Genetic Variants in the Promoter Region of the Macrophage Migration Inhibitory Factor are Associated with the Severity of Hepatitis C Virus-Induced Liver Fibrosis
Abstract
1. Introduction
2. Results
2.1. Cohort Characteristics of the HCV 2008 Cohort
2.2. Genotype Frequency of -173 G/C SNP and -794 CATT5–8 Microsatellite in the HCV 2008 Cohort
2.3. The G/C and C/C Genotypes in the -173 G/C SNP and High Microsatellite Repeat Numbers at -794 CATT5–8 Significantly Correlate with Lower Fibrosis Stages Independent of Inflammatory Activity in the HCV 2008 Cohort
2.4. Cohort Characteristics of the HCV 2018 Cohort
2.5. Genotype Frequency of -173 G/C SNP and -794 CATT5–8 Microsatellite in the HCV 2018 Cohort
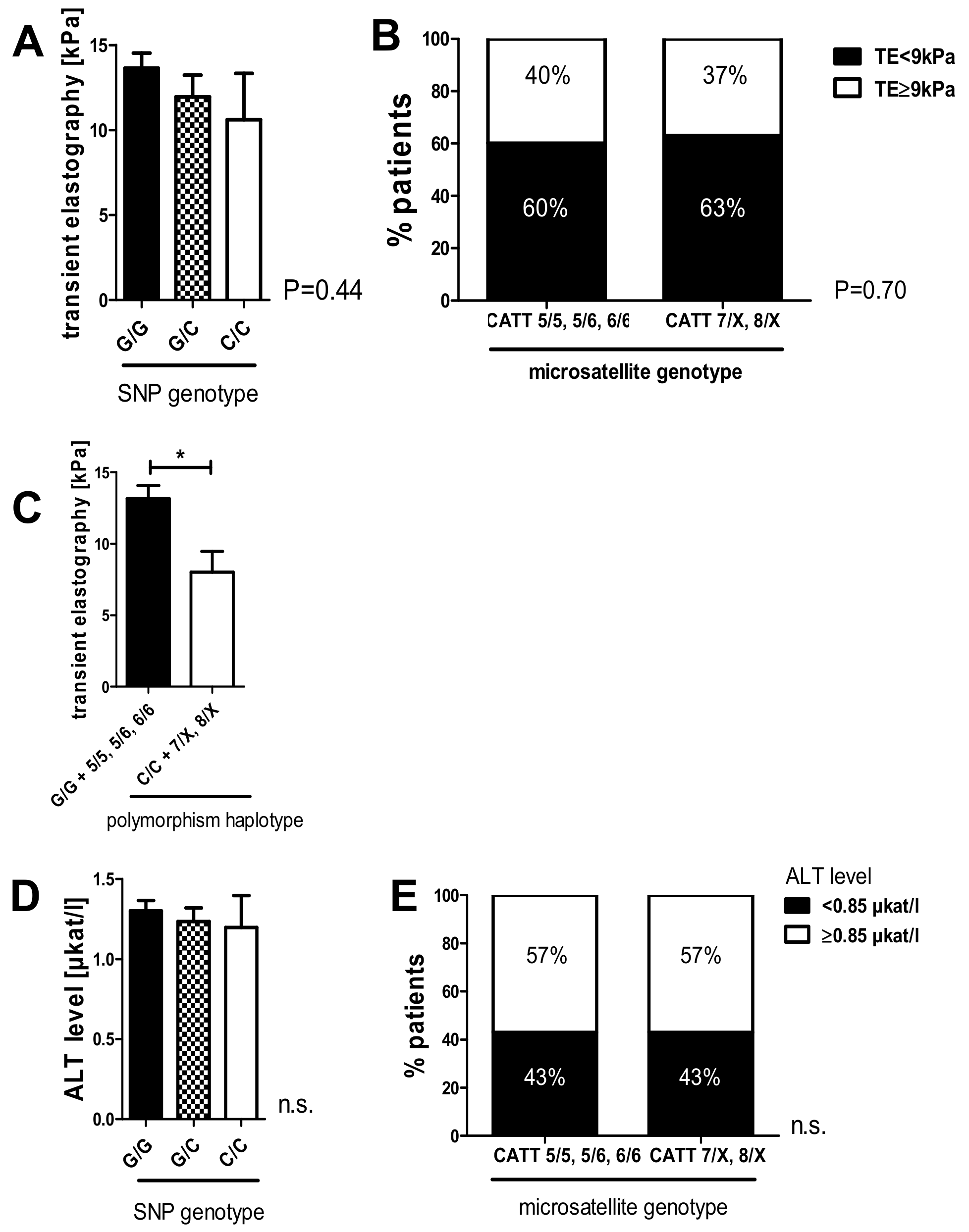
2.6. The C/C Genotype in the -173 G/C SNP and High Microsatellite Repeat Counts in the -794 CATT5–8 Showed a Tendency Toward Lower Liver Stiffness Measurements in the HCV 2018 Cohort
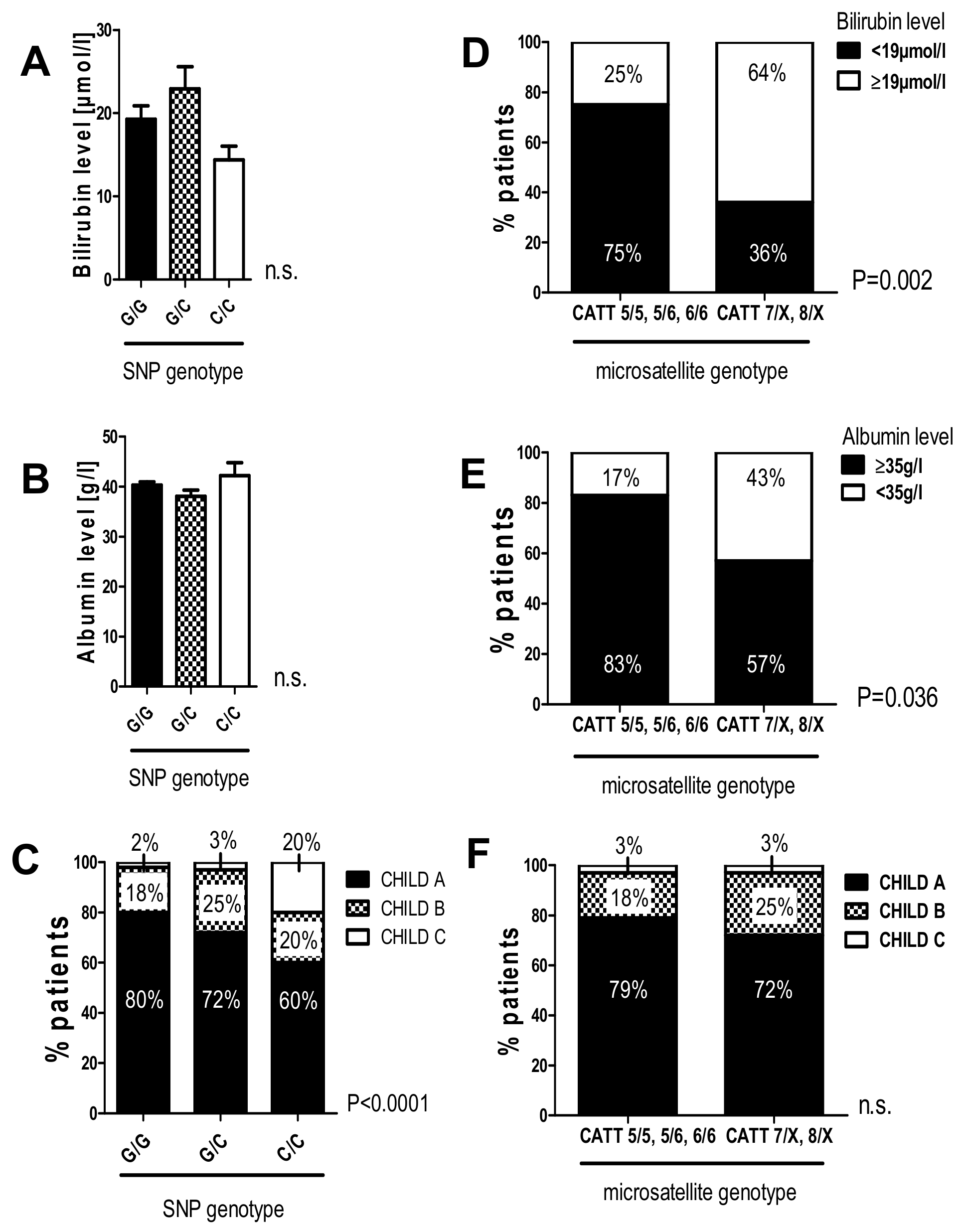
2.7. The G/C and C/C Genotypes in the -173 G/C SNP and High Microsatellite Repeat Counts in the -794 CATT5-8 Were Associated with Worse Liver Function in Patients with HCV-Induced Liver Cirrhosis
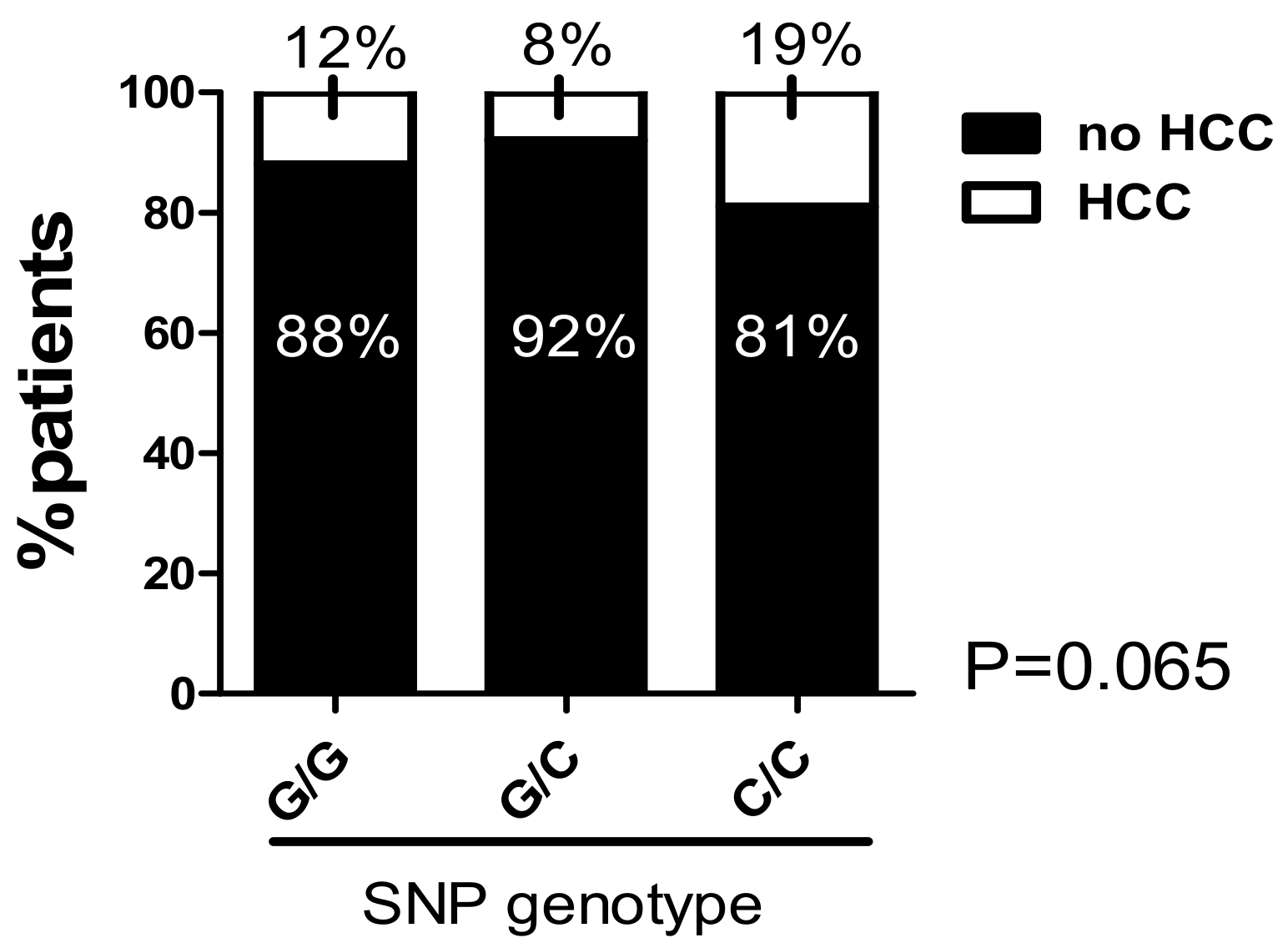
2.8. The C/C Genotype in the -173 G/C SNP and High Microsatellite Repeat Counts in the -794 CATT5–8 Were Associated with a Higher Prevalence of Hepatocellular Carcinoma
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Ethical Approval
4.3. Determination of MIF Promoter Polymorphisms -173 G/C (rs755622) and -794 CATT5–8 (rs5844572)
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MIF | Macrophage migration inhibitory factor |
| SNP | Single nucleotide polymorphism |
| HCC | Hepatocellular carcinoma |
| FS | Fibroscan® |
| DAA | Direct-acting antivirals |
| ALT | Alaninaminotransferase |
| SVR | Sustained virological response |
| AIH | Autoimmune hepatitis |
| TE | Transient elastography |
References
- Chen, S.L.; Morgan, T.R. The natural history of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 2006, 3, 47–52. [Google Scholar]
