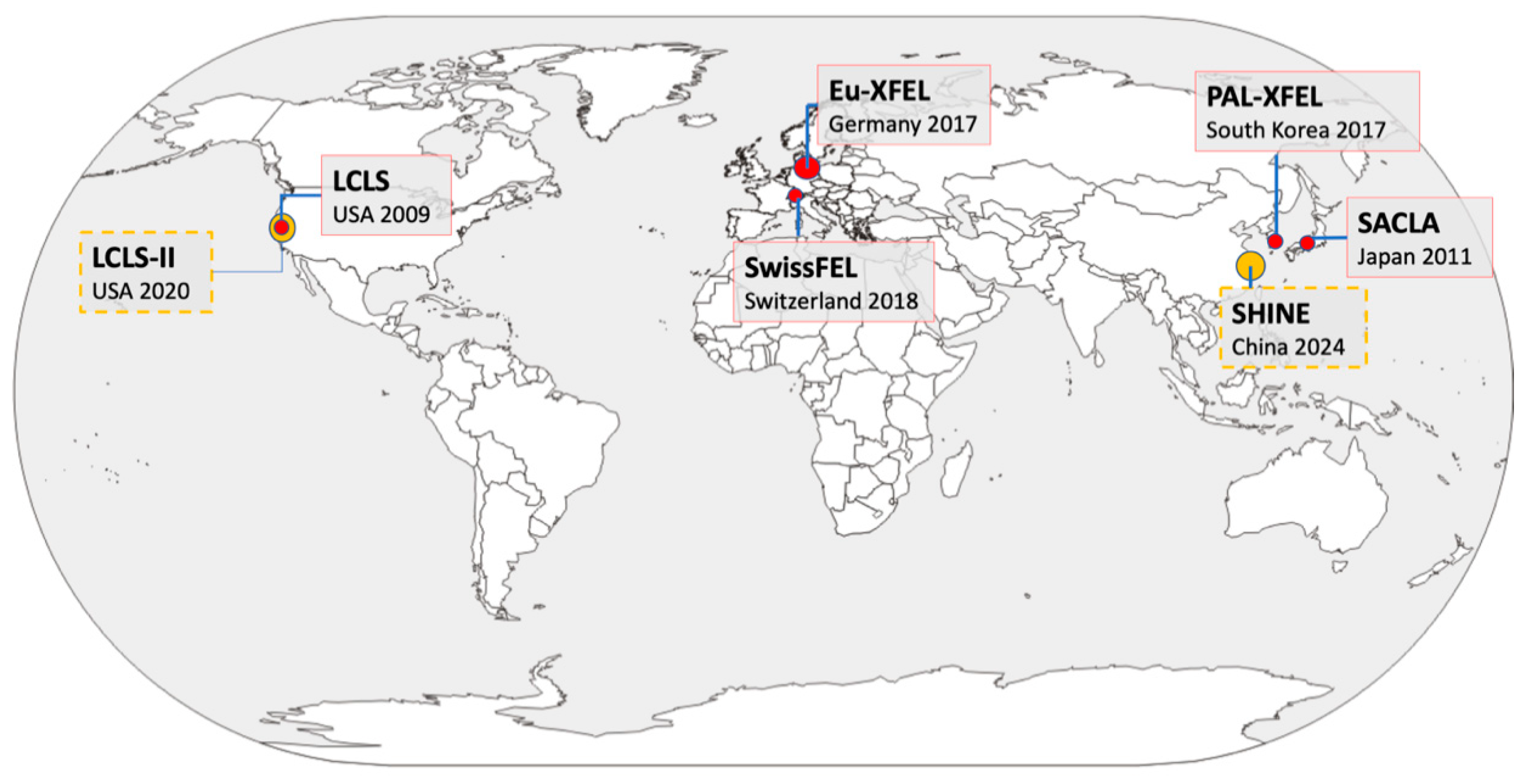
The XFEL Protein Crystallography: Developments and Perspectives
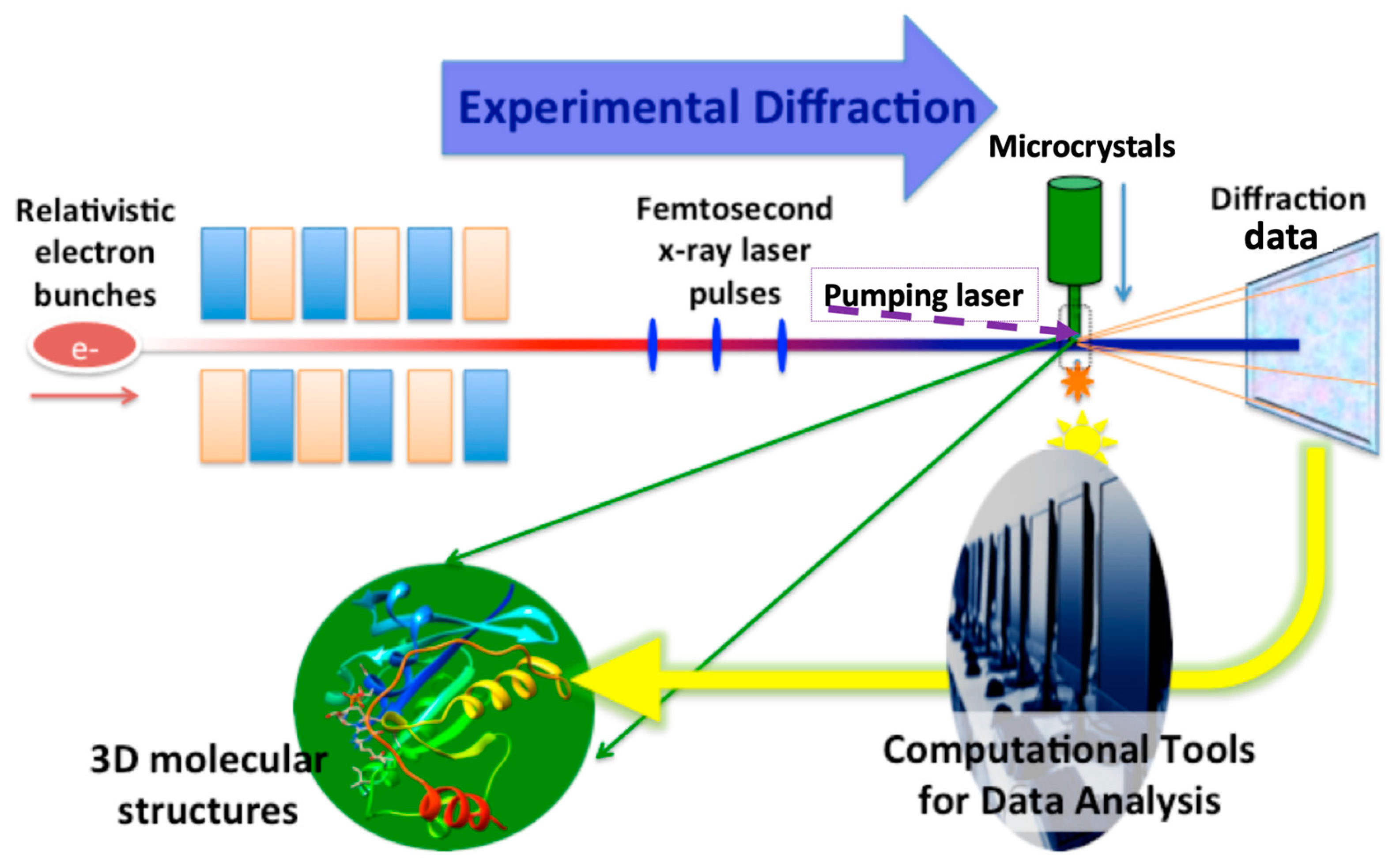
- Each crystal usually results from a single diffraction pattern within tens of femtoseconds of exposure time, so the measured intensity for each reflection in a single pattern is incomplete (inaccurate) due to the excited volume defined by the overlapped region between the thin Ewald shell and the full reflection volume. Therefore, a high measurement redundancy is desirable for accurate diffraction intensity. This can be achieved by the high repetition rates of XFEL pulses. The processing of large data volumes requires special software for screening [18,19,20], auto-indexing [21,22,23], merging and also post-refinement, as summarized in [24].
- With the ‘diffraction-before-destruction’ approach, the cryogenic protection of crystals is not required, so the experiments can be done with the samples at room temperature (if the temperature changes during the injection can be neglected). Therefore, the structures might be different from those determined at synchrotrons in some regions, as discussed by Park et al. in the HIV-CCD structure analysis [15]. Such comparisons may provide important clues for the understanding of protein functions and how they may apply to drug development [25].
- With the femtosecond XFEL pulses, the temporal resolution can also reach the femtosecond time scale in theory, making it possible to study ultrafast dynamics. Light-triggered reactions are very suitable for time resolved SFX, revealing conformational changes down to a sub-picosecond time scale [26,27]. Enzymatic reactions or receptor conformational changes triggered by substrate binding are possible by using a fast mixing device or premixed solution of engineered substrates that are photocaged, although the time resolution will be limited to a longer time scale [28,29].
- The ultimate goal of structure studies is to probe structure information from single molecules using XFELs, similar to the cryogenic electron microscopy method. Even with the most powerful XFELs, the resolution for single particle imaging can only reach a few nanometers for viruses [30,31]. Nonetheless, the ensemble measurement can yield interpretable solution scattering signals, providing dynamics information when combined with pump–probe technology [32,33].
