Prioritization of Variants for Investigation of Genotype-Directed Nutrition in Human Superpopulations
Abstract
1. Introduction
2. Results
2.1. Literature Annotation for Nutrigenetic Database
2.2. Summary Statistics for Nutrigenetic Variants
2.3. Genotype-Directed Nutrition Prioritization for Superpopulations Based on Nutrigenetic Variants
3. Discussion
3.1. Variant Quality
3.2. Nutrigenetics Database
3.3. Genotype-Directed Nutrition for Populations
3.4. Limitations
4. Material and Methods
4.1. Data Sources for Nutrigenetic Variants
4.2. Population Frequencies and FST Values of Nutrigenetic Variants
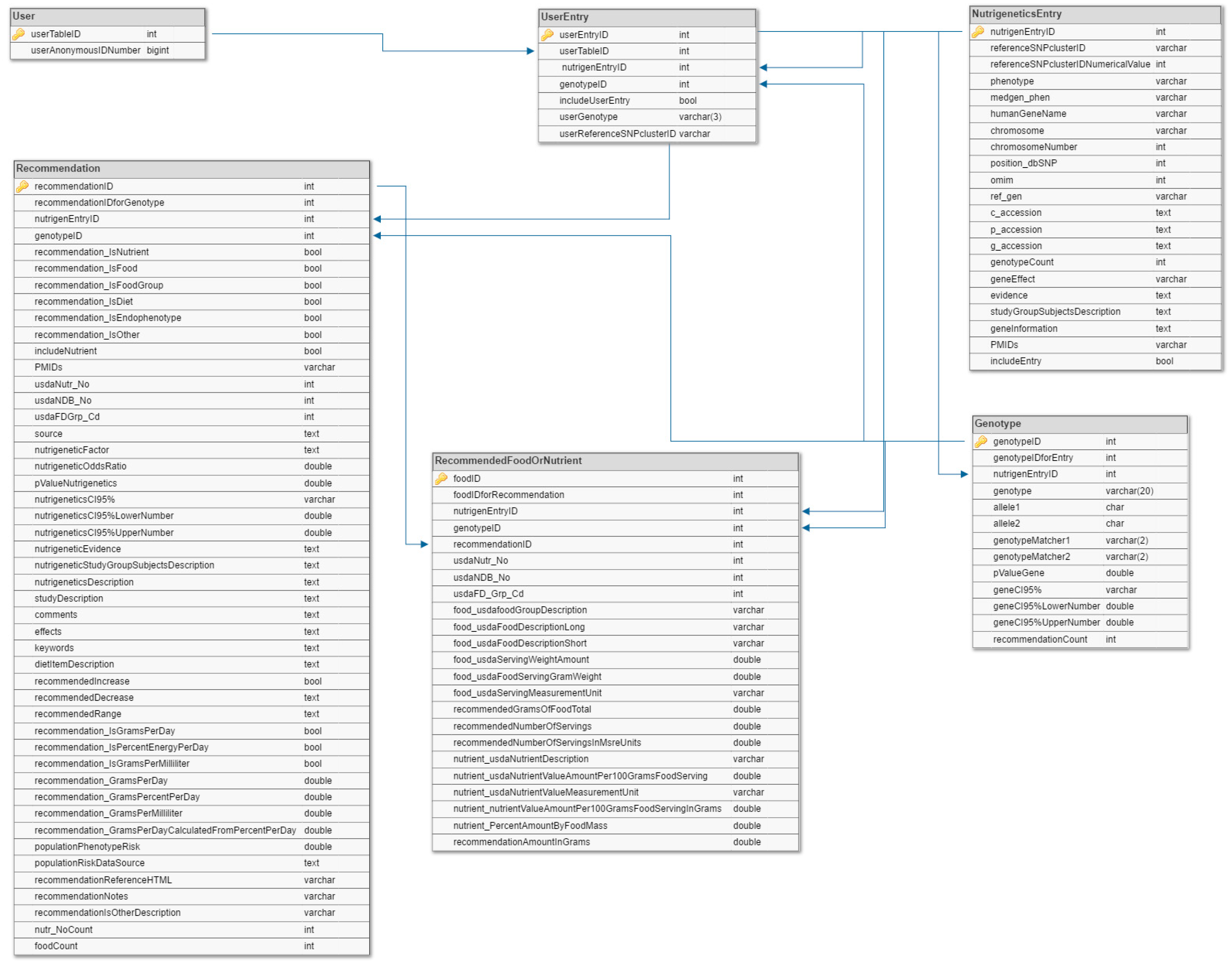
4.3. Data Model
5. Conclusions
- Nutrigenetic variants with high superpopulation frequencies can be used to prioritize dietary modifications for the purpose of reducing disease risk for human superpopulations with the potential for widespread health benefits.