- Seeff, L.B.; Hoofnagle, J.H. National institutes of health consensus development conference statement: Management of hepatitis C: 2002—June 10–12, 2002. Hepatology 2002, 36, S3–S20. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, H.; Nalpas, B.; Poulet, B.; Carnot, F.; Zylberberg, H.; Brechot, C.; Pol, S. Hepatitis activity index is a key factor in determining the natural history of chronic hepatitis C. Hum. Pathol. 2001, 32, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Bochud, P.Y.; Cai, T.; Overbeck, K.; Bochud, M.; Dufour, J.F.; Müllhaupt, B.; Borovicka, J.; Heim, M.; Moradpour, D.; Cerny, A.; et al. Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J. Hepatol. 2009, 51, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, E.S.; Graham, C.S. Price and affordability of direct-acting antiviral regimens for hepatitis C virus in the United States. Infect. Agent. Cancer 2016, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, R.; Aghemo, A.; Rumi, M.G.; Ronchi, G.; Donato, M.F.; Paradis, V.; Colombo, M.; Bedossa, P. A morphometric and immunohistochemical study to assess the benefit of a sustained virological response in hepatitis C virus patients with cirrhosis. Hepatology 2012, 56, 532–543. [Google Scholar] [CrossRef]
- Welsch, C.; Efinger, M.; von Wagner, M.; Herrmann, E.; Zeuzem, S.; Welzel, T.M.; Lange, C.M. Ongoing liver inflammation in patients with chronic hepatitis C and sustained virological response. PLoS ONE 2017, 12, e0171755. [Google Scholar] [CrossRef]
- Rout, G.; Nayak, B.; Patel, A.H.; Gunjan, D.; Singh, V.; Kedia, S. Shalimar Therapy with oral directly acting agents in hepatitis c infection is associated with reduction in fibrosis and increase in hepatic steatosis on transient elastography. J. Clin. Exp. Hepatol. 2019, 9, 207–214. [Google Scholar] [CrossRef]
- Calandra, T.; Bernhagen, J.; Metz, C.N.; Spiegel, L.A.; Bacher, M.; Donnelly, T.; Cerami, A.; Bucala, R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature 1995, 377, 68–71. [Google Scholar] [CrossRef]
