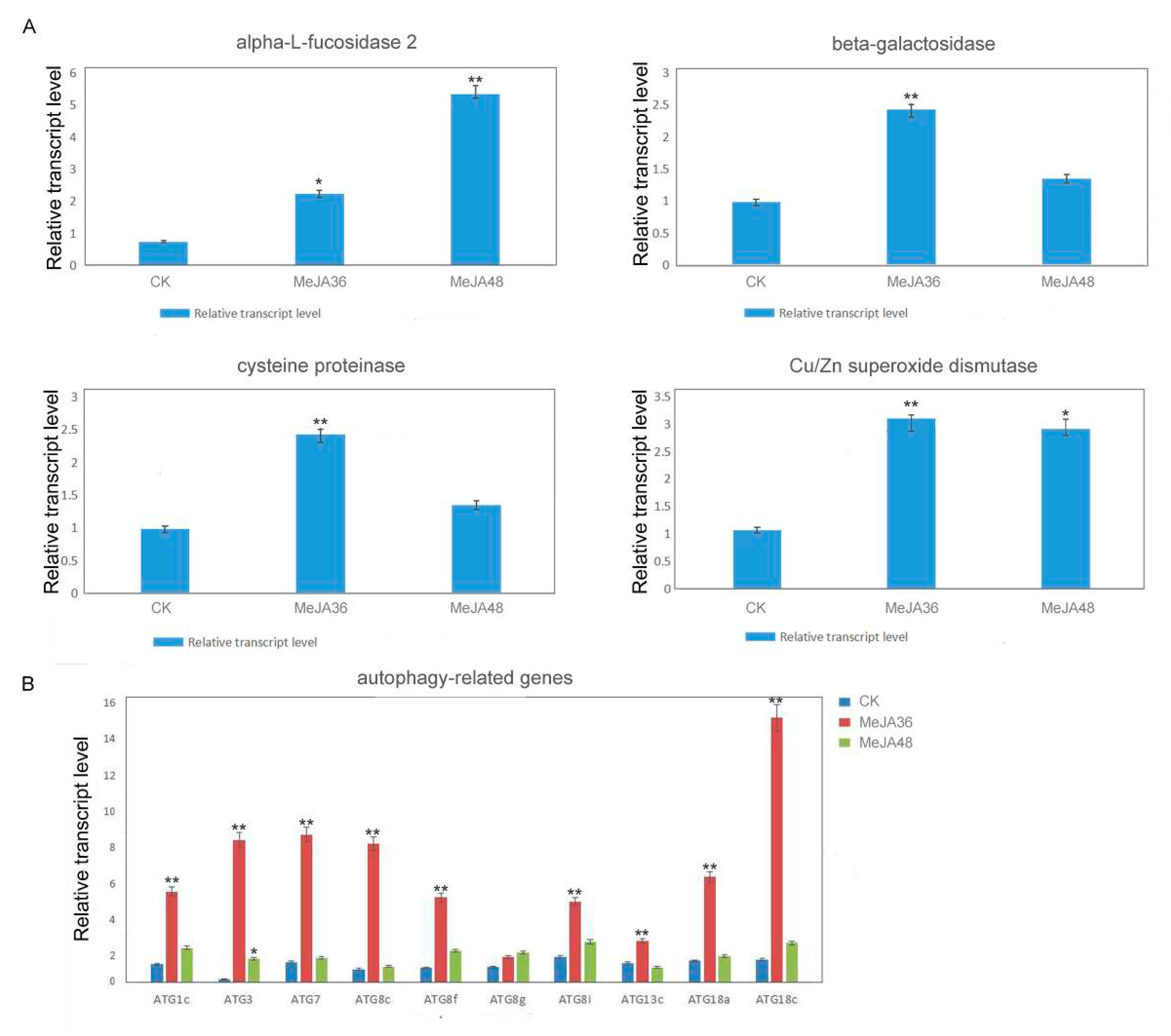
2.4. Quantitative RT-PCR (qRT-PCR) Analysis on Expression of Genes of Significantly Differentially Expressed Proteins under MeJA Stress
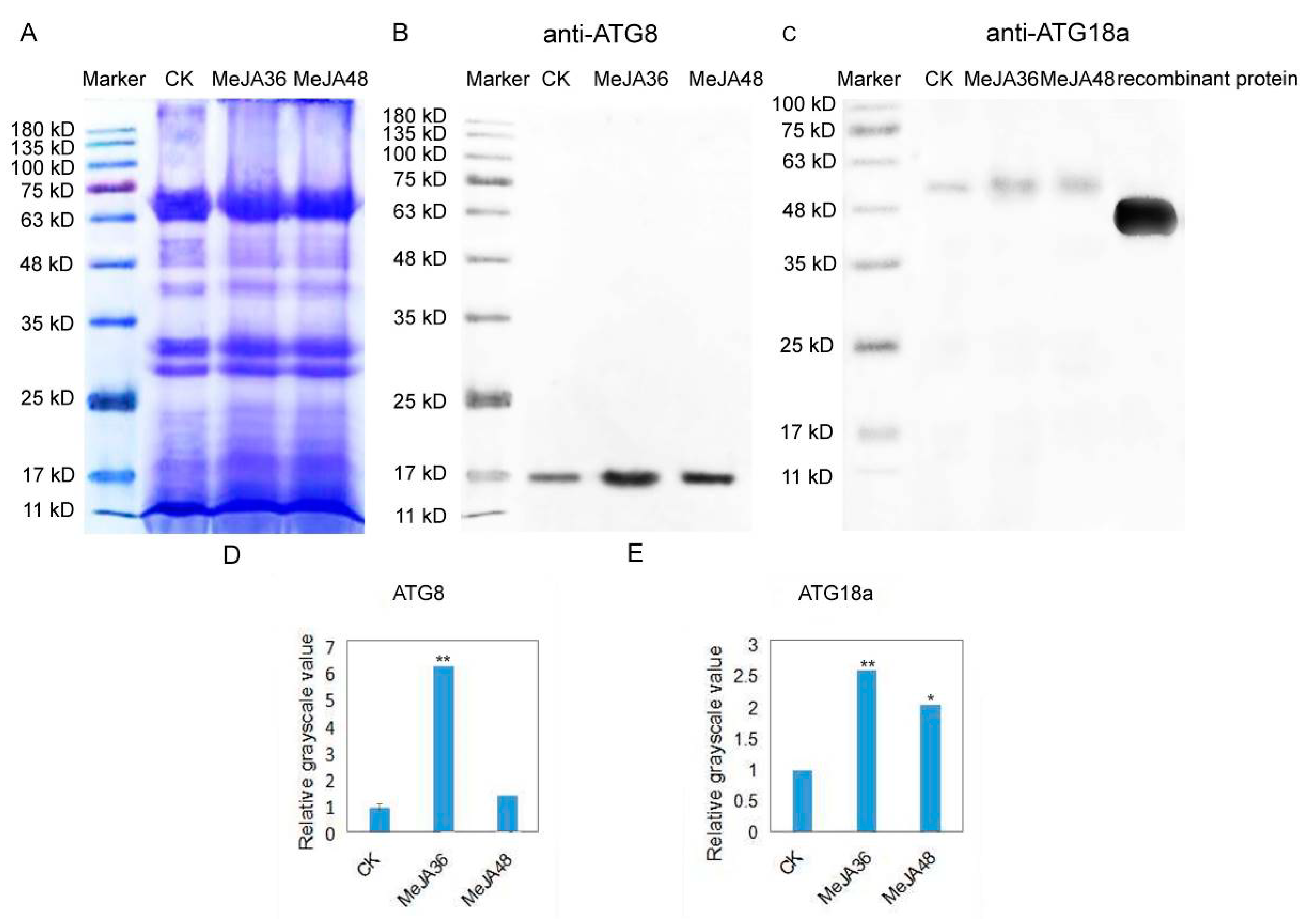
2.5. Immunoblotting Analysis of ATG8 and ATG18a in Laticifers under MeJA Treatment
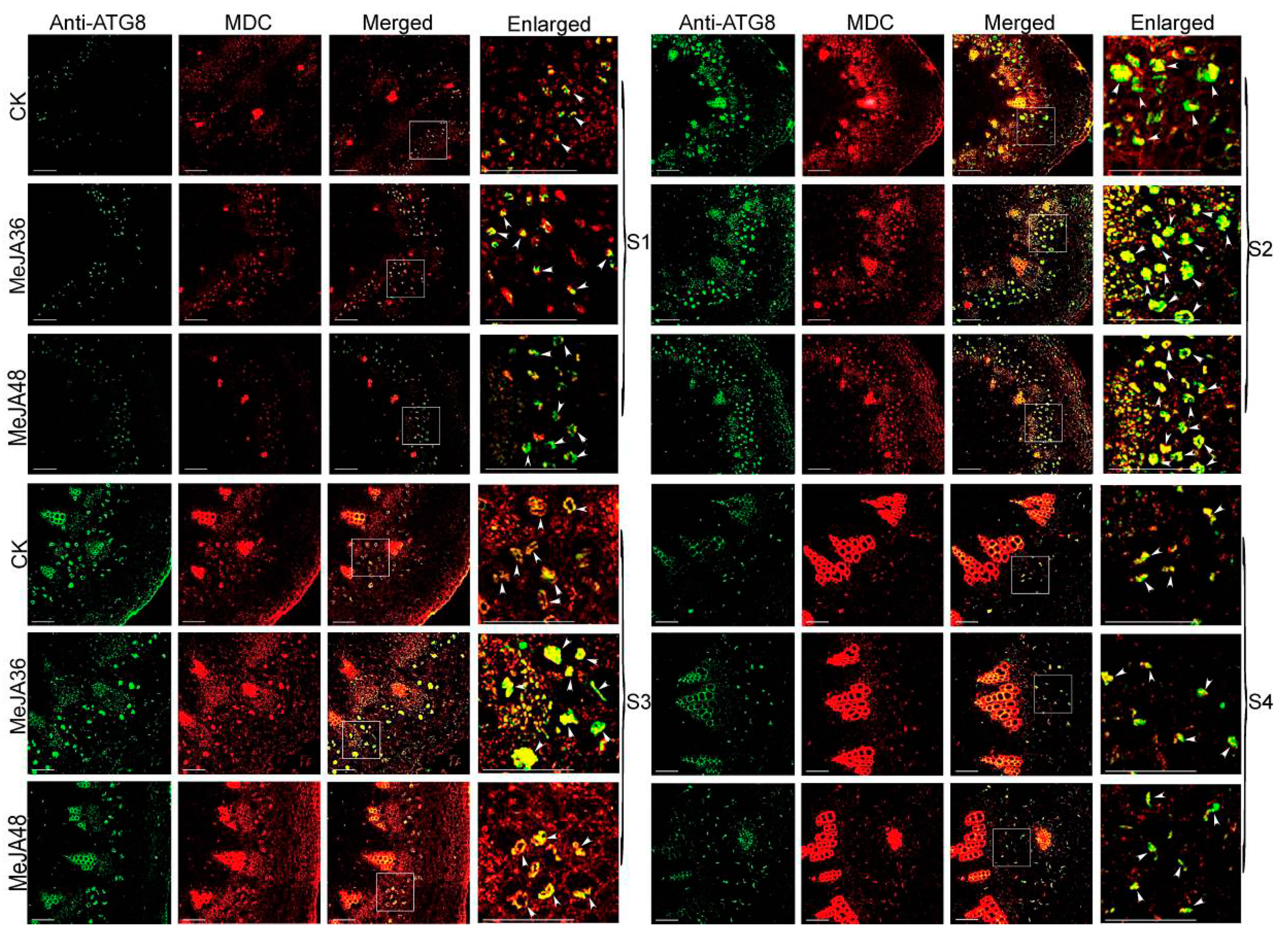
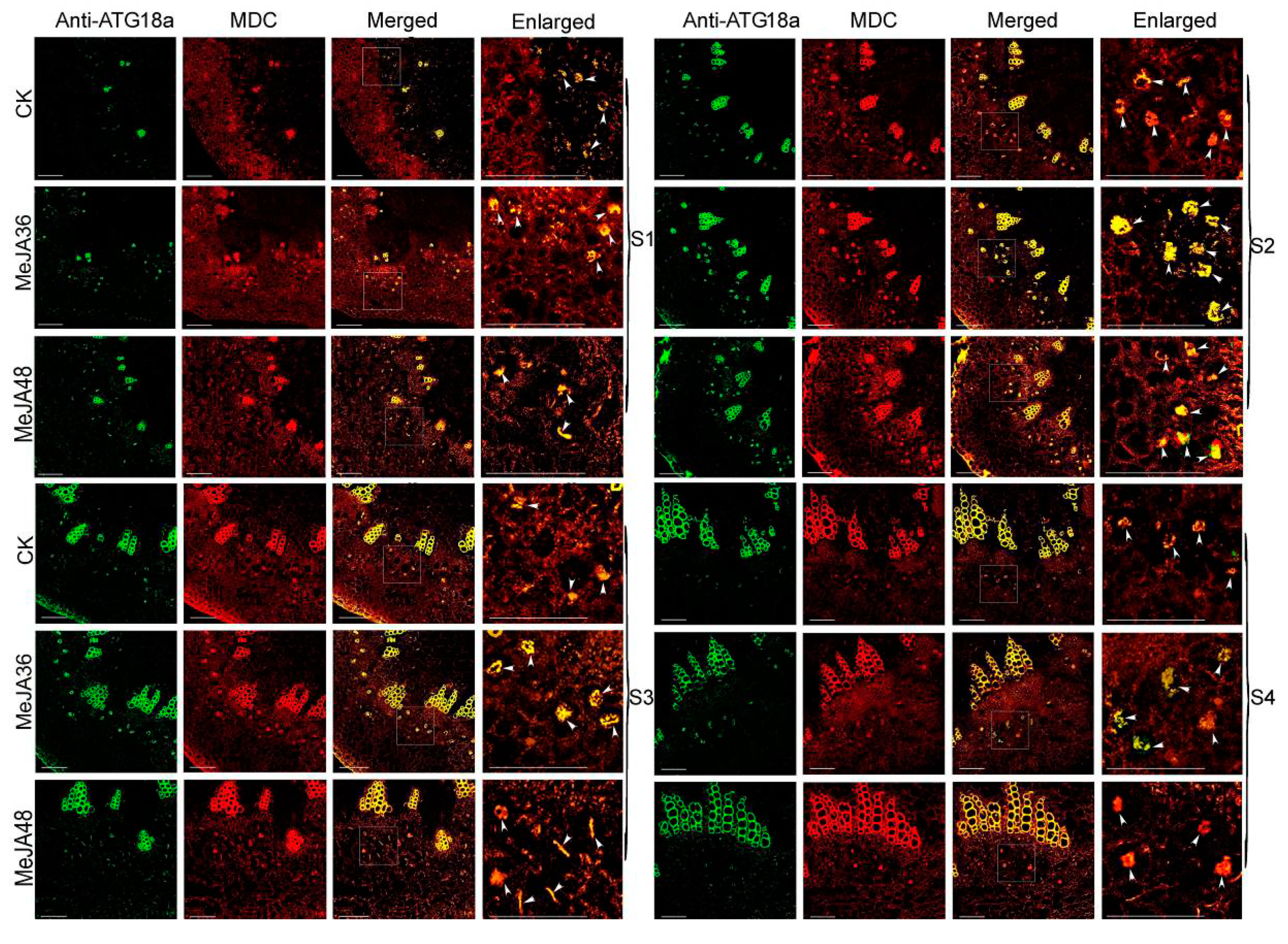
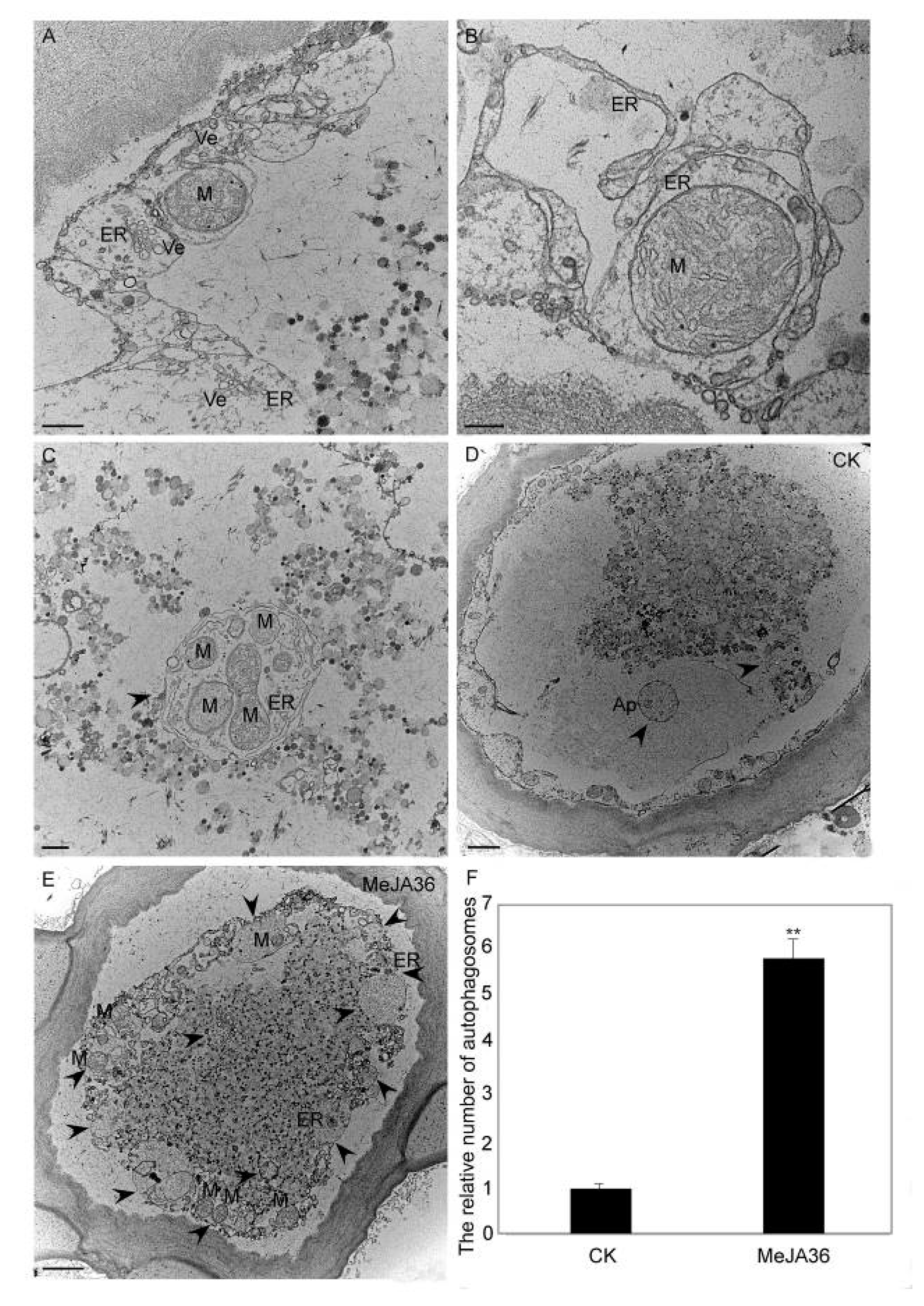
2.6. Confocal Immunofluorescence Analysis and TEM Analysis of Autophagy in Laticifers with MeJA Treatment
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Protein Preparation and iTRAQ Labeling
4.3. LC-MS/MS Analysis
4.4. Protein Identification and Quantification
4.5. Bioinformatics Analysis
4.6. Validation by Real-Time qPCR Analysis
4.7. Antibodies and Western Blot Analysis
4.8. Confocal Immunofluorescence Studies
4.9. Statistical Analysis
4.10. Transmission Electron Microscopy
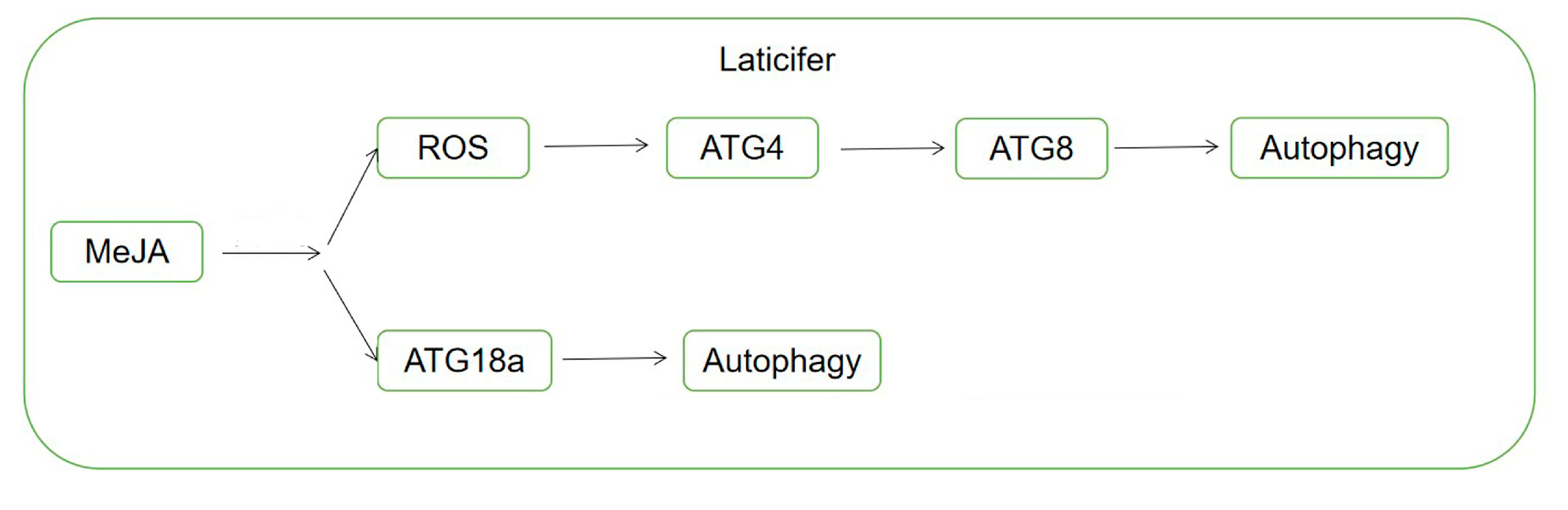
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MeJA | Methyl-jasmonate |
| AP | Autophagosome |
| ER | Endoplasmic Reticulum |
| M | Mitochondria |
| Ve | Vesicle |
References
- Wasternack, C.; Parthier, B. Jasmonate-signaled plant gene expression. Trends Plant Sci. 1997, 2, 302–307. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mullet, J.E. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Biol. 1997, 48, 355–381. [Google Scholar] [CrossRef] [PubMed]
