Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Clonogenic Assays
4.3. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NSCLC | non-small cell lung carcinoma |
| LQ model | linear quadratic model |
| MID | mean inactivation dose |
| DNA-PKcs | DNA-dependent protein kinase, catalytic subunit |
| NHEJ | non-homologous end joining |
References
- Okunieff, P.; Morgan, D.; Niemierko, A.; Suit, H.D. Radiation dose-response of human tumors. Int. J. Radiat. Oncol. Biol. Phys. 1995, 32, 1227–1237. [Google Scholar] [CrossRef]
- Hinkson, I.V.; Davidsen, T.M.; Klemm, J.D.; Kerlavage, A.R.; Kibbe, W.A. A Comprehensive Infrastructure for Big Data in Cancer Research: Accelerating Cancer Research and Precision Medicine. Front. Cell Dev. Biol. 2017, 2, 83. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.K.; Chauhan, L.; Shin, M.; Loken, M.R.; Pollard, J.A.; Wang, Y.C.; Ries, R.E.; Aplenc, R.; Hirsch, B.A.; Raimondi, S.C.; et al. CD33 Splicing Polymorphism Determines Gemtuzumab Ozogamicin Response in De Novo Acute Myeloid Leukemia: Report from Randomized Phase III Children’s Oncology Group Trial AAML0531. J. Clin. Oncol. 2017, 35, 2674–2682. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.H.; DuBois, S.G.; Glade Bender, J.L.; Kim, A.; Crompton, B.D.; Parker, E.; Dumont, I.P.; Hong, A.L.; Guo, D.; Church, A.; et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol. 2016, 2, 608–615. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 2, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Fertil, B.; Malaise, E.P. Inherent cellular radiosensitivity as a basic concept for human tumor radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 1981, 7, 621–629. [Google Scholar] [CrossRef]
