RETRACTED: AEG-1/miR-221 Axis Cooperatively Regulates the Progression of Hepatocellular Carcinoma by Targeting PTEN/PI3K/AKT Signaling Pathway
Abstract
1. Introduction
2. Results
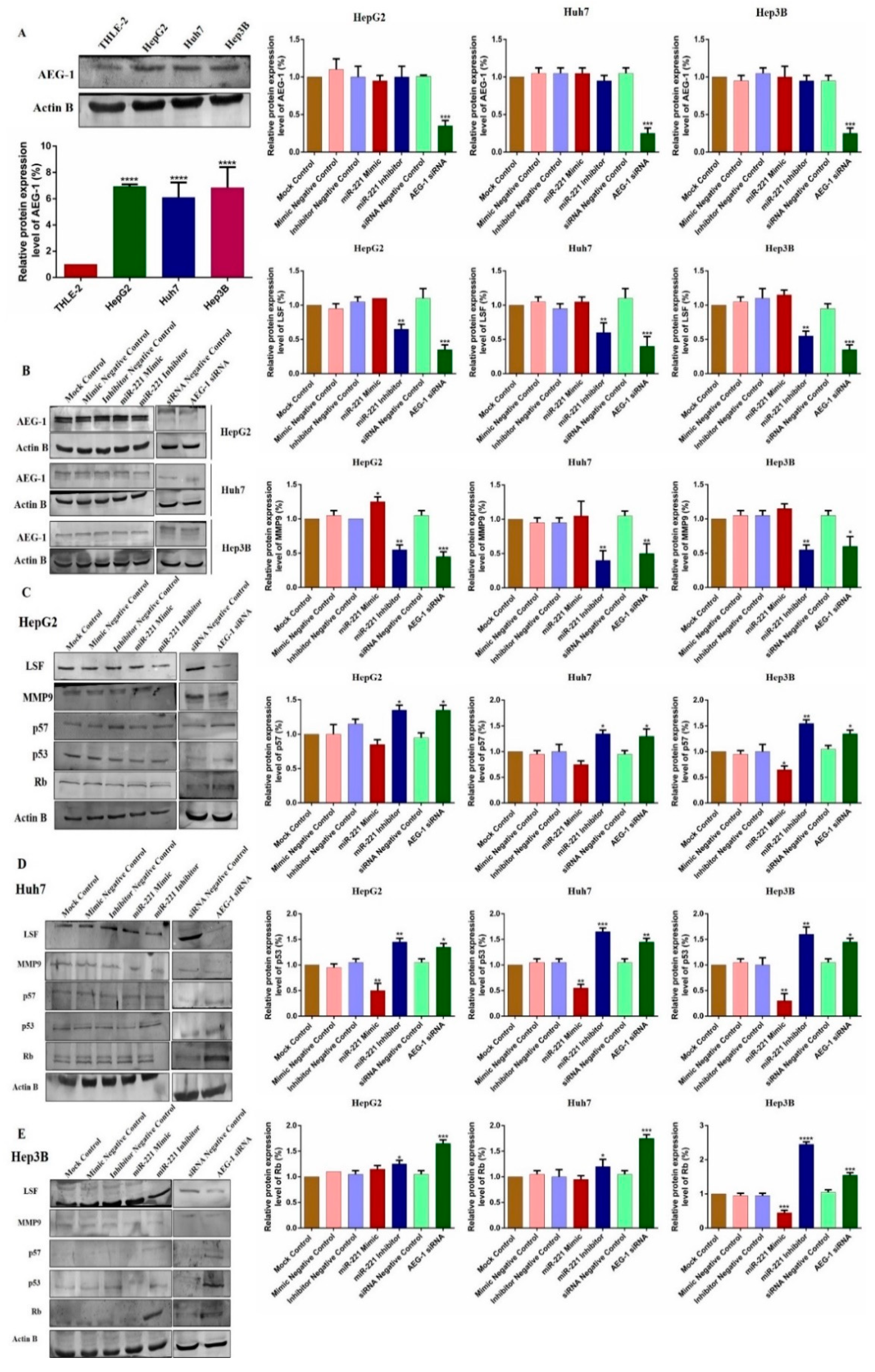
2.1. miR-221/AEG-1 Overexpressed in HCC Cell Lines
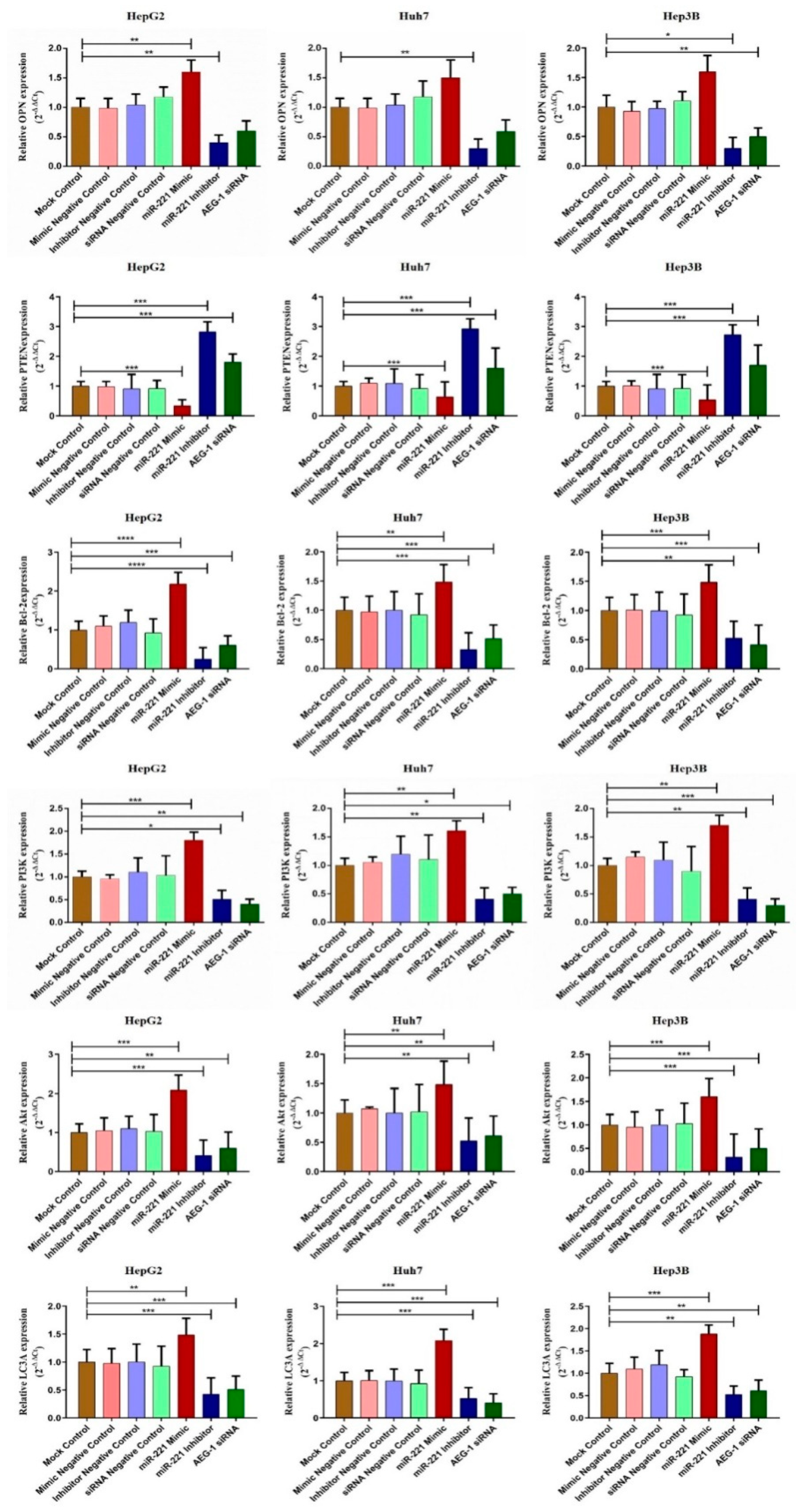
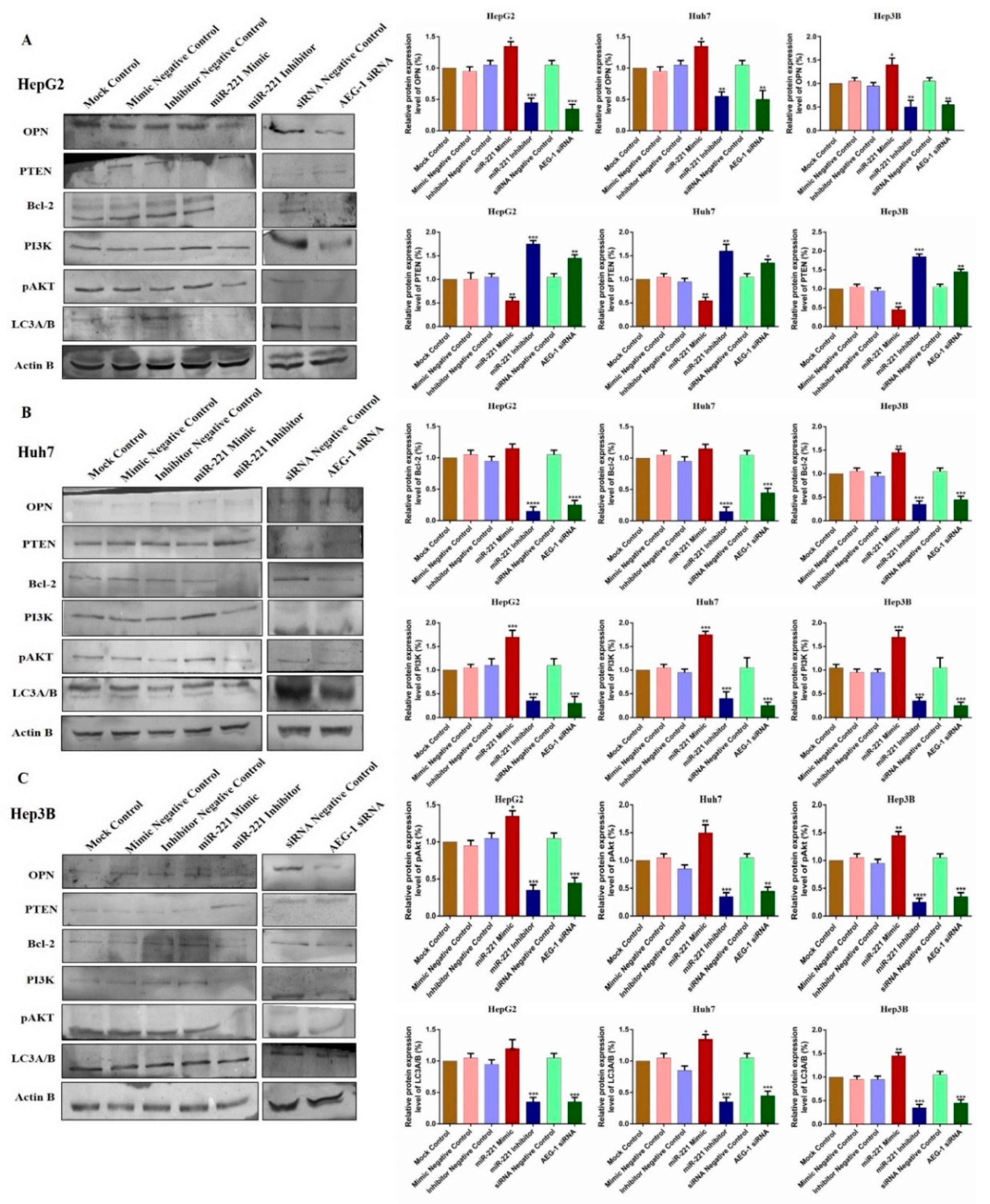
2.2. miR-221/AEG-1 Axis Regulates Apoptosis, Cell Cycle, Angiogenesis and Autophagy Mechanism by the Activation of Regulatory Genes in HCC Cells In Vitro
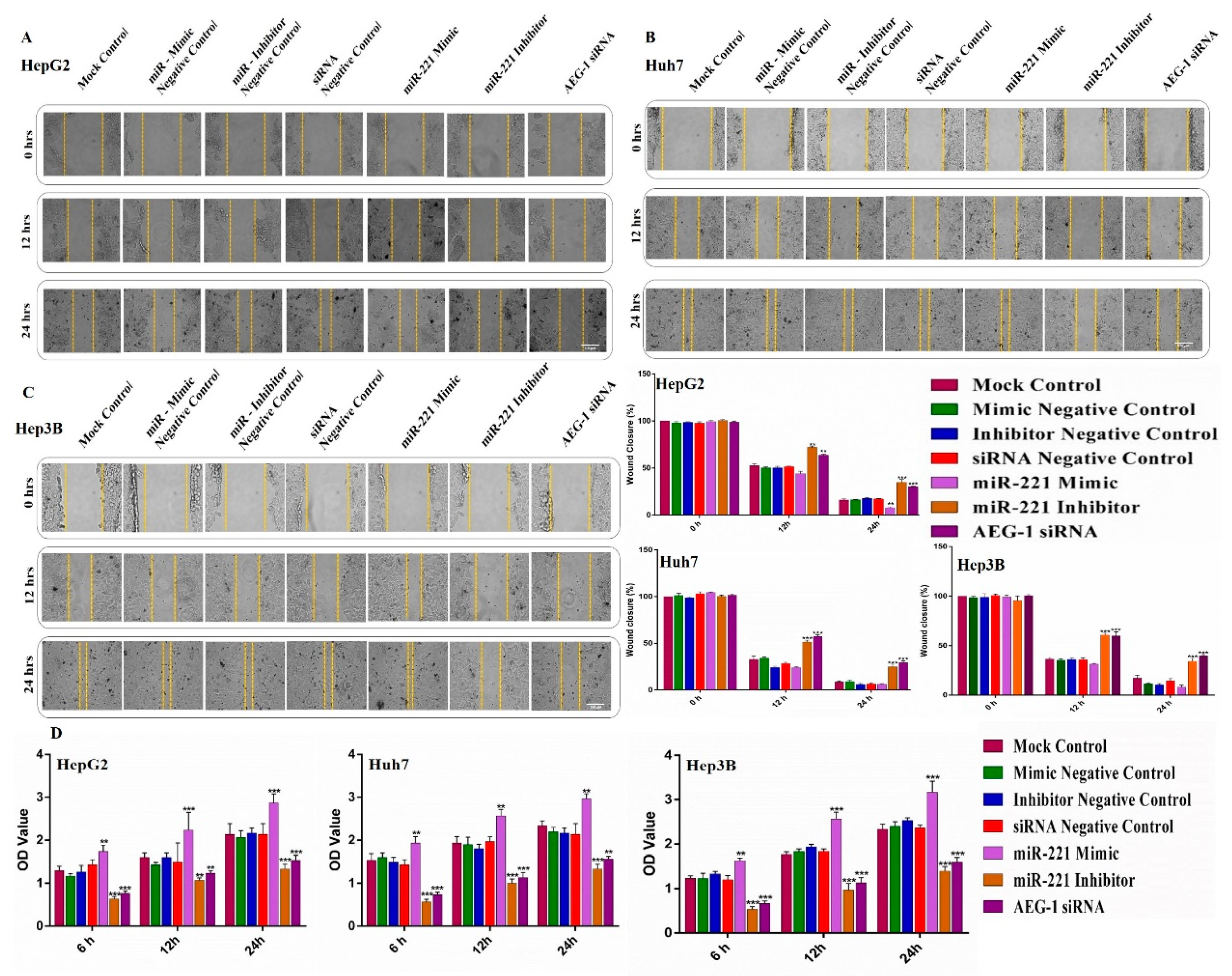
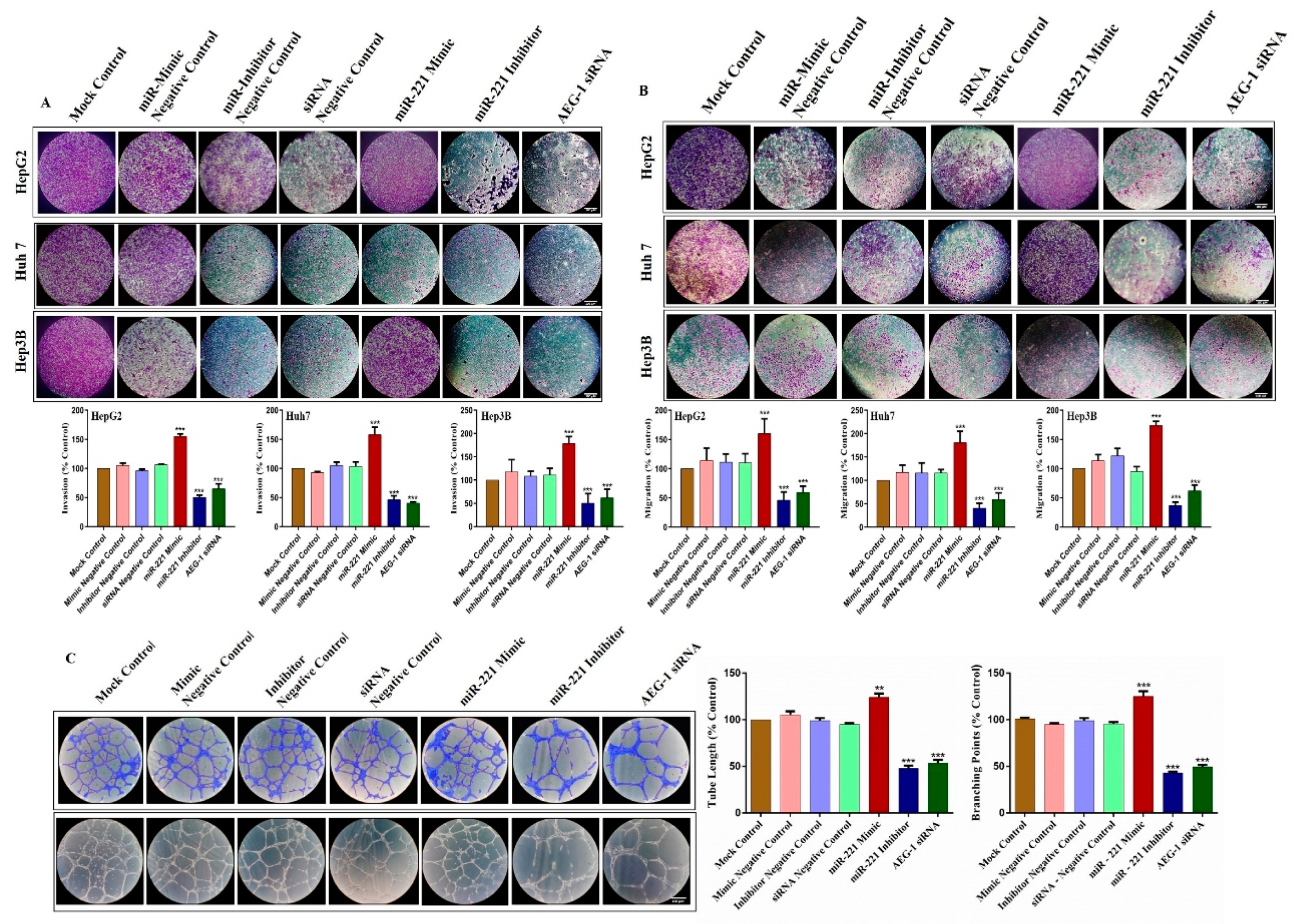
2.3. Knockdown of miR-221 and AEG-1 Inhibits Cellular Proliferation, Migration, Invasion and Tube Formation In Vitro
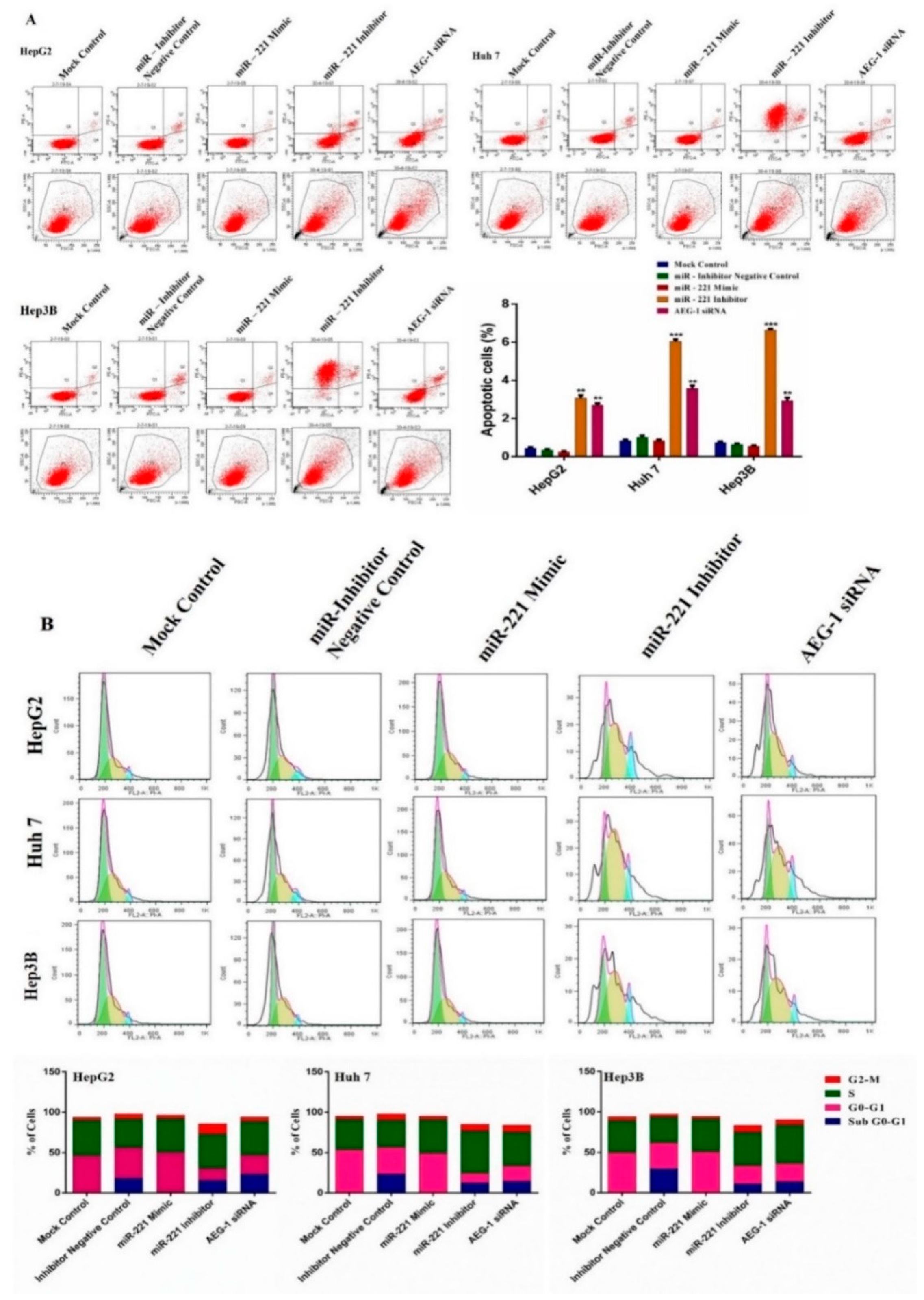
2.4. Downregulation of miR-221 and AEG-1 Promotes Apoptosis and Cell Cycle Arrest in HCC Cells In Vitro
2.5. Downregulation of miR-221 and AEG-1 Inhibits Angiogenesis, and Enhance Apoptosis, Cell Cycle Arrest by Modulating Regulatory Proteins In Vitro
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Cell Culture
4.2. miRNA and siRNA Transfection
4.3. Quantitative Real-Time PCR
4.4. Transwell Migration and Invasion Assay
4.5. Annexin V-FITC/PI Double Staining of Cell Apoptosis
4.6. Wound-Healing Assay
4.7. Propidium Iodide Staining of Cell Cycle Analysis
4.8. MTT Assay
4.9. Capillary Tube Formation Assay
4.10. Western Blotting Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mor tality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Di-Bisceglie, A.M. Hepatitis B and hepatocellular carcinoma. Hepatology 2009, 49, 56–60. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Duseja, A. Nonalcoholic Fatty Liver Disease--The Clinician’s Perspective. Trop. Gastroenterol. 2014, 35, 212–221. [Google Scholar] [CrossRef]