- Arimura, G.; Ozawa, R.; Shimoda, T.; Nishioka, T.; Boland, W.; Takabayashi, J. Herbivory-induced volatiles elicit defence genes in Lima bean leaves. Nature 2000, 406, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.J.; Choi, Y.D. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003, 19, 409–413. [Google Scholar] [CrossRef]
- Ueda, J.; Kato, J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Huang, J.; Zheng, C.C.; Cai, C.X.; Wang, Q.M.; Wu, C.A. Salt and methyl jasmonate aggravate growth inhibition and senescence in Arabidopsis seedlings via the ja signaling pathway. Plant Sci. 2017, 261, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rossato, L.; Macduff, J.H.; Laine, P.; Le, D.E.; Ourry, A. Nitrogen storage and remobilization in Brassica napus L. during the growth cycle: Effects of methyl jasmonate on nitrate uptake, senescence, growth, and vsp accumulation. J. Exp. Bot. 2002, 53, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates-biosynthesis and role in stress responses and developmental processes. In Plant Cell Death Processes; Nooden, L.D., Ed.; Elsevier/Academic Press: New York, NY, USA, 2004; pp. 143–155. [Google Scholar]
- Wasternack, C. Oxilipins: Biosynthesis, signal transduction and action. In Annual Plant Reviews, Plant Hormone Signaling; Hedden, P., Thomas, S., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 185–228. [Google Scholar]
- Deng, X.; Guo, D.; Yang, S.; Shi, M.; Chao, J.; Peng, S.; Tian, W. Jasmonate signalling in regulation of rubber biosynthesis in laticifer cells of rubber tree (Hevea brasiliensis Muell. Arg.). J. Exp. Bot. 2018, 69, 3559–3571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shaohua, W.U.; Tian, W. The secondary laticifer differentiation in rubber tree is induced by trichostatin an inhibitor of histone acetylation. Front. Agric. Sci. Eng. 2016, 4, 77–82. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Vierstra, R.D. Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Toyooka, K.; Moriyasu, Y.; Goto, Y.; Takeuchi, M.; Fukuda, H.; Matsuoka, K. Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2006, 2, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.T.; Warr, M.R.; Adelman, E.R.; Lansinger, O.M.; Flach, J.; Verovskaya, E.V. Autophagy maintains the metabolism and function of young and old stem cells. Nature 2017, 543, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Contento, A.L.; Nguyen, P.Q.; Bassham, D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007, 143, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Yoshimoto, K.; Ohsumi, Y.; Jeon, J.S.; An, G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 2009, 27, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wu, J.; Sahu, S.K.; Zeng, H.; Huang, L.; Liu, Z. Loss of alkaline ceramidase inhibits autophagy in Arabidopsis and plays an important role during environmental stress response. Plant Cell Environ. 2018, 41, 4. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.P.; Dinesh-Kumar, S.P. What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 2011, 49, 557–576. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Jingquan, Y.U. The critical role of autophagy in plant responses to abiotic stresses. Front. Agric. Sci. Eng. 2017, 4, 29–36. [Google Scholar] [CrossRef]
- Fingrut, O.; Reischer, D.; Rotem, R.; Goldin, N.; Altboum, I.; Zan-Bar, I.; Flescher, E. Jasmonates induce nonapoptotic death in high-resistance mutant p53-expressing B-lymphoma cells. Br. J. Pharmacol. 2005, 146, 800–808. [Google Scholar] [CrossRef]
- Tong, Q.S.; Jiang, G.S.; Zheng, L.D.; Tang, S.T.; Cai, J.B.; Liu, Y.; Zeng, F.Q.; Dong, J.H. Methyl jasmonate downregulates expression of proliferating cell nuclear antigen and induces apoptosis in human neuroblastoma cell lines. Anti Cancer Drug 2008, 19, 573–581. [Google Scholar] [CrossRef]
- Kniazhanski, T.; Jackman, A.; Heyfets, A.; Gonen, P.; Flescher, E.; Sherman, L. Methyl jasmonate induces cell death with mixed characteristics of apoptosis and necrosis in cervical cancer cells. Cancer Lett. 2008, 271, 34–46. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, D.; Zhang, H.; Wang, M.; Li, P.; Fang, X.A.; Cai, X. Detection of autophagy processes during the development of nonarticulated laticifers in Euphorbia kansui Liou. Planta 2018, 247, 845–861. [Google Scholar] [CrossRef]
- Fang, X.A.; Zhang, Y.; Wang, M.; Li, P.; Zhang, Q.; Si, J.J.; Wei, B.F.; Miao, Y.; Tian, L.T.; Cai, X. Lysosome and proteasome pathways are distributed in laticifers of Euphorbia helioscopia L. Physiol. Plant. 2019, 166, 1026–1038. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Honig, A.; Galili, G. Variations on a theme: Plant autophagy in comparison to yeast and mammals. Protoplasma 2012, 249, 285–299. [Google Scholar] [CrossRef]
- Ryabovol, V.V.; Minibayeva, F.V. Molecular mechanisms of autophagy in plants: Role of ATG8 proteins in formation and functioning of autophagosomes. Biochemistry (Moscow) 2016, 81, 348–363. [Google Scholar] [CrossRef]