- The proposed superpopulation genotype-directed nutrition modifications will need to be validated in a research study.
6. Data Availability
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lai, C.-Q. How much of human height is genetic and how much is due to nutrition? Sci. Am. 2006. [Google Scholar]
- van den Berg, L.; Henneman, P.; Willems van Dijk, K.; Delemarre-van de Waal, H.A.; Oostra, B.A.; van Duijn, C.M.; Janssens, A.C.J.W. Heritability of dietary food intake patterns. Acta Diabetol. 2013, 50, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, H.; Beaty, T.H.; Chen, H.; Caballero, B.; Wang, Y. Heritability of Children’s Dietary Intakes: A Population-Based Twin Study in China. Twin Res. Hum. Genet. 2016, 19, 472–484. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mozaffarian, D.; Dashti, H.S.; Wojczynski, M.K.; Chu, A.Y.; Nettleton, J.A.; Männistö, S.; Kristiansson, K.; Reedik, M.; Lahti, J.; Houston, D.K.; et al. Genome-wide association meta-analysis of fish and EPA+DHA consumption in 17 US and European cohorts. PLoS ONE 2017, 12, e0186456. [Google Scholar] [CrossRef] [PubMed]
- Harris, W.S.; Pottala, J.V.; Lacey, S.M.; Vasan, R.S.; Larson, M.G.; Robins, S.J. Clinical correlates and heritability of erythrocyte eicosapentaenoic and docosahexaenoic acid content in the Framingham Heart Study. Atherosclerosis 2012, 225, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Hasselbalch, A.L.; Silventoinen, K.; Keskitalo, K.; Pietiläinen, K.H.; Rissanen, A.; Heitmann, B.L.; Kyvik, K.O.; Sørensen, T.I.A.; Kaprio, J. Twin study of heritability of eating bread in Danish and Finnish men and women. Twin Res. Hum. Genet. 2010, 13, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Steck, S.E.; Keku, T.; Butler, L.M.; Galanko, J.; Massa, B.; Millikan, R.C.; Sandler, R.S. Polymorphisms in methionine synthase, methionine synthase reductase and serine hydroxymethyltransferase, folate and alcohol intake, and colon cancer risk. Lifestyle Genomics 2008, 1, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Abdelmagid, S.A.; Clarke, S.E.; Roke, K.; Nielsen, D.E.; Badawi, A.; El-Sohemy, A.; Mutch, D.M.; Ma, D.W. Ethnicity, sex, FADS genetic variation, and hormonal contraceptive use influence delta-5- and delta-6-desaturase indices and plasma docosahexaenoic acid concentration in young Canadian adults: A cross-sectional study. Nutr. Metab. 2015, 12, 14. [Google Scholar] [CrossRef]
- Merritt, D.C.; Jamnik, J.; El-Sohemy, A. FTO genotype, dietary protein intake, and body weight in a multiethnic population of young adults: A cross-sectional study. Genes Nutr. 2018, 13, 4. [Google Scholar] [CrossRef]
- García-Bailo, B.; Brenner, D.R.; Nielsen, D.; Lee, H.-J.; Domanski, D.; Kuzyk, M.; Borchers, C.H.; Badawi, A.; Karmali, M.A.; El-Sohemy, A. Dietary patterns and ethnicity are associated with distinct plasma proteomic groups. Am. J. Clin. Nutr. 2012, 95, 352–361. [Google Scholar] [CrossRef]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014, 42, D1001–D1006. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef] [PubMed]