- Jindal, A.; Thadi, A.; Shailubhai, K. Hepatocellular carcinoma: Etiology, current and future drugs. J. Clin. Exp. Hepatology 2019, 9, 221–232. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet. 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, N.; Li, S.; Fang, J.H.; Chen, M.X.; Yang, J.; Jia, W.H.; Yuan, Y.; Zhuang, S.M. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology 2013, 58, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Kang, N.; Chen, Y.; Liu, L.; Zhang, Y. Oncogene miR-106a promotes proliferation and metastasis of prostate cancer cells by directly targeting PTEN in vivo and in vitro. Minerva. Med. 2018, 109, 24–30. [Google Scholar]
- Xue, Y.; Xu, W.; Zhao, W.; Wang, W.; Zhang, D.; Wu, P. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed. Pharmacother. 2017, 86, 426–433. [Google Scholar] [CrossRef]
- Jayamohan, S.; Kannan, M.; Moorthi, R.K.; Rajasekaran, N.; Jung, H.S.; Shin, Y.K.; Arockiam, A.J. Dysregulation of miR-375/AEG-1 axis by Human Papillomavirus 16/18-E6/E7 Promotes Cellular Proliferation, Migration and Invasion in Cervical Cancer. Front. Oncol. 2019, 9, 847. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.C.; Wolff, R.K.; Stevens, J.R.; Samowitz, W.S.; Herrick, J.S. The PI3K/AKT signaling pathway: Associations of miRNAs with dysregulated gene expression in colorectal cancer. Mol. Carcinog. 2018, 57, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Amini-Farsani, Z.; Sangtarash, M.H.; Shamsara, M.; Teimori, H. MiR-221/222 promote chemoresistance to cisplatin in ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway. Cytotechnology 2018, 70, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Liu, J.; Tycksen, E.; Nunley, R.; McAlinden, A. MicroRNA-181a/b-1 over-expression enhances osteogenesis by modulating PTEN/PI3K/AKT signaling and mitochondrial metabolism. Bone 2019, 123, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Moya, J.; Wert-Lamas, L.; Santisteban, P. MicroRNA-146b promotes PI3K/AKT pathway hyperactivation and thyroid cancer progression by targeting PTEN. Oncogene 2018, 37, 3369–3383. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhou, J.; Dong, M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019, 39, BSR20190249. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Xie, D.; Yuan, P.; Wang, D.; Jin, H.; Chen, H. Expression and prognostic significance of miR-375 and miR-221 in liver cancer. Oncol. Lett. 2017, 14, 2305–2309. [Google Scholar] [CrossRef]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef]
- Britt, D.E.; Yang, D.F.; Yang, D.Q.; Flanagan, D.; Callanan, H.; Lim, Y.P.; Lin, S.H.; Hixson, D.C. Identification of a novel protein, LYRIC, localized to tight junctions of polarized epithelial cells. Exp. Cell. Res. 2004, 300, 134–148. [Google Scholar] [CrossRef]
- Kang, D.C.; Su, Z.Z.; Sarkar, D.; Emdad, L.; Volsky, D.J.; Fisher, P.B. Cloning and characterization of HIV-1- inducible astrocyte elevated gene-1, AEG-1. Gene 2005, 353, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Reghupaty, S.C.; Sarkar, D. Current Status of Gene Therapy in Hepatocellular Carcinoma. Cancers 2019, 11, 1265. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Emdad, L.; Bacolod, M.D.; Kegelman, T.P.; Shen, X.N.; Alzubi, M.A.; Das, S.K.; Sarkar, D.; Fisher, P.B. Astrocyte elevated gene-1 interacts with Akt isoform 2 to control glioma growth, survival, and pathogenesis. Cancer Res. 2014, 74, 7321–7332. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.K.; Santhekadur, P.K.; Gredler, R.; Chen, D.; Emdad, L.; Bhutia, S.; Pannell, L.; Fisher, P.B.; Sarkar, D. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 2011, 53, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, M.; Li, F.; Qian, L.; Zhang, P.; Lv, F.; Cheng, W.; Hou, R. MiR-221 Promotes Hepatocellular Carcinoma Cells Migration via Targeting PHF2. Biomed. Res. Int. 2019, 