- Obara, K.; Noda, T.; Niimi, K.; Ohsumi, Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes. Genes Cells 2010, 13, 537–547. [Google Scholar] [CrossRef]
- Farmer, E.E.; Ryan, C.A. Interplant Communication: Airborne Methyl Jasmonate Induces Synthesis of Proteinase Inhibitors in Plant Leaves. Proc. Natl. Acad. Sci. USA 1990, 87, 7713–7716. [Google Scholar] [CrossRef]
- Chen, L.R.; Chen, Y.J.; Lee, C.Y.; Lin, T.Y. Meja-induced transcriptional changes in adventitious roots of bupleurum kaoi. Plant Sci. 2007, 173, 12–24. [Google Scholar] [CrossRef]
- Soares, A.M.D.S.; Souza, T.F.D.; Jacinto, T.; Machado, O.L.T. Effect of methyl jasmonate on antioxidative enzyme activities and on the contents of ros and H2O2 in Ricinus communis leaves. J. Plant Physiol. 2010, 22, 151–158. [Google Scholar] [CrossRef]
- Gibson, S.B.; Azad, M.B.; Chen, Y. Regulation of Autophagy by Reactive Oxygen Species (ROS): Implications for Cancer Progression and Treatment. Antioxid. Redox. Signal. 2009, 2011, 777–790. [Google Scholar]
- Moore, M.N. Autophagy as a second level protective process in conferring resistance to environmentally-induced oxidative stress. Autophagy 2008, 4, 254–256. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007, 17, 422–427. [Google Scholar] [CrossRef]
- Gurusamy, N. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. J. Mol. Cell Biol. 2009, 13, 373–387. [Google Scholar] [CrossRef]