- Bacher, M.; Metz, C.N.; Calandra, T.; Mayer, K.; Chesney, J.; Lohoff, M.; Gemsa, D.; Donnelly, T.; Bucala, R. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc. Natl. Acad. Sci. USA 1996, 93, 7849–7854. [Google Scholar] [CrossRef]
- Radstake, T.R.; Sweep, F.C.; Welsing, P.; Franke, B.; Vermeulen, S.H.; Geurts-Moespot, A.; Calandra, T.; Donn, R.; van Riel, P.L. Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum. 2005, 52, 3020–3029. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.P.; Leng, L.; Feng, Z.; Liu, N.; Zhao, H.; McDonald, C.; Lee, A.; Arnett, F.C.; Gregersen, P.K.; Mayes, M.D. Macrophage migration inhibitory factor promoter polymorphisms and the clinical expression of scleroderma. Arthritis Rheum. 2006, 54, 3661–3669. [Google Scholar] [CrossRef] [PubMed]
- Nohara, H.; Okayama, N.; Inoue, N.; Koike, Y.; Fujimura, K.; Suehiro, Y.; Hamanaka, Y.; Higaki, S.; Yanai, H.; Yoshida, H.; et al. Association of the −173 G/C polymorphism of the macrophage migration inhibitory factor gene with ulcerative colitis. J. Gastroenterol. 2004, 39, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.A.; McMullen, M.R.; Roychowdhury, S.; Pisano, S.G.; Liu, X.; Stavitsky, A.B.; Bucala, R.; Nagy, L.E. Macrophage migration inhibitory factor contributes to ethanol-induced liver injury by mediating cell injury, steatohepatitis, and steatosis. Hepatology 2013, 57, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Assis, D.N.; Leng, L.; Du, X.; Zhang, C.K.; Grieb, G.; Merk, M.; Garcia, A.B.; McCrann, C.; Chapiro, J.; Meinhardt, A.; et al. The role of macrophage migration inhibitory factor in autoimmune liver disease. Hepatology 2014, 59, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Sadek, K.H.; Ezzat, S.; Abdel-Aziz, S.A.; Alaraby, H.; Mosbeh, A.; Abdel-Rahman, M.H. Macrophage Migration Inhibitory Factor (MIF) Gene Promotor Polymorphism Is Associated with Increased Fibrosis in Biliary Atresia Patients, but Not with Disease Susceptibility. Ann. Hum. Genet. 2017, 81, 177–183. [Google Scholar] [CrossRef]