- Deacon, J.; Peckham, M.J.; Steel, G.G. The radioresponsiveness of human tumours and the initial slope of the cell survival curve. Radiother. Oncol. 1984, 2, 317–323. [Google Scholar] [CrossRef]
- Fertil, B.; Dertinger, H.; Courdi, A.; Malaise, E.P. Mean inactivation dose: A useful concept for intercomparison of human cell survival curves. Radiat. Res. 1984, 99, 73–84. [Google Scholar] [CrossRef]
- Fertil, B.; Malaise, E.P. Intrinsic radiosensitivity of human cell lines is correlated with radioresponsiveness of human tumors: Analysis of 101 published survival curves. Int. J. Radiat. Oncol. Biol. Phys. 1985, 11, 1699–1707. [Google Scholar] [CrossRef]
- Steel, G.G. The radiobiology of tumours. In Basic Clinical Radiobiology, 3rd ed.; Steel, G.G., Ed.; CRC Press: Boca Raton, FL, USA, 2002; pp. 182–191. [Google Scholar]
- Yagishita, S.; Horinouchi, H.; Taniyama, K.T.; Nakamichi, S.; Kitazono, S.; Mizugaki, H.; Kanda, S.; Fujiwara, Y.; Nokihara, H.; Yamamoto, N.; et al. Epidermal growth factor receptor mutation is associated with longer local control after definitive chemoradiotherapy in patients with stage III nonsquamous non-small-cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2015, 91, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Amornwichet, N.; Oike, T.; Shibata, A.; Nirodi, C.S.; Ogiwara, H.; Makino, H.; Kimura, Y.; Hirota, Y.; Isono, M.; Yoshida, Y.; et al. The EGFR mutation status affects the relative biological effectiveness of carbon-ion beams in non-small cell lung carcinoma cells. Sci. Rep. 2015, 5, 11305. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Chen, B.P.; Story, M.D.; Sato, M.; Minna, J.D.; Chen, D.J.; Nirodi, C.S. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007, 67, 5267–5274. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, K.; Mayer, C.; Fehrenbacher, B.; Schaller, M.; Raju, U.; Milas, L.; Chen, D.J.; Kehlbach, R.; Rodemann, H.P. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J. Biol. Chem. 2005, 280, 31182–31189. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.M.; Harari, P.M. Modulation of radiation response after epidermal growth factor receptor blockade in squamous cell carcinomas: Inhibition of damage repair, cell cycle kinetics, and tumor angiogenesis. Clin. Cancer Res. 2000, 6, 2166–2174. [Google Scholar]
- Strom, T.; Harrison, L.B.; Giuliano, A.R.; Schell, M.J.; Eschrich, S.A.; Berglund, A.; Fulp, W.; Thapa, R.; Coppola, D.; Kim, S.; et al. Tumour radiosensitivity is associated with immune activation in solid tumours. Eur. J. Cancer 2017, 84, 304–314. [Google Scholar] [CrossRef]
- Torres-Roca, J.F.; Fulp, W.J.; Caudell, J.J.; Servant, N.; Bollet, M.A.; van de Vijver, M.; Naghavi, A.O.; Harris, E.E.; Eschrich, S.A. Integration of a radiosensitivity molecular signature into the assessment of local recurrence risk in breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 631–638. [Google Scholar] [CrossRef]
- Ahmed, K.A.; Chinnaiyan, P.; Fulp, W.J.; Eschrich, S.; Torres-Roca, J.F.; Caudell, J.J. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget 2015, 6, 34414–34422. [Google Scholar] [CrossRef]
- Strom, T.; Hoffe, S.E.; Fulp, W.; Frakes, J.; Coppola, D.; Springett, G.M.; Malafa, M.P.; Harris, C.L.; Eschrich, S.A.; Torres-Roca, J.F.; et al. Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother. Oncol. 2015, 117, 159–164. [Google Scholar] [CrossRef]