Author Contributions
Funding
Conflicts of Interest
References
- Bostedt, C.; Boutet, S.; Fritz, D.M.; Huang, Z.; Lee, H.J.; Lemke, H.T.; Robert, A.; Schlotter, W.F.; Turner, J.J.; Williams, G.J. Linac Coherent Light Source: The first five years. Rev. Mod. Phys. 2016, 88, 015007. [Google Scholar] [CrossRef]
- Madey, J.M.J. Stimulated Emission of Bremsstrahlung in a Periodic Magnetic Field. J. Appl. Phys. 1971, 42, 1906–1913. [Google Scholar] [CrossRef]
- Emma, P.; Akre, R.; Arthur, J.; Bionta, R.; Bostedt, C.; Bozek, J.; Brachmann, A.; Bucksbaum, P.; Coffee, R.; Decker, F.J.; et al. First lasing and operation of an angstrom-wavelength free-electron laser. Nat. Photonics. 2010, 4, 641–647. [Google Scholar] [CrossRef]
- Ball, P. Europe’s X-ray laser fires up. Nature 2017, 548, 507–508. [Google Scholar] [CrossRef] [PubMed]
- Solem, J.C. Imaging biological specimens with high-intensity soft X rays. J. Opt. Soc. Am. B 1986, 3, 1551–1565. [Google Scholar] [CrossRef]
- Neutze, R.; Wouts, R.; van der Spoel, D.; Weckert, E.; Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Chapman, H.N.; Barty, A.; Bogan, M.J.; Boutet, S.; Frank, M.; Hau-Riege, S.P.; Marchesini, S.; Woods, B.W.; Bajt, S.; Benner, W.H.; et al. Femtosecond diffractive imaging with a soft-X-ray free-electron laser. Nat. Phys. 2006, 2, 839–843. [Google Scholar] [CrossRef]
- Chapman, H.N.; Fromme, P.; Barty, A.; White, T.A.; Kirian, R.A.; Aquila, A.; Hunter, M.S.; Schulz, J.; DePonte, D.P.; Weierstall, U.; et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [Google Scholar] [CrossRef]
- Barty, A.; Caleman, C.; Aquila, A.; Timneanu, N.; Lomb, L.; White, T.A.; Andreasson, J.; Arnlund, D.; Bajt, S.; Barends, T.R.M.; et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat. Photonics 2012, 6, 35–40. [Google Scholar] [CrossRef]
- Hara, T.; Inubushi, Y.; Katayama, T.; Sato, T.; Tanaka, H.; Tanaka, T.; Togashi, T.; Togawa, K.; Tono, K.; Yabashi, M.; et al. Two-colour hard X-ray free-electron laser with wide tunability. Nat. Commun. 2013, 4, 2919. [Google Scholar] [CrossRef]
- Barends, T.R.M.; Foucar, L.; Botha, S.; Doak, R.B.; Shoeman, R.L.; Nass, K.; Koglin, J.E.; Williams, G.J.; Boutet, S.; Messerschmidt, M.; et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature 2014, 505, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wacker, D.; Gati, C.; Han, G.W.; James, D.; Wang, D.; Nelson, G.; Weierstall, U.; Katritch, V.; Barty, A.; et al. Serial femtosecond crystallography of G protein-coupled receptors. Science 2013, 342, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhou, X.E.; Gao, X.; He, Y.; Liu, W.; Ishchenko, A.; Barty, A.; White, T.A.; Yefanov, O.; Han, G.W.; et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Nam, K. Sample Delivery Media for Serial Crystallography. Int. J. Mol. Sci. 2019, 20, 1094. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Yun, J.H.; Shi, Y.; Han, J.; Li, X.; Jin, Z.; Kim, T.; Park, J.; Park, S.; Liu, H.; et al. Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers. Int. J. Mol. Sci. 2019, 20, 1943. [Google Scholar] [CrossRef]
- Ciftci, H.I.; Sierra, R.G.; Yoon, C.; Su, Z.; Tateishi, H.; Koga, R.; Kotaro, K.; Yumoto, F.; Senda, T.; Liang, M.; et al. Serial Femtosecond X-Ray Diffraction of HIV-1 Gag MA-IP6 Microcrystals at Ambient Temperature. Int. J. Mol. Sci. 2019, 20, 1675. [Google Scholar] [CrossRef]
- Schmidt, M. Time-Resolved Macromolecular Crystallography at Pulsed X-ray Sources. Int. J. Mol. Sci. 2019, 20, 1401. [Google Scholar] [CrossRef] [PubMed]
- Barty, A.; Kirian, R.A.; Maia, F.R.N.C.; Hantke, M.; Yoon, C.H.; White, T.A.; Chapman, H. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014, 47, 1118–1131. [Google Scholar] [CrossRef]
- Foucar, L.; Barty, A.; Coppola, N.; Hartmann, R.; Holl, P.; Hoppe, U.; Kassemeyer, S.; Kimmel, N.; Küpper, J.; Scholz, M.; et al. CASS-CFEL-ASG software suite. Comput. Phys. Commun. 2012, 183, 2207–2213. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Liu, H. ClickX: A visualization-based program for preprocessing of serial crystallography data. J. Appl. Crystallogr. 2019, 52, 674–682. [Google Scholar]