- Jain, A. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-derived bioactive peptides on inflammation and oxidative stress. J. Biomed. Biotechnol. 2014, 2014, 608979. [Google Scholar] [CrossRef]
- Fandy, T.E.; Jiemjit, A.; Thakar, M.; Rhoden, P.; Suarez, L.; Gore, S.D. Decitabine induces delayed reactive oxygen species (ROS) accumulation in leukemia cells and induces the expression of ROS generating enzymes. Clin. Cancer Res. 2014, 20, 1249–1258. [Google Scholar]
- Zhang, H.; Kong, X.; Kang, J. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol. Sci. 2009, 110, 376–388. [Google Scholar] [CrossRef]
- Li, L.L.; Zhang, Q.; Tan, J.; Fang, X. Autophagy and hippocampal neuronal injury. Sleep Breath. 2014, 18, 243–249. [Google Scholar] [CrossRef]
- Li, J.; Liu, R.; Lei, Y.L.; Wang, K.; Lau, Q.C.; Xie, N.; Zhou, S.T.; Nei, C.L.; Chen, L.J.; Wei, Y.Q.; et al. Proteomic analysis revealed association of aberrant ROS signaling with suberoylanilide hydroxamic acid-induced autophagy in Jurkat T-leukemia cells. Autophagy 2015, 6, 711–724. [Google Scholar] [CrossRef]
- Chen, Y.; Azad, M.B.; Gibson, S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009, 16, 1040–1052. [Google Scholar] [CrossRef]
- Alphalfucosidase, C.S.H. Cell surface human αl-fucosidase. FEBS J. 2010, 268, 3321–3331. [Google Scholar]
- Mohammed, W.S.; Ziganshina, E.E.; Shagimardanova, E.I.; Gogoleva, N.E.; Ziganshin, A.M. Comparison of intestinal bacterial and fungal communities across various xylophagous beetle larvae (coleoptera: Cerambycidae). Sci. Rep. 2018, 8, 10073–10082. [Google Scholar] [CrossRef]