- Nuryadi, E.; Permata, T.B.M.; Komatsu, S.; Oike, T.; Nakano, T. Inter-assay presicion of clonogenic assay for radiosensitivity in cancer cell line A549. Oncotarget 2018, 9, 13706–13712. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Iwakawa, R.; Tang, M.; Kohno, T.; Angulo, B.; Pio, R.; Montuenga, L.M.; Minna, J.D.; Yokota, J.; Sanchez-Cespedes, M. A gene-alteration profile of human lung cancer cell lines. Hum. Mutat. 2009, 30, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, B.A.; Raben, D.; Varella-Garcia, M.; Gustafson, D.; Chan, D.C.; Bemis, L.; Coldren, C.; Barón, A.; Zeng, C.; Franklin, W.A.; et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin. Cancer Res. 2006, 12, 7117–7125. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Yokote, H.; Murakami-Murofushi, K.; Tamura, T.; Saijo, N.; Nishio, K. Pertuzumab, a novel HER dimerization inhibitor.; inhibits the growth of human lung cancer cells mediated by the HER3 signaling pathway. Cancer Sci. 2007, 98, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, A.; Yamamoto, C.; Nakashima, K.; Azuma, K.; Hattori, S.; Kashihara, M.; Aizawa, H.; Basaki, Y.; Kuwano, M.; Kage, M.; et al. Molecular diagnosis of activating EGFR mutations in non-small cell lung cancer using mutation-specific antibodies for immunohistochemical analysis. Clin. Cancer Res. 2010, 16, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Spoerke, J.M.; O’Brien, C.; Huw, L.; Koeppen, H.; Fridlyand, J.; Brachmann, R.K.; Haverty, P.M.; Pandita, A.; Mohan, S.; Sampath, D.; et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin. Cancer Res. 2012, 18, 6771–6783. [Google Scholar] [CrossRef] [PubMed]