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, B.; Ding, M.; Lu, S.; Zhou, H.; Sun, D.; Wu, G.; Gan, X. C1QTNF1-AS1 regulates the occurrence and development of hepatocellular carcinoma by regulating miR-221-3p/SOCS3. Hepatol. Int. 2019, 13, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.H.; Chen, H.Y.; He, Z.L.; Lv, Z.; Xu, X.F.; Qian, Y.G.; Zheng, S.S. Silencing of microRNA-375 affects immune function in mice with liver failure by upregulating astrocyte elevated gene-1 through reducing apoptosis of Kupffer cells. J. Cell. Biochem. 2019, 120, 253–263. [Google Scholar] [CrossRef]
- He, X.X.; Chang, Y.; Meng, F.Y.; Wang, M.Y.; Xie, Q.H.; Tang, F.; Li, P.Y.; Song, Y.H.; Lin, J.S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012, 31, 3357–3369. [Google Scholar] [CrossRef]
- Yan, J.J.; Chang, Y.; Zhang, Y.N.; Lin, J.S.; He, X.X.; Huang, H.J. miR-195 inhibits cell proliferation via targeting AEG-1 in hepatocellular carcinoma. Oncol. Lett. 2017, 13, 3118–3126. [Google Scholar] [CrossRef]
- Yarchoan, M.; Johnson, B.A.; Lutz, E.R.; Laheru, D.A.; Jaffee, E.M. Targeting neoantigens to augment antitumor immunity. Nat. Rev. Cancer 2017, 17, 209. [Google Scholar]
- Liu, S.; Koh, S.S.; Lee, C.G. Hepatitis B Virus X Protein and Hepatocarcinogenesis. Int. J. Mol. Sci. 2016, 17, 940. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Tang, Y.S.; Huang, S.F.; Ai, J.G.; Wang, H.X.; Zhang, L.P. Hbx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBv related hepatocellular carcinoma by targeting estrogen receptor-α. Oncol. Rep. 2015, 33, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, W.; Huang, Y.; Wu, J.; Chen, M.; Cui, P.; Zhang, W.; Zhang, Y. HBx elevates oncoprotein AEG-1 expression to promote cell migration by downregulating miR-375 and miR-136 in malignant hepatocytes. DNA Cell Biol. 2014, 33, 715–722. [Google Scholar] [CrossRef]
