Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973
Abstract
1. Introduction
2. Results
2.1. REPSA Selection of TTHA0973-Binding DNAs
2.2. Identification and Characterization of a TTHA0973-Binding Consensus Sequence
2.3. Identification of Potential TTHA0973-Binding Sites Within the T. thermophilus HB8 Genome
2.4. Validation of Potential TTHA0973-Regulated Genes in T. thermophilus HB8
2.5. Postulated Biological Role for TTHA0973 in T. thermophilus HB8
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. Protein Expression and Purification
4.3. Transcription Factor Consensus Sequence Determination
4.4. Protein-DNA Binding Assays
4.5. Bioinformatic Determination of Candidate Regulated Genes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BLI | Biolayer interferometry |
| EMSA | Electrophoretic mobility shift assay |
| FIMO | Find individual motif occurrences |
| GEO | Gene expression omnibus repository |
| IISRE | Type IIS restriction endonuclease |
| MEME | Multiple Em for motif elicitation |
| REPSA | Restriction endonuclease protection, selection, and amplification |
| SELEX | Systematic evolution of ligands by exponential enrichment |
References
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Katta, H.Y.; Mojica, A.; Chen, I.-M.A.; Kyrpides, N.C.; Reddy, T.B.K. Genomes OnLine Database (GOLD) v.7: Updates and new features. Nucleic Acids Res. 2019, 47, D649–D659. [Google Scholar] [CrossRef] [PubMed]
- Gama-Castro, S.; Salgado, H.; Santos-Zavaleta, A.; Ledezma-Tejeida, D.; Muñiz-Rascado, L.; García-Sotelo, J.S.; Alquicira-Hernández, K.; Martínez-Flores, I.; Pannier, L.; Castro-Mondragón, J.A.; et al. RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016, 44, D133–D143. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Imahori, K. Description of Thermus thermophilus (Yoshida and Oshima) comb. Nov., a non-sporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 1974, 24, 102–112. [Google Scholar] [CrossRef]
- Yokoyama, S.; Hirota, H.; Kigawa, T.; Yabuki, T.; Shirouzu, M.; Terada, T.; Ito, Y.; Matsuo, Y.; Kuroda, Y.; Nishimura, Y.; et al. Structural genomics projects in Japan. Nat. Struct. Mol. Biol. 2000, 7, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Tomita, M.; Itaya, M. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J. Bacteriol. 2010, 192, 5499–5505. [Google Scholar] [CrossRef] [PubMed]
- Tonthat, N.K.; Arold, S.T.; Pickering, B.F.; Van Dyke, M.W.; Liang, S.; Lu, Y.; Beuria, T.K.; Margolin, W.; Schumacher, M.A. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011, 30, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Van Dyke, M.W.; Beyer, M.D.; Clay, E.; Hiam, K.J.; McMurry, J.L.; Xie, Y. Identification of preferred DNA-binding for the Thermus thermophilus transcriptional regulator SbtR by the combinatorial approach REPSA. PLoS ONE 2016, 11, e0159408. [Google Scholar] [CrossRef]