- Lorenzo, G.D.; Velho, R.V.; Winter, D.; Thelen, M.; Ahmadi, S.; Schweizer, M. Lysosomal proteome and secretome analysis identifies missorted enzymes and their non-degraded substrates in mucolipidosis iii mouse cells. Mol. Cell. Proteom. 2018, 17, 1612–1626. [Google Scholar] [CrossRef]
- Ohto, U.; Usui, K.; Ochi, T.; Yuki, K.; Satow, Y.; Shimizu, T. Crystal structure of human β-galactosidase. J. Biol. Chem. 2012, 287, 1801–1812. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Turk, V. Lysosomal cysteine proteases: More than scavengers. Biochim. Biophys. Acta Mol. Cell Res. 2000, 1477, 98–111. [Google Scholar] [CrossRef]
- Cavalli, S.E.V.; Arribére, M.A.; Caffini, N.O.; Priolo, N.S. Morrenain b I, a papain-like endopeptidase from the latex of Morrenia brachystephana Griseb. J. Protein Chem. 2003, 22, 15–22. [Google Scholar] [CrossRef]
- Liggieri, C.; Arribére, S.A.; Trejo, F.; Canals, F.; Aviles, F.; Priolo, N. Purification and biochemical characterization of Asclepain c I from the latex of Asclepias curassavica L. Protein J. 2004, 23, 403–411. [Google Scholar] [CrossRef]
- Mathew, R.; Khor, S.; Hackett, S.R. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol. Cell. 2014, 55, 916–930. [Google Scholar] [CrossRef]
- Wang, F.X.; Luo, Y.M.; Ye, Z.Q.; Cao, X.; Liang, J.N.; Wang, Q.; Wu, Y.; Wu, J.H.; Wang, H.Y.; Zhang, M.; et al. Itraq-based proteomics analysis of autophagy-mediated immune responses against the vascular fungal pathogen verticillium dahliae in Arabidopsis. Autophagy 2018, 14, 598–618. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Yue, Z.; Liang, D.; Wang, N.; Ma, F. Isolation and characterization of mdatg18a, a wd40-repeat autophagy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. 2014, 165, 51–61. [Google Scholar] [CrossRef]