- White, T.A.; Kirian, R.A.; Martin, A.V.; Aquila, A.; Nass, K.; Barty, A.; Chapman, H.N. CrystFEL: A software suite for snapshot serial crystallography. J. Appl. Crystallogr. 2012, 45, 335–341. [Google Scholar] [CrossRef]
- Hattne, J.; Echols, N.; Tran, R.; Kern, J.; Gildea, R.J.; Brewster, A.S.; Alonso-Mori, R.; Glöckner, C.; Hellmich, J.; Laksmono, H.; et al. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nat. Methods 2014, 11, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, X.; Kirian, R.; Spence, J.C.H.; Liu, H.; Zatsepin, N.A. SPIND: A reference-based auto-indexing algorithm for sparse serial crystallography data. IUCrJ 2019, 6, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Spence, J.C.H. XFEL data analysis for structural biology. Quant. Biol. 2016, 4, 159–176. [Google Scholar] [CrossRef]
- Yun, J.H.; Li, X.; Park, J.H.; Wang, Y.; Ohki, M.; Jin, Z.; Lee, W.; Park, S.Y.; Hu, H.; Li, C.; et al. Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency. J. Biol. Chem. 2019, 294, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Pande, K.; Hutchison, C.D.M.; Groenhof, G.; Aquila, A.; Robinson, J.S.; Tenboer, J.; Basu, S.; Boutet, S.; DePonte, D.P.; Liang, M.; et al. Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science 2016, 352, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Panneels, V.; Tanaka, T.; Jaeger, K.; Carbajo, S.; Furrer, A.; White, T.; Båth, P.; Standfuss, J.; Lane, T.; Wu, W.; et al. Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser. Science 2018, 361, eaat0094. [Google Scholar]
- Wang, D.; Weierstall, U.; Pollack, L.; Spence, J. Double-focusing mixing jet for XFEL study of chemical kinetics. J. Synchrotron Radiat. 2014, 21, 1364–1366. [Google Scholar] [CrossRef] [PubMed]
- Stagno, J.R.; Liu, Y.; Bhandari, Y.R.; Conrad, C.E.; Panja, S.; Swain, M.; Fan, L.; Nelson, G.; Li, C.; Wendel, D.R.; et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature 2017, 541, 242–246. [Google Scholar] [CrossRef]
- Shi, Y.; Yin, K.; Tai, X.; DeMirci, H.; Hosseinizadeh, A.; Hogue, B.G.; Li, H.; Ourmazd, A.; Schwander, P.; Vartanyants, I.A.; et al. Evaluation of the performance of classification algorithms for XFEL single-particle imaging data. IUCrJ 2019, 6, 331–340. [Google Scholar] [CrossRef]
- Rose, M.; Bobkov, S.; Ayyer, K.; Kurta, R.P.; Dzhigaev, D.; Kim, Y.Y.; Morgan, A.J.; Yoon, C.H.; Westphal, D.; Bielecki, J.; et al. Single-particle imaging without symmetry constraints at an X-ray free-electron laser. IUCrJ 2018, 5, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Neutze, R.; Moffat, K. Time-resolved structural studies at synchrotrons and X-ray free electron lasers: Opportunities and challenges. Curr. Opin. Struct. Biol. 2012, 22, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Arnlund, D.; Johansson, L.C.; Wickstrand, C.; Barty, A.; Williams, G.J.; Malmerberg, E.; Davidsson, J.; Milathianaki, D.; DePonte, D.P.; Shoeman, R.L.; et al. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat. Methods 2014, 11, 923–926. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Lee, W. The XFEL Protein Crystallography: Developments and Perspectives. Int. J. Mol. Sci. 2019, 20, 3421. https://doi.org/10.3390/ijms20143421
Liu H, Lee W. The XFEL Protein Crystallography: Developments and Perspectives. International Journal of Molecular Sciences. 2019; 20(14):3421. https://doi.org/10.3390/ijms20143421
Chicago/Turabian StyleLiu, Haiguang, and Weontae Lee. 2019. "The XFEL Protein Crystallography: Developments and Perspectives" International Journal of Molecular Sciences 20, no. 14: 3421. https://doi.org/10.3390/ijms20143421
APA StyleLiu, H., & Lee, W. (2019). The XFEL Protein Crystallography: Developments and Perspectives. International Journal of Molecular Sciences, 20(14), 3421. https://doi.org/10.3390/ijms20143421