- Oike, T.; Ogiwara, H.; Tominaga, Y.; Ito, K.; Ando, O.; Tsuta, K.; Mizukami, T.; Shimada, Y.; Isomura, H.; Komachi, M.; et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013, 73, 5508–5518. [Google Scholar] [CrossRef] [PubMed]
- Oike, T.; Ogiwara, H.; Torikai, K.; Nakano, T.; Yokota, J.; Kohno, T. Garcinol, a histone acetyltransferase inhibitor, radiosensitizes cancer cells by inhibiting non-homologous end joining. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, 815–821. [Google Scholar] [CrossRef]
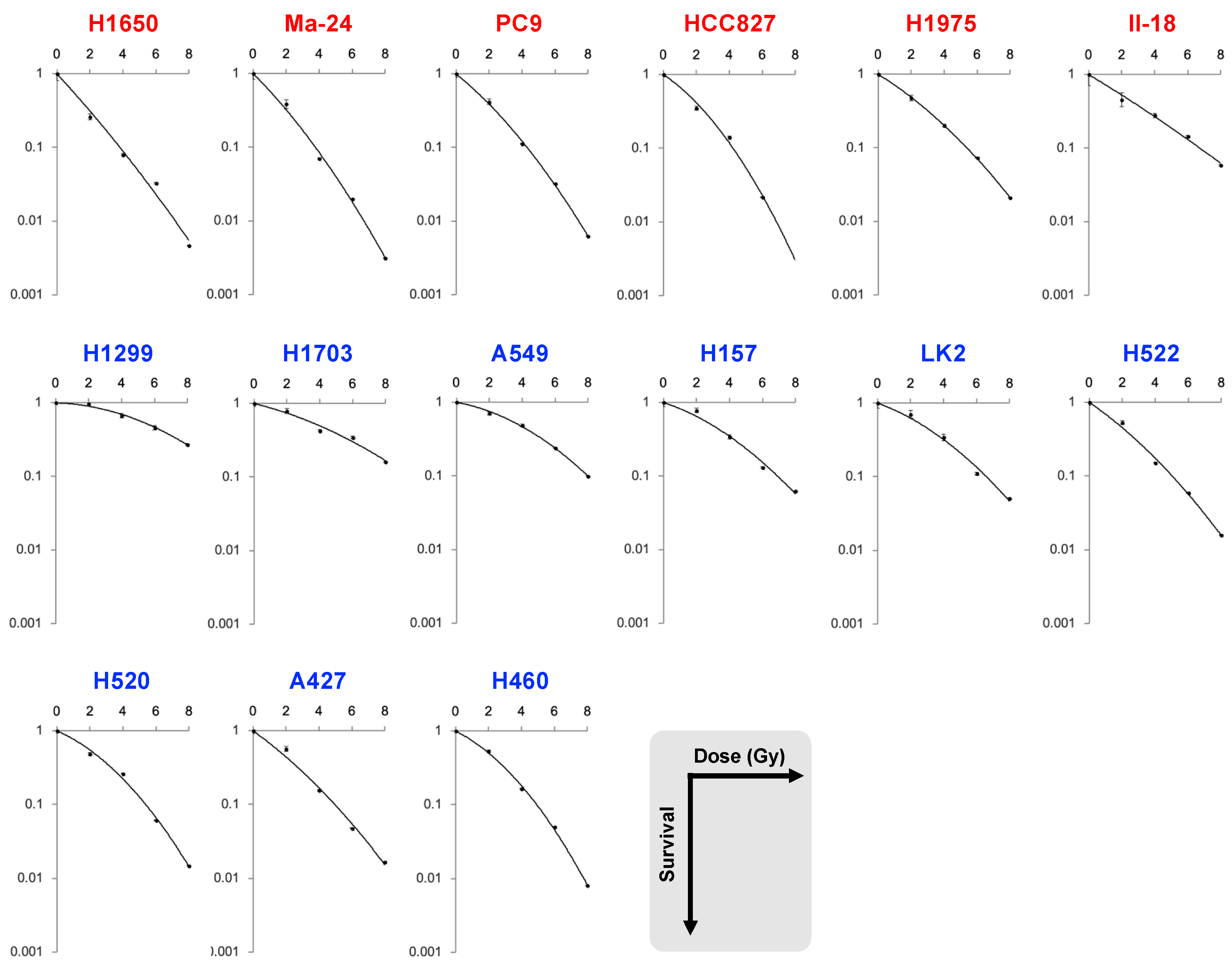
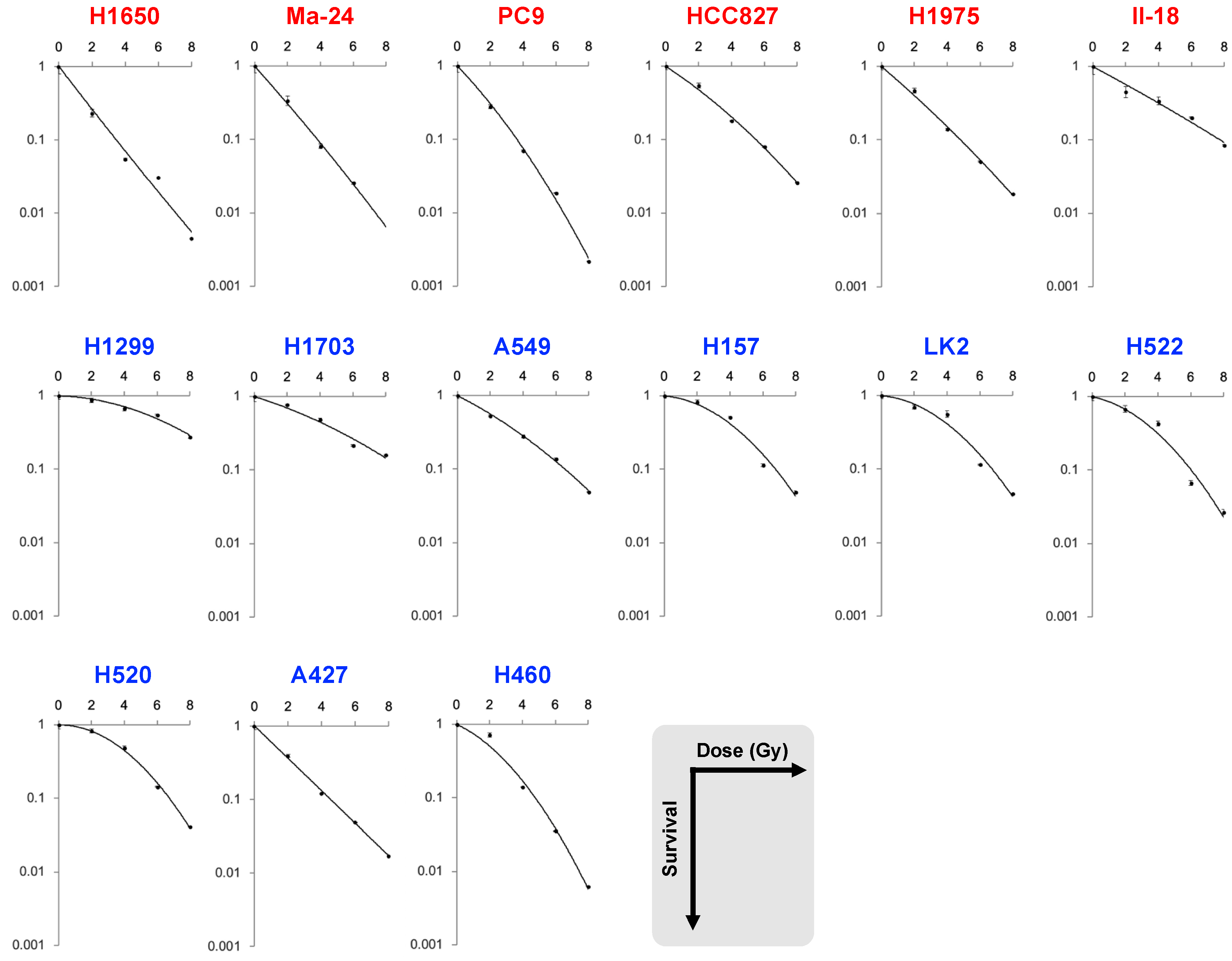
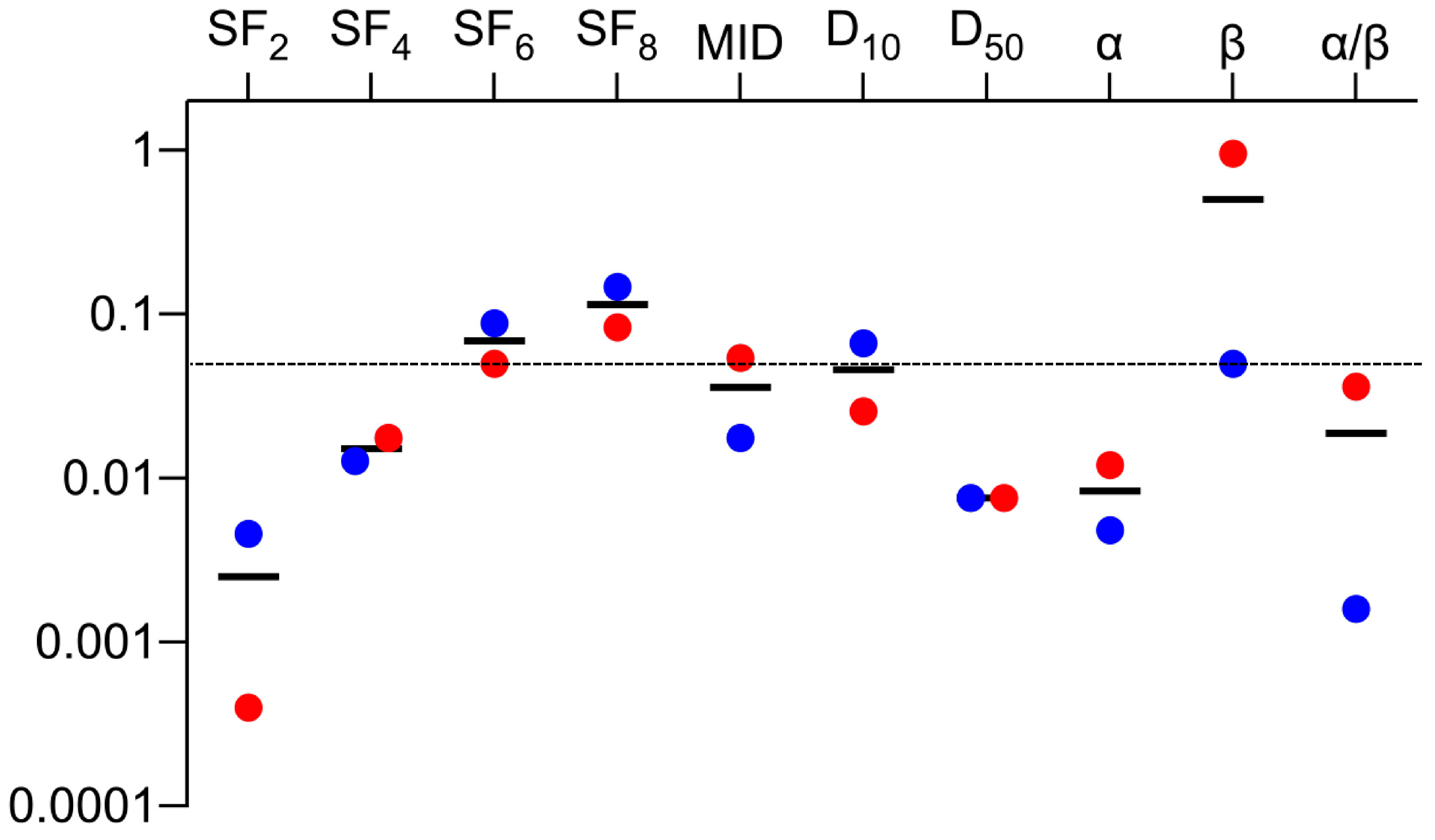
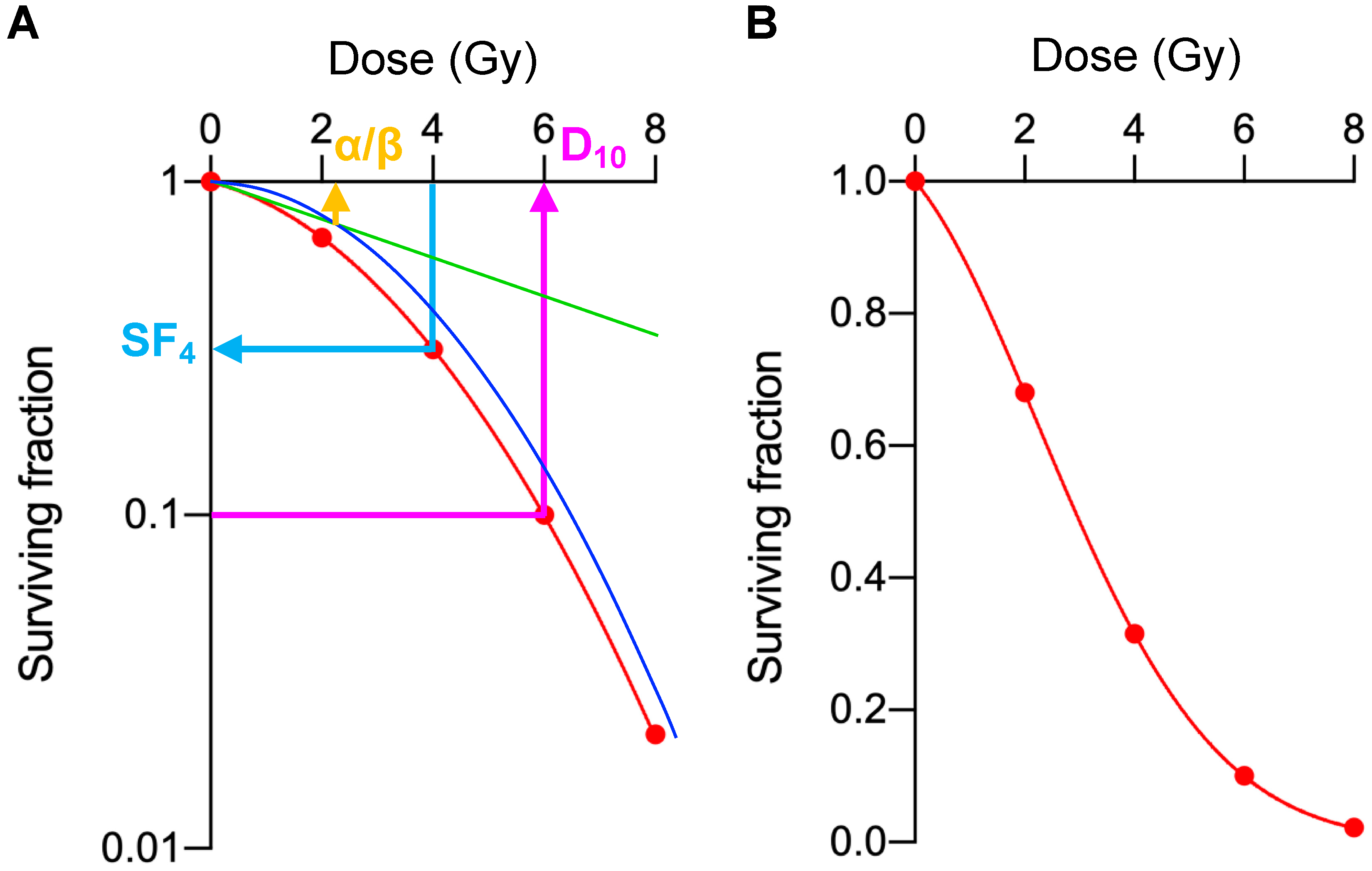
| Dataset | Cell Line | EGFR | SF2 | SF4 | SF6 | SF8 | MID | D10 | D50 | α | β | α/β |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | H1650 | mutant | 0.26 | 0.079 | 0.033 | 0.0047 | 1.7 | 3.8 | 1.2 | 0.56 | 0.012 | 46.4 |
| Ma-24 | 0.39 | 0.070 | 0.020 | 0.0031 | 1.7 | 3.8 | 1.3 | 0.51 | 0.026 | 19.7 | ||
| PC9 | 0.42 | 0.11 | 0.032 | 0.0063 | 2.0 | 4.3 | 1.5 | 0.42 | 0.027 | 15.4 | ||
| HCC827 | 0.35 | 0.14 | 0.022 | NA | 2.0 | 4.2 | 1.6 | 0.34 | 0.048 | 7.2 | ||
| H1975 | 0.47 | 0.20 | 0.073 | 0.021 | 2.5 | 5.4 | 1.9 | 0.32 | 0.021 | 15.5 | ||
| II-18 | 0.45 | 0.28 | 0.15 | 0.058 | 3.0 | 6.7 | 2.2 | 0.31 | 0.0047 | 65.9 | ||
| H1299 | wild-type | 0.97 | 0.67 | 0.46 | 0.27 | 6.1 | 10.7 | 5.7 | 0.015 | 0.019 | 0.82 | |
| H1703 | 0.79 | 0.43 | 0.35 | 0.16 | 4.7 | 9.5 | 4.0 | 0.13 | 0.012 | 10.3 | ||
| A549 | 0.71 | 0.49 | 0.24 | 0.098 | 4.2 | 8.0 | 3.8 | 0.092 | 0.025 | 3.7 | ||