- Brown, D.M.; Ruoslahti, E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 2004, 5, 365–374. [Google Scholar] [CrossRef]
- Santhekadur, P.K.; Das, S.K.; Gredler, R.; Chen, D.; Srivastava, J.; Robertson, C.; Baldwin, A.S.; Fisher, P.B.; Sarkar, D. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J. Biol. Chem. 2012, 287, 13952–13958. [Google Scholar] [CrossRef]
- Caixia, H.; Shichang, C.; Zheng, J.; Yin, T.; Junsheng, L.; Long, J.; Zhang, W.; Wang, X.; Sheng, S.; Cong, L.; et al. MiR-875-5p inhibits hepatocellular carcinoma cell proliferation and migration by repressing astrocyte elevated gene-1 (AEG-1) expression. Transl. Cancer Res. 2018, 7, 158–169. [Google Scholar]
- Kim, H.; Huang, W.; Jiang, X.; Pennicooke, B.; Park, P.J.; Johnson, M.D. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc. Natl. Acad. Sci. USA. 2009, 107, 2183–2188. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.K.; Emdad, L.; Gredler, R.; Fuller, C.; Dumur, C.I.; Jones, K.H.; Jackson-Cook, C.; Su, Z.Z.; Chen, D.; Sarkar, D.; et al. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2010, 107, 8357–8362. [Google Scholar] [CrossRef]
- Yoo, B.K.; Gredler, R.; Chen, D.; Santhekadur, P.K.; Fisher, P.B.; Sarkar, D. c-Met activation through a novel pathway involving osteopontin mediates oncogenesis by the transcription factor LSF. J. Hepatol. 2011, 55, 1317–1324. [Google Scholar] [CrossRef]
- Santhekadur, P.K.; Gredler, R.; Chen, D.; Siddiq, A.; Shen, X.N.; Das, S.K.; Emdad, L.; Fisher, P.B.; Sarkar, D. Late SV40 factor (LSF) enhances angiogenesis by transcriptionally up-regulating matrix metalloproteinase-9 (MMP-9). J. Biol. Chem. 2012, 287, 3425–3432. [Google Scholar] [CrossRef]
- Yoo, B.K.; Emdad, L.; Su, Z.Z.; Villanueva, A.; Chiang, D.Y.; Mukhopadhyay, N.D.; Mills, A.S.; Waxman, S.; Fisher, R.A.; Sarkar, D.; et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J. Clin. Invest. 2009, 119, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Dai, J.; Zhang, H.; Ge, Z. MicroRNA-221 promotes cell proliferation, migration, and differentiation by regulation of ZFPM2 in osteoblasts. Braz. J. Med. Biol. Res. 2018, 51, e7574. [Google Scholar]