- Fölling, A. Über Ausscheidung von Phenylbrenztraubensäure in den Harn als Stoffwechselanomalie in Verbindung mit Imbezillität. Hoppe Seyler’s Z. Physiol. Chem. 1934, 227, 169–181. [Google Scholar] [CrossRef]
- Penrose, L.; Quastel, J.H. Metabolic studies in phenylketonuria. Biochem. J. 1937, 31, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Scriver, C.R.; Byck, S.; Prevost, L.; Hoang, L. The phenylalanine hydroxylase locus: A marker for the history of phenylketonuria and human genetic diversity. PAH Mutation Analysis Consortium. Var. Hum. Genome 1996, 197, 73–90. [Google Scholar]
- Woolf, L.I.; Griffiths, R.; Moncrieff, A. Treatment of phenylketonuria with a diet low in phenylalanine. Br. Med. J. 1955, 1, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Novelli, G.; Reichardt, J.K. Molecular basis of disorders of human galactose metabolism: Past, present, and future. Mol. Genet. Metab. 2000, 71, 62–65. [Google Scholar] [CrossRef]
- Bouteldja, N.; Timson, D.J. The biochemical basis of hereditary fructose intolerance. J. Inherit. Metab. Dis. 2010, 33, 105–112. [Google Scholar] [CrossRef]
- Peregrin, T. The new frontier of nutrition science: Nutrigenomics. J. Am. Diet Assoc. 2001, 101, 1306. [Google Scholar] [CrossRef]
- Vargas, A.J.; Wertheim, B.C.; Gerner, E.W.; Thomson, C.A.; Rock, C.L.; Thompson, P.A. Dietary polyamine intake and risk of colorectal adenomatous polyps. Am. J. Clin. Nutr. 2012, 96, 133–141. [Google Scholar] [CrossRef]
- Alfredo Martínez, J. Perspectives on personalized nutrition for obesity. Lifestyle Genomics 2014, 7, 1–3. [Google Scholar]
- Phillips, C.M.; Goumidi, L.; Bertrais, S.; Field, M.R.; Cupples, L.A.; Ordovas, J.M.; Defoort, C.; Lovegrove, J.A.; Drevon, C.A.; Gibney, M.J.; et al. Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J. Lipid Res. 2010, 51, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.-S.; Huang, T.; Li, K.; Chen, Y.; Xie, H.; Xu, D.; Sun, J.; Li, D. Modulation of the Association between the PEPD Variant and the Risk of Type 2 Diabetes by n-3 Fatty Acids in Chinese Hans. Lifestyle Genomics 2015, 8, 36–43. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Cooper, D.N.; Ball, E.V.; Krawczak, M. The human gene mutation database. Nucleic Acids Res. 1998, 26, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Küntzer, J.; Eggle, D.; Klostermann, S.; Burtscher, H. Human variation databases. Database 2010, 2010, baq015. [Google Scholar] [CrossRef] [PubMed]
- Fokkema, I.F.A.C.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; den Dunnen, J.T. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Smith, G. At Home—DNA Tests: Marketing Scam or Medical Breakthrough; US Government Printong Office: Wshington, DC, USA, 2006; pp. 109–707.
- Grimaldi, K.A.; van Ommen, B.; Ordovas, J.M.; Parnell, L.D.; Mathers, J.C.; Bendik, I.; Brennan, L.; Celis-Morales, C.; Cirillo, E.; Daniel, H.; et al. Proposed guidelines to evaluate scientific validity and evidence for genotype-based dietary advice. Genes Nutr. 2017, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Kutz, G. Nutrigenetic Testing: Tests Purchased from Four Web Sites Mislead Consumers; GAO: Washington, DC, USA, 2006.