- Baugh, J.A.; Chitnis, S.; Donnelly, S.C.; Monteiro, J.; Lin, X.; Plant, B.J.; Wolfe, F.; Gregersen, P.K.; Bucala, R. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002, 3, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Calandra, T.; Roger, T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef]
- De la Cruz-Mosso, U.; Bucala, R.; Palafox-Sánchez, C.A.; Parra-Rojas, I.; Padilla-Gutiérrez, J.R.; Pereira-Suárez, A.L.; Rangel-Villalobos, H.; Vázquez-Villamar, M.; Angel-Chávez, L.I.; Muñoz-Valle, J.F. Macrophage migration inhibitory factor: Association of −794 CATT5-8 and −173 G>C polymorphisms with TNF-alpha in systemic lupus erythematosus. Hum. Immunol. 2014, 75, 433–439. [Google Scholar] [CrossRef]
- Oliver, J.; Bentley, R.W.; Merriman, T.R.; Hampton, M.B.; Barclay, M.L.; Gearry, R.B.; Roberts, R.L. Association of the macrophage migration inhibitory factor gene polymorphisms with inflammatory bowel disease. World J. Gastroenterol. 2007, 56, 150–151. [Google Scholar] [CrossRef]
- Banos-Hernandez, C.J.; Navarro-Zarza, J.E.; Bucala, R.; Hernández-Bello, J.; Parra-Rojas, I.; Ramírez-Dueñas, M.G.; García-Arellano, S.; Hernández-Palma, L.A.; Machado-Sulbarán, A.C.; Muñoz-Valle, J.F. Macrophage migration inhibitory factor polymorphisms are a potential susceptibility marker in systemic sclerosis from southern Mexican population: Association with MIF mRNA expression and cytokine profile. Clin. Rheumatol. 2019, 38, 1643–1654. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.B.; Leng, L.; Beitin, A.; Chen, R.; McDonald, C.; Hsiao, B.; Jenison, R.D.; Kang, I.; Park, S.H.; Lee, A.; et al. Simultaneous detection of microsatellite repeats and SNPs in the macrophage migration inhibitory factor (MIF) gene by thin-film biosensor chips and application to rural field studies. Nucleic Acids Res. 2005, 33, e121. [Google Scholar] [CrossRef] [PubMed]