- Huang, L.L.; Wang, Z.; Cao, C.J.; Ke, Z.F.; Wang, F.; Wang, R.; Luo, C.Q.; Lu, X.; Wang, L.T. AEG-1 associates with metastasis in papillary thyroid cancer through upregulation of MMP2/9. Int. J. Oncol. 2017, 51, 812–822. [Google Scholar] [CrossRef]
- Xu, Q.; Li, P.; Chen, X.; Zong, L.; Jiang, Z.; Nan, L.; Lei, J.; Duan, W.; Zhang, D.; Wang, Z.; et al. miR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget 2015, 6, 14153–14164. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhu, C.; Xu, Y.; Qian, W.; Zheng, M. Negative Regulation of PTEN by MicroRNA-221 and Its Association with Drug Resistance and Cellular Senescence in Lung Cancer Cells. Biomed. Res. Int. 2018, 2018, 7908950. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Negrini, M.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ding, J.; Yang, J.; Guo, X.; Zheng, Y. MicroRNA Roles in the Nuclear Factor Kappa B Signaling Pathway in Cancer. Front. Immunol. 2018, 9, 546. [Google Scholar] [CrossRef]
- Zhao, D.; Zhuang, N.; Ding, Y.; Kang, Y.; Shi, L. miR-221 activates the NF-kB pathway by targeting A20. Biochm. Biophys. Res. Comm. 2016, 472, 11–18. [Google Scholar] [CrossRef]
- Emdad, L.; Sarkar, D.; Su, Z.; Randolph, A.; Boukerche, H.; Fisher, P.B. Activation of the Nuclear Factor KB Pathway by Astrocyte Elevated Gene-1: Implications for Tumor Progression and Metastasis. Cancer Res. 2006, 66, 1509–1516. [Google Scholar] [CrossRef]
- Zhang, W.; Kone, B.C. NF-kappaB inhibits transcription of the H(+)-K(+)-ATPase alpha(2)-subunit gene: Role of histone deacetylases. Am. J. Physiol. Renal. Physiol. 2002, 283, 904–911. [Google Scholar] [CrossRef]