- Castle, D. Genomic Nutritional Profiling: Innovation and Regulation in Nutrigenomics. Minn. J. Law Sci. Technol. 2008, 9, 37–60. [Google Scholar]
- Castle, D.; Ries, N.M. Ethical, legal and social issues in nutrigenomics: The challenges of regulating service delivery and building health professional capacity. Mutat. Res. 2007, 622, 138–143. [Google Scholar] [CrossRef]
- Clarke, L.; Fairley, S.; Zheng-Bradley, X.; Streeter, I.; Perry, E.; Lowy, E.; Tassé, A.-M.; Flicek, P. The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res. 2017, 45, D854–D859. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R.; De Caterina, R.; Görman, U.; Allayee, H.; Kohlmeier, M.; Prasad, C.; Choi, M.S.; Curi, R.; de Luis, D.A.; Gil, Á.; et al. Guide and Position of the International Society of Nutrigenetics/Nutrigenomics on Personalised Nutrition: Part 1—Fields of Precision Nutrition. Lifestyle Genomics 2016, 9, 12–27. [Google Scholar] [CrossRef] [PubMed]
- du Prel, J.-B.; Hommel, G.; Röhrig, B.; Blettner, M. Confidence interval or p-value? Part 4 of a series on evaluation of scientific publications. Dtsch. Arztebl. Int. 2009, 106, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, Y.A.; Kim, D.-H.; Lee, B.-H.; Hwang, D.-Y.; Jeong, J.; Lee, H.-J.; Matsuo, K.; Tajima, K.; Ahn, Y.-O. Dietary intake of folate and alcohol, MTHFR C677T polymorphism, and colorectal cancer risk in Korea. Am. J. Clin. Nutr. 2012, 95, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Abecasis, G.R.; Auton, A.; Brooks, L.D.; DePristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A. An integrated map of genetic variation from 1,092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [PubMed]
- Junyent, M.; Parnell, L.D.; Lai, C.-Q.; Arnett, D.K.; Tsai, M.Y.; Kabagambe, E.K.; Straka, R.J.; Province, M.; An, P.; Smith, C.E.; et al. ADAM17_i33708A>G polymorphism interacts with dietary n-6 polyunsaturated fatty acids to modulate obesity risk in the Genetics of Lipid Lowering Drugs and Diet Network study. Nutr. Metab. Cardiovasc. Dis. 2010, 20, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Teutsch, S.M.; Bradley, L.A.; Palomaki, G.E.; Haddow, J.E.; Piper, M.; Calonge, N.; Dotson, W.D.; Douglas, M.P.; Berg, A.O. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: Methods of the EGAPP Working Group. Genet. Med. 2009, 11, 3–14. [Google Scholar] [CrossRef]
- Little, J.; Higgins, J.P.T.; Ioannidis, J.P.A.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. STrengthening the REporting of Genetic Association Studies (STREGA)—An Extension of the STROBE Statement. PLoS Med. 2009, 6, e1000022. [Google Scholar] [CrossRef]
- Rafiq, M.; Boccia, S. Application of the GRADE Approach in the Development of Guidelines and Recommendations in Genomic Medicine. Genomics Insights 2018, 11, 117863101775336. [Google Scholar] [CrossRef]
- Conran, C.; Na, R.; Chen, H.; Jiang, D.; Lin, X.; Zheng, S.l.; Brendler, C.; Xu, J. Population-standardized genetic risk score: The SNP-based method of choice for inherited risk assessment of prostate cancer. Asian J. Androl. 2016, 18, 520. [Google Scholar]