- Lee, M.; Um, H.; Van Dyke, M.W. Identification and characterization of preferred DNA-binding sites for the Thermus thermophilus transcriptional regulator FadR. PLoS ONE 2017, 12, e0184796. [Google Scholar] [CrossRef]
- Agari, Y.; Agari, K.; Sakamoto, K.; Kuramitsu, S.; Shinkai, A. TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation. Microbiology 2011, 157, 1589–1601. [Google Scholar] [CrossRef]
- Agari, Y.; Sakamoto, K.; Yutani, K.; Kuramitsu, S.; Shinkai, A. Structure and function of a TetR family transcriptional regulator, SbtR, from Thermus thermophilus HB8. Proteins 2013, 81, 1166–1178. [Google Scholar] [CrossRef]
- Sakamoto, K.; Agari, Y.; Kuramitsu, S.; Shinkai, A. Phenylacetyl Coenzyme A Is an Effector Molecule of the TetR Family Transcriptional Repressor PaaR from Thermus thermophilus HB8. J. Bacteriol. 2011, 193, 4388–4395. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agari, Y.; Sakamoto, K.; Kuramitsu, S.; Shinkai, A. Transcriptional Repression Mediated by a TetR Family Protein, PfmR, from Thermus thermophilus HB8. J. Bacteriol. 2012, 194, 4630–4641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agari, Y.; Kuramitsu, S.; Shinkai, A. Identification of novel genes regulated by the oxidative stress-responsive transcriptional activator SdrP in Thermus thermophilus HB8. FEMS Microbiol. Lett. 2010, 313, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Fujita, N.; Maeda, M.; Ishihama, A. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells 2005, 10, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Van Dyke, M.W.; Van Dyke, N.; Sunavala-Dossabhoy, G. REPSA: General combinatorial approach for identifying preferred ligand-DNA binding sequences. Methods 2007, 42, 118–127. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Van Dyke, M.W.; Cox, J. Electrophoretic mobility shift assays using infrared-fluorescent DNA probes. Protocols.io 2018, mbdc2i6. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Li, R.W., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Baerends, R.J.; Smits, W.K.; de Jong, A.; Hamoen, L.W.; Kok, J.; Kuipers, O.P. Genome2D: A visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 2004, 5, R37. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Ciria, R.; Martinez-Guerrero, C.E.; Merino, E. ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res. 2012, 40, D627–D631. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2017, bbx085. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
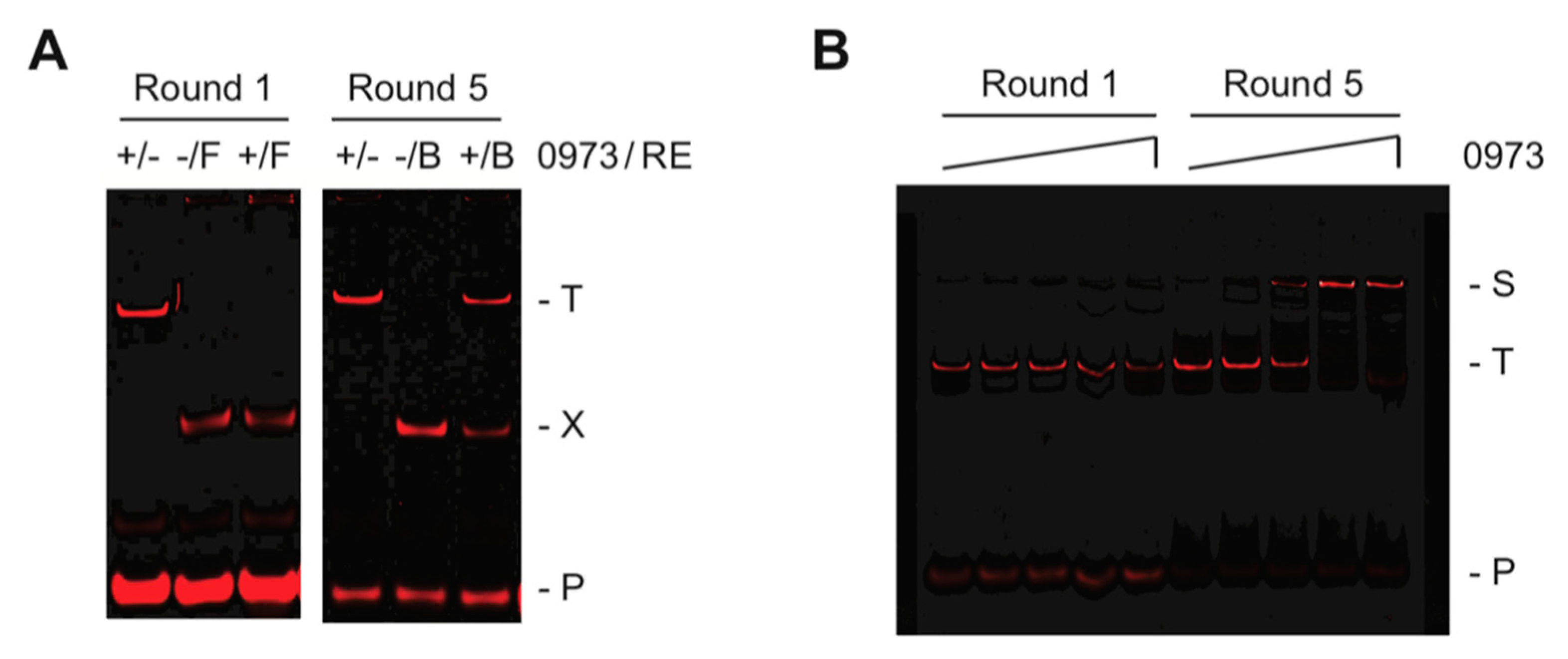