- Catz, S.D.; Johnson, J.L. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer. Oncogene 2001, 20, 7342–7351. [Google Scholar] [CrossRef]
- Zhou, Z.; Deng, H.; Yan, W.; Luo, M.; Tu, W.; Xia, Y.; He, J.; Han, P.; Fu, Y.; Tian, D. AEG-1 promotes anoikis resistance and orientation chemotaxis in hepatocellular carcinoma cells. PLoS ONE 2014, 9, e100372. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Wang, Y.; Zhang, X.; Lang, B.; Zhou, X.; Xu, X.; Zeng, T.; Liu, W.; Zhang, X.; Wang, G.; et al. MiR-221-induced PUMA silencing mediates immune evasion of bladder cancer cells. Int. J. Oncol. 2015, 46, 1169–1180. [Google Scholar] [CrossRef]
- Wang, S.Q.; Wang, C.; Chang, L.M.; Zhou, K.R.; Wang, J.W.; Ke, Y.; Yang, D.X.; Shi, H.G.; Wang, R.; Liu, H.M.; et al. Geridonin and paclitaxel act synergistically to inhibit the proliferation of gastric cancer cells through ROS-mediated regulation of the PTEN/PI3K/Akt pathway. Oncotarget 2016, 7, 72990–73002. [Google Scholar] [CrossRef]
- Paramio, J.M.; Navarro, M.; Segrelles, C.; Gómez-Casero, E.; Jorcano, J.L. PTEN tumour suppressor is linked to the cell cycle control through the retinoblastoma protein. Oncogene 1999, 18, 7462–7468. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Ding, D.; Yan, Y.; Li, H.; Wang, B.; Ma, L.; Ye, Z.; Ma, T.; Wu, Q.; Huang, H.; et al. Phosphorylated RB Promotes Cancer Immunity by Inhibiting NF-κB Activation and PD-L1 Expression. Mol. Cell. 2019, 73, 22–35. [Google Scholar] [CrossRef]
- Peng, X.; Wang, Y.; Li, H.; Fan, J.; Shen, J.; Yu, X.; Zhou, Y.; Mao, H. ATG5-mediated autophagy suppresses NF-κB signaling to limit epithelial inflammatory response to kidney injury. Cell Death Dis. 2019, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Jeyamohan, S.; Moorthy, R.K.; Kannan, M.K.; Arockiam, A.J. Parthenolide induces apoptosis and autophagy through the suppression of PI3K/Akt signaling pathway in cervical cancer. Biotech. Lett. 2016, 38, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Galluzzi, L.; Morselli, E.; Kepp, O.; Malik, S.A.; Kroemer, G. Autophagy regulation by p53. Curr. Opin. Cell. Biol. 2010, 22, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.F.; Sebti, S.; Wei, Y.; Zou, Z.; Shi, M.; McMillan, K.L.; He, C.; Ting, T.; Liu, Y.; Levine, B.; et al. Disruption of the beclin 1/Bcl-2 autophagy regulatory complex promotes longevity in mice. Nature 2018, 558, 136–140. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kannan, M.; Jayamohan, S.; Moorthy, R.K.; Chabattula, S.C.; Ganeshan, M.; Arockiam, A.J.V. RETRACTED: AEG-1/miR-221 Axis Cooperatively Regulates the Progression of Hepatocellular Carcinoma by Targeting PTEN/PI3K/AKT Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 5526. https://doi.org/10.3390/ijms20225526
Kannan M, Jayamohan S, Moorthy RK, Chabattula SC, Ganeshan M, Arockiam AJV. RETRACTED: AEG-1/miR-221 Axis Cooperatively Regulates the Progression of Hepatocellular Carcinoma by Targeting PTEN/PI3K/AKT Signaling Pathway. International Journal of Molecular Sciences. 2019; 20(22):5526. https://doi.org/10.3390/ijms20225526
Chicago/Turabian StyleKannan, Maheshkumar, Sridharan Jayamohan, Rajesh Kannan Moorthy, Siva Chander Chabattula, Mathan Ganeshan, and Antony Joseph Velanganni Arockiam. 2019. "RETRACTED: AEG-1/miR-221 Axis Cooperatively Regulates the Progression of Hepatocellular Carcinoma by Targeting PTEN/PI3K/AKT Signaling Pathway" International Journal of Molecular Sciences 20, no. 22: 5526. https://doi.org/10.3390/ijms20225526
APA StyleKannan, M., Jayamohan, S., Moorthy, R. K., Chabattula, S. C., Ganeshan, M., & Arockiam, A. J. V. (2019). RETRACTED: AEG-1/miR-221 Axis Cooperatively Regulates the Progression of Hepatocellular Carcinoma by Targeting PTEN/PI3K/AKT Signaling Pathway. International Journal of Molecular Sciences, 20(22), 5526. https://doi.org/10.3390/ijms20225526