| H157 | 0.78 | 0.34 | 0.13 | 0.062 | 3.4 | 6.9 | 2.9 | 0.17 | 0.024 | 7.2 | ||
| LK2 | 0.70 | 0.34 | 0.11 | 0.051 | 3.2 | 6.6 | 2.7 | 0.19 | 0.025 | 7.5 | ||
| H522 | 0.53 | 0.15 | 0.059 | 0.016 | 2.3 | 5.0 | 1.8 | 0.35 | 0.021 | 17.0 | ||
| H520 | 0.49 | 0.26 | 0.061 | 0.015 | 2.6 | 5.4 | 2.3 | 0.22 | 0.038 | 5.8 | ||
| A427 | 0.57 | 0.15 | 0.047 | 0.017 | 2.2 | 5.0 | 1.7 | 0.37 | 0.019 | 19.3 | ||
| H460 | 0.53 | 0.16 | 0.050 | 0.0081 | 2.4 | 4.9 | 2.0 | 0.26 | 0.043 | 6.0 | ||
| p value | <0.001 | 0.018 | 0.049 | 0.082 | 0.054 | 0.025 | 0.0076 | 0.012 | 0.95 | 0.036 | ||
| B | H1650 | mutant | 0.23 | 0.054 | 0.031 | 0.0072 | 1.3 | 3.4 | 0.95 | 0.75 | −0.018 | NA |
| Ma-24 | 0.34 | 0.081 | 0.026 | NA | 1.7 | 3.8 | 1.2 | 0.58 | 0.0068 | 84.4 | ||
| PC9 | 0.28 | 0.070 | 0.019 | 0.0022 | 1.6 | 3.6 | 1.2 | 0.53 | 0.028 | 19.2 | ||
| HCC827 | 0.55 | 0.18 | 0.081 | 0.026 | 2.5 | 5.5 | 1.9 | 0.34 | 0.015 | 22.8 | ||
| H1975 | 0.47 | 0.14 | 0.051 | 0.018 | 2.1 | 4.8 | 1.5 | 0.44 | 0.0082 | 53.8 | ||
| II-18 | 0.45 | 0.34 | 0.20 | 0.085 | 3.4 | 7.8 | 2.5 | 0.27 | 0.0029 | 93.3 | ||
| H1299 | wild-type | 0.87 | 0.67 | 0.55 | 0.28 | 6.3 | 11.1 | 5.9 | 0.015 | 0.017 | 0.85 | |
| H1703 | 0.77 | 0.49 | 0.22 | 0.16 | 4.3 | 9.1 | 3.6 | 0.16 | 0.010 | 15.5 | ||
| A549 | 0.53 | 0.28 | 0.14 | 0.049 | 3.0 | 6.6 | 2.4 | 0.26 | 0.014 | 17.9 | ||
| H157 | 0.83 | 0.51 | 0.11 | 0.049 | 3.8 | 6.8 | 3.5 | 0.042 | 0.044 | 1.0 | ||
| LK2 | 0.70 | 0.56 | 0.12 | 0.046 | 3.8 | 6.7 | 3.5 | 0.044 | 0.044 | 1.0 | ||
| H522 | 0.68 | 0.43 | 0.066 | 0.027 | 3.2 | 6.0 | 2.9 | 0.11 | 0.046 | 2.3 | ||
| H520 | 0.83 | 0.49 | 0.14 | 0.041 | 3.9 | 6.8 | 3.7 | −0.005 | 0.051 | NA | ||