- Donn, R.; Alourfi, Z.; Zeggini, E.; Lamb, R.; Jury, F.; Lunt, M.; Meazza, C.; De Benedetti, F.; Thomson, W.; Ray, D. A functional promoter haplotype of macrophage migration inhibitory factor is linked and associated with juvenile idiopathic arthritis. Arthritis Rheum. 2004, 50, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Bedossa, P.; Poynard, T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996, 24, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Friedrich-Rust, M.; Ong, M.F.; Martens, S.; Sarrazin, C.; Bojunga, J.; Zeuzem, S.; Herrmann, E. Performance of transient elastography for the staging of liver fibrosis: A meta-analysis. Gastroenterology 2008, 134, 960–974. [Google Scholar] [CrossRef] [PubMed]
- Castera, L.; Vergniol, J.; Foucher, J.; Le Bail, B.; Chanteloup, E.; Haaser, M.; Darriet, M.; Couzigou, P.; De Lédinghen, V. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology 2005, 128, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Wang, X.Y.; Thapa, S.; Tao, L.Y.; Wu, S.Z.; Luo, G.J.; Wang, L.P.; Wang, J.N.; Wang, J.; Li, J.; et al. Macrophage migration inhibitory factor promoter polymorphisms (−794 CATT5-8): Relationship with soluble MIF levels in coronary atherosclerotic disease subjects. BMC Cardiovasc. Disord. 2017, 17, 144. [Google Scholar] [CrossRef]
- Heinrichs, D.; Knauel, M.; Offermanns, C.; Berres, M.L.; Nellen, A.; Leng, L.; Schmitz, P.; Bucala, R.; Trautwein, C.; Weber, C.; et al. Macrophage migration inhibitory factor (MIF) exerts antifibrotic effects in experimental liver fibrosis via CD74. Proc. Natl. Acad. Sci. USA 2011, 108, 17444–17449. [Google Scholar] [CrossRef]
- Wang, D.; Luo, L.; Chen, W.; Chen, L.Z.; Zeng, W.T.; Li, W.; Huang, X.H. Significance of the vascular endothelial growth factor and the macrophage migration inhibitory factor in the progression of hepatocellular carcinoma. Oncol. Rep. 2014, 31, 1199–1204. [Google Scholar] [CrossRef]
- Laleman, W.; Claria, J.; Van der Merwe, S.; Moreau, R.; Trebicka, J. Systemic Inflammation and Acute-on-Chronic Liver Failure: Too Much, Not Enough. Can. J. Gastroenterol. Hepatol. 2018, 2018, 1027152. [Google Scholar] [CrossRef]