- Dudbridge, F. Power and Predictive Accuracy of Polygenic Risk Scores. PLoS Genet. 2013, 9, e1003348. [Google Scholar] [CrossRef]
- Wray, N.R.; Goddard, M.E.; Visscher, P.M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007, 17, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Start, K. Treating phenylketonuria by a phenylalanine-free diet. Prof. Care Mother Child 1998, 8, 109–110. [Google Scholar] [PubMed]
- Schakel, S.F.; Sievert, Y.A.; Buzzard, I.M. Sources of data for developing and maintaining a nutrient database. J. Am. Diet Assoc. 1988, 88, 1268–1271. [Google Scholar] [PubMed]
- Neveu, V.; Perez-Jimenez, J.; Vos, F.; Crespy, V.; du Chaffaut, L.; Mennen, L.; Knox, C.; Eisner, R.; Cruz, J.; Wishart, D.; et al. Phenol-Explorer: An online comprehensive database on polyphenol contents in foods. Database 2010, 2010, bap024. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Yang, J.; Hayes, B.J.; Price, A.L.; Goddard, M.E.; Visscher, P.M. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 2013, 14, 507–515. [Google Scholar] [CrossRef]
- Guerreiro, C.S.; Ferreira, P.; Tavares, L.; Santos, P.M.; Neves, M.; Brito, M.; Cravo, M. Fatty Acids, IL6, and TNFα Polymorphisms: An Example of Nutrigenetics in Crohn’s Disease. Am. J. Gastroenterol. 2009, 104, 2241–2249. [Google Scholar] [CrossRef]
- Matullo, G.; Di Gaetano, C.; Guarrera, S. Next generation sequencing and rare genetic variants: From human population studies to medical genetics. Environ. Mol. Mutagen. 2013, 54, 518–532. [Google Scholar] [CrossRef]
- Wasserstein, R.L.; Schirm, A.L.; Lazar, N.A. Moving to a World Beyond “ p <0.05.”. Am. Stat. 2019, 73, 1–19. [Google Scholar]



| Category | 2Number |
|---|---|
| Articles | 67 |
| Annotations | 156 |
| Phenotypes | 36 |
| Genes | 84 |
| 1SNPs | 101 |
| Protective | 52 |
| Risk | 104 |
| 1OR range (Avg) | 0.07–35 (2.17) |
| P-value range (Avg) | 3.5 × 10−5 – 0.05 (0.018) |
| Diet types | 106 |
| Participants range (Avg) | 1–16,624 (1106) |
| 2dbSNP ID | Gene | Disease | Dietary Change | 1,2,3Superpopulation SNP Frequency | |||||
|---|---|---|---|---|---|---|---|---|---|
| ALL | AFR | AMR | EAS | EUR | SAS | ||||
| rs9997745 | ACSL1 | Metabolic Syndrome | 1Low-fat (<35% energy), high-PUFA diet (>5.5% energy) | 78 | 40 | 87 | 100 | 85 | 93 |
| rs6008259 | PPARA | Hypercholesterolemia | Low n–6 fatty Acid (≤7.99 g/day) | 73 | 86 | 24 | 100 | 82 | 92 |
| rs6087990 | DNMT3B | Colorectal Cancer | 1,4High RBC folate | 68 | 76 | 63 | 92 | 37 | 68 |
| rs3790433 | LEPR | Metabolic Syndrome | 1,5Low n-6 PUFA, high n-3 PUFA | 59 | 23 | 67 | 84 | 77 | 58 |
| rs11568820 | VDR | Prostate Cancer | Low calcium (<680 mg/day) | 54 | 11 | 82 | 60 | 77 | 64 |
| rs512535 | APOB | Metabolic Syndrome | Low fat (<35% energy) | 53 | 19 | 51 | 81 | 51 | 73 |
| rs10495563 | ADAM17 | Obesity | 6Low n-6 fatty Acid | 52 | 30 | 56 | 90 | 34 | 58 |
| rs2287161 | CRY1 | Metabolic Syndrome | Low carbohydrate (% of energy intake <41.7%) | 46 | 64 | 52 | 13 | 45 | 54 |