| A427 | 0.39 | 0.12 | 0.049 | 0.021 | 1.9 | 4.5 | 1.3 | 0.54 | −0.0068 | NA | ||
| H460 | 0.72 | 0.14 | 0.036 | 0.0062 | 2.4 | 4.8 | 2.0 | 0.24 | 0.051 | 4.7 | ||
| p value | 0.005 | 0.012 | 0.087 | 0.14 | 0.017 | 0.0660 | 0.0076 | 0.0048 | 0.049 | 0.0016 |
| Parameters | R Values | p Values |
|---|---|---|
| SF2 | 0.73 | 0.003 |
| SF4 | 0.81 | <0.001 |
| SF6 | 0.86 | <0.001 |
| SF8 | 0.91 | <0.001 |
| MID | 0.87 | <0.001 |
| D10 | 0.90 | <0.001 |
| D50 | 0.88 | <0.001 |
| α | 0.79 | <0.001 |
| β | 0.63 | 0.014 |
| α/β | 0.62 | 0.037 |
| Cell Line | Histopathology | EGFR Status | Reference |
|---|---|---|---|
| H1650 | Adenocarcinoma | ΔE746_A750 | [23,24] |
| Ma-24 | Adenocarcinoma | L858R, E709G | [23,25] |
| PC9 | Adenocarcinoma | ΔE746_A750 | [23,26] |
| HCC827 | Adenocarcinoma | ΔE746_A750 | [24,27] |
| H1975 | Adenocarcinoma | L858R, T790M | [24,26,27] |
| II-18 | Adenocarcinoma | L858R | [23,26,28] |
| H1299 | Large cell carcinoma | Wild-type | [23] |
| H1703 | Adenocarcinoma | Wild-type | [23,24] |
| A549 | Adenocarcinoma | Wild-type | [23,24,27,28] |
| H157 | Squamous cell carcinoma | Wild-type | [23,24,28] |
| LK2 | Squamous cell carcinoma | Wild-type | [23,26] |
| H522 | Adenocarcinoma | Wild-type | [23] |
| H520 | Squamous cell carcinoma | Wild-type | [23,24,27] |
| A427 | Adenocarcinoma | Wild-type | [23,28] |
| H460 | Large cell carcinoma | Wild-type | [23,24,27,28] |
| Cell Line | PE (%) | 0 Gy | 2 Gy | 4 Gy | 6 Gy | 8 Gy |
|---|---|---|---|---|---|---|
| H1650 | 12 | 500 | 500 | 1000 | 2000 | 3000 |
| Ma-24 | 24 | 500 | 500 | 1000 | 2000 | 3000 |
| PC9 | 25 | 500 | 500 | 1000 | 2000 | 3000 |
| HCC827 | 39 | 300 | 300 | 500 | 1000 | 2000 |