- Li, L.; Tan, J.; Miao, Y.; Lei, P.; Zhang, Q. ROS and autophagy: Interactions and molecular regulatory mechanisms. Cell Mol. Neurobiol. 2015, 35, 615–621. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Czymmek, K.; Zsolt, T.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Yao, X.; Zhang, Y.; Tian, Z.; Wang, M.; Li, P.; Cai, X. iTRAQ-Based Proteomics Analysis of Autophagy-Mediated Responses against MeJA in Laticifers of Euphorbia kansui L. Int. J. Mol. Sci. 2019, 20, 3770. https://doi.org/10.3390/ijms20153770
Fang X, Yao X, Zhang Y, Tian Z, Wang M, Li P, Cai X. iTRAQ-Based Proteomics Analysis of Autophagy-Mediated Responses against MeJA in Laticifers of Euphorbia kansui L. International Journal of Molecular Sciences. 2019; 20(15):3770. https://doi.org/10.3390/ijms20153770
Chicago/Turabian StyleFang, Xiaoai, Xiangyu Yao, Yue Zhang, Zheni Tian, Meng Wang, Peng Li, and Xia Cai. 2019. "iTRAQ-Based Proteomics Analysis of Autophagy-Mediated Responses against MeJA in Laticifers of Euphorbia kansui L." International Journal of Molecular Sciences 20, no. 15: 3770. https://doi.org/10.3390/ijms20153770
APA StyleFang, X., Yao, X., Zhang, Y., Tian, Z., Wang, M., Li, P., & Cai, X. (2019). iTRAQ-Based Proteomics Analysis of Autophagy-Mediated Responses against MeJA in Laticifers of Euphorbia kansui L. International Journal of Molecular Sciences, 20(15), 3770. https://doi.org/10.3390/ijms20153770