| rs3827730 | FAF1 | Alcohol Dependence | 7Low amounts of alcohol | 38 | 7 | 52 | 79 | 35 | 28 |
| rs2424913 | DNMT3B | Adenoma, Colorectal Cancer | No alcohol | 31 | 33 | 36 | 1 | 59 | 29 |
| rs1801181 | CBS | Colorectal Cancer | 1,4High RBC folate | 30 | 2 | 19 | 57 | 39 | 36 |
| rs2424909 | DNMT3B | Colorectal Cancer | Moderate alcohol >0 and <1.7 drinks/week | 28 | 8 | 36 | 8 | 63 | 31 |
| rs1378942 | CSK | Hypertension | 11.8 g/day of EPA and DHA | 24 | 3 | 33 | 18 | 61 | 16 |
| rs2168784 | (Intergenic) | Alcohol dependence | no alcoholic drinks/week | 24 | 62 | 10 | 9 | 10 | 13 |
| rs1229984 | ADH1B | Alcohol dependence | no alcoholic drinks/week | 16 | 0 | 6 | 70 | 3 | 2 |
| rs75038630 | NADSYN1 | Abnormal Eating Behavior | High vitamin D (>75 nmol/L) | 2 | 0 | 4 | 100 | 6 | 3 |
| Category | 1Diseases | 1,2Dietary Suggestion |
|---|---|---|
| All | Metabolic Syndrome, Hypercholesterolemia, Colorectal Cancer, Prostate Cancer, Obesity | Low-fat (<35% energy), High-PUFA diet (>5.5% energy), Low n–6 Fatty Acid (≤7.99 g/day), Low Calcium (<680 mg/day) |
| AFR | Hypercholesterolemia, Alcohol dependence | Low n–6 Fatty Acid (≤7.99 g/day), 0 alcoholic drinks/week |
| AMR | Colorectal Cancer, Prostate Cancer, Obesity, Alcohol Dependence | High PUFA, Low Calcium (<680 mg/day), 3Low n-6 Fatty Acid |
| EAS | Hypercholesterolemia, Prostate Cancer, Obesity, Alcohol Dependence, Abnormal Eating Behavior | Low n–6 Fatty Acid (≤7.99 g/day), Low Calcium (<680 mg/day), 3Low n-6 Fatty Acid, High vitamin D (>75 nmol/L) |
| EUR | Hypercholesterolemia, Prostate Cancer, Adenoma, Hypertension | Low n–6 Fatty Acid (≤7.99 g/day), Low Calcium (<680 mg/day), 1.8 g/day of EPA and DHA |
| SAS | Hypercholesterolemia, Prostate Cancer, Obesity | Low n–6 Fatty Acid (≤7.99 g/day), Low Calcium (<680 mg/day), 3Low n-6 Fatty Acid |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nilsson, P.D.; Newsome, J.M.; Santos, H.M.; Schiller, M.R. Prioritization of Variants for Investigation of Genotype-Directed Nutrition in Human Superpopulations. Int. J. Mol. Sci. 2019, 20, 3516. https://doi.org/10.3390/ijms20143516
Nilsson PD, Newsome JM, Santos HM, Schiller MR. Prioritization of Variants for Investigation of Genotype-Directed Nutrition in Human Superpopulations. International Journal of Molecular Sciences. 2019; 20(14):3516. https://doi.org/10.3390/ijms20143516
Chicago/Turabian StyleNilsson, Pascal D., Jacklyn M. Newsome, Henry M. Santos, and Martin R. Schiller. 2019. "Prioritization of Variants for Investigation of Genotype-Directed Nutrition in Human Superpopulations" International Journal of Molecular Sciences 20, no. 14: 3516. https://doi.org/10.3390/ijms20143516
APA StyleNilsson, P. D., Newsome, J. M., Santos, H. M., & Schiller, M. R. (2019). Prioritization of Variants for Investigation of Genotype-Directed Nutrition in Human Superpopulations. International Journal of Molecular Sciences, 20(14), 3516. https://doi.org/10.3390/ijms20143516