| H1975 | 35 | 300 | 300 | 500 | 1000 | 2000 |
| II-18 | 21 | 500 | 500 | 1000 | 2000 | 3000 |
| H1299 | 71 | 200 | 200 | 300 | 500 | 500 |
| H1703 | 34 | 300 | 300 | 500 | 1000 | 2000 |
| A549 | 72 | 200 | 200 | 300 | 500 | 500 |
| H157 | 65 | 200 | 200 | 300 | 500 | 500 |
| LK2 | 40 | 300 | 300 | 500 | 1000 | 2000 |
| H522 | 43 | 300 | 300 | 500 | 1000 | 2000 |
| H520 | 28 | 500 | 500 | 1000 | 2000 | 3000 |
| A427 | 32 | 300 | 300 | 500 | 1000 | 2000 |
| H460 | 58 | 300 | 300 | 500 | 1000 | 2000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anakura, M.; Nachankar, A.; Kobayashi, D.; Amornwichet, N.; Hirota, Y.; Shibata, A.; Oike, T.; Nakano, T. Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses. Int. J. Mol. Sci. 2019, 20, 3635. https://doi.org/10.3390/ijms20153635
Anakura M, Nachankar A, Kobayashi D, Amornwichet N, Hirota Y, Shibata A, Oike T, Nakano T. Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses. International Journal of Molecular Sciences. 2019; 20(15):3635. https://doi.org/10.3390/ijms20153635
Chicago/Turabian StyleAnakura, Mai, Ankita Nachankar, Daijiro Kobayashi, Napapat Amornwichet, Yuka Hirota, Atsushi Shibata, Takahiro Oike, and Takashi Nakano. 2019. "Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses" International Journal of Molecular Sciences 20, no. 15: 3635. https://doi.org/10.3390/ijms20153635
APA StyleAnakura, M., Nachankar, A., Kobayashi, D., Amornwichet, N., Hirota, Y., Shibata, A., Oike, T., & Nakano, T. (2019). Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses. International Journal of Molecular Sciences, 20(15), 3635. https://doi.org/10.3390/ijms20153635