- Trebicka, J.; Amoros, A.; Pitarch, C.; Titos, E.; Alcaraz-Quiles, J.; Schierwagen, R.; Deulofeu, C.; Fernandez-Gomez, J.; Piano, S.; Caraceni, P.; et al. Addressing Profiles of Systemic Inflammation Across the Different Clinical Phenotypes of Acutely Decompensated Cirrhosis. Front. Immunol. 2019, 10, 476. [Google Scholar] [CrossRef] [PubMed]

| n = 473 | |
|---|---|
| Age (years) | 54 (26–87) |
| Metavir Staging | n (%) |
| 5 (1) 193 (41) 168 (36) 67 (14) 40 (8) |
| Metavir Grading | n (%) |
| 8 (2) 210 (44) 227 (48) 26 (5.5) 2 (0.5) |
| SNP -173 G/C (rs755622) | Allele Frequency Difference | Linear-by-Linear Association | ||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype (n) | G/G | G/C | C/C | Freq. (C) | Odds Ratio (95% CI) | p-value | X² | p-value |
| <F2 (198) | 120 (61%) | 69 (35%) | 9 (5%) | 0.20 | 0.66 (0.48–0.93) | 0.015 | 6.07 | 0.014 |
| ≥F2 (273) | 192 (70%) | 76 (28%) | 5 (2%) | 0.16 | ||||
| Microsatellite CATT5–8 (rs5844572) | ||||
|---|---|---|---|---|
| Phenotype (n) | CATT 5/5, 5/5, 6/6 | CATT 7/X, 8/X | Odds Ratio (95% CI) | p-value |
| <F2 (197) | 138 (70%) | 59 (30%) | 0.64 (0.42–0.97) | 0.04 |
| ≥F2 (271) | 213 (79%) | 58 (21%) | ||
| n = 443 | |
| Age (years) | 52 (18–86) |
| Gender | n (%) |
| 214 (48) 229 (52) |
| Transient Elastography (Fibroscan®) | n (%) |
| 144 (38) 120 (32) 113 (30) |
| Liver Cirrhosis | n (%) |
| 290 (66) 152 (34) |
| Child–Pugh Stadium | n (%) |
| 93 (76) 24 (20) 4 (3) |
| HCC | n (%) |
| 394 (89) 49 (11) |
| Liver transplant | n (%) |
| 417 (94) 26 (6) |
| HCV Genotype | n (%) |
| 350 (83) 7 (2) 50 (12) 13 (3) |
| SNP -173 G/C (rs755622) | Allele Frequency Difference | Linear-by-Linear Association | ||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype (n) | G/G | G/C | C/C | Freq. (C) | Odds Ratio (95% CI) | p-value | X² | p-value |
| TE < 9 kPa (228) | 149 (65%) | 72 (31%) | 7 (3%) | 0.19 | 0.73 (0.49–1.08) | 0.11 | 2.49 | 0.14 |
| TE ≥ 9 kPa (149) | 110 (74%) | 35 (23%) | 4 (3%) | 0.14 | ||||
| Microsatellite CATT5–8 (rs5844572) | ||||
|---|---|---|---|---|
| Phenotype (n) | CATT 5/5, 5/5, 6/6 | CATT 7/X, 8/X | Odds Ratio (95% CI) | p-value |
| TE <9 kPa (212) | 159 (75%) | 53 (25%) | 0.89 (0.53–1.47) | 0.70 |
| TE ≥9 kPa (136) | 105 (77%) | 31 (23%) | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wirtz, T.H.; Fischer, P.; Backhaus, C.; Bergmann, I.; Brandt, E.F.; Heinrichs, D.; Koenen, M.T.; Schneider, K.M.; Eggermann, T.; Kurth, I.; et al. Genetic Variants in the Promoter Region of the Macrophage Migration Inhibitory Factor are Associated with the Severity of Hepatitis C Virus-Induced Liver Fibrosis. Int. J. Mol. Sci. 2019, 20, 3753. https://doi.org/10.3390/ijms20153753
Wirtz TH, Fischer P, Backhaus C, Bergmann I, Brandt EF, Heinrichs D, Koenen MT, Schneider KM, Eggermann T, Kurth I, et al. Genetic Variants in the Promoter Region of the Macrophage Migration Inhibitory Factor are Associated with the Severity of Hepatitis C Virus-Induced Liver Fibrosis. International Journal of Molecular Sciences. 2019; 20(15):3753. https://doi.org/10.3390/ijms20153753
Chicago/Turabian StyleWirtz, Theresa Hildegard, Petra Fischer, Christina Backhaus, Irina Bergmann, Elisa Fabiana Brandt, Daniel Heinrichs, Maria Teresa Koenen, Kai Markus Schneider, Thomas Eggermann, Ingo Kurth, and et al. 2019. "Genetic Variants in the Promoter Region of the Macrophage Migration Inhibitory Factor are Associated with the Severity of Hepatitis C Virus-Induced Liver Fibrosis" International Journal of Molecular Sciences 20, no. 15: 3753. https://doi.org/10.3390/ijms20153753
APA StyleWirtz, T. H., Fischer, P., Backhaus, C., Bergmann, I., Brandt, E. F., Heinrichs, D., Koenen, M. T., Schneider, K. M., Eggermann, T., Kurth, I., Stoppe, C., Bernhagen, J., Bruns, T., Fischer, J., Berg, T., Trautwein, C., & Berres, M.-L. (2019). Genetic Variants in the Promoter Region of the Macrophage Migration Inhibitory Factor are Associated with the Severity of Hepatitis C Virus-Induced Liver Fibrosis. International Journal of Molecular Sciences, 20(15), 3753. https://doi.org/10.3390/ijms20153753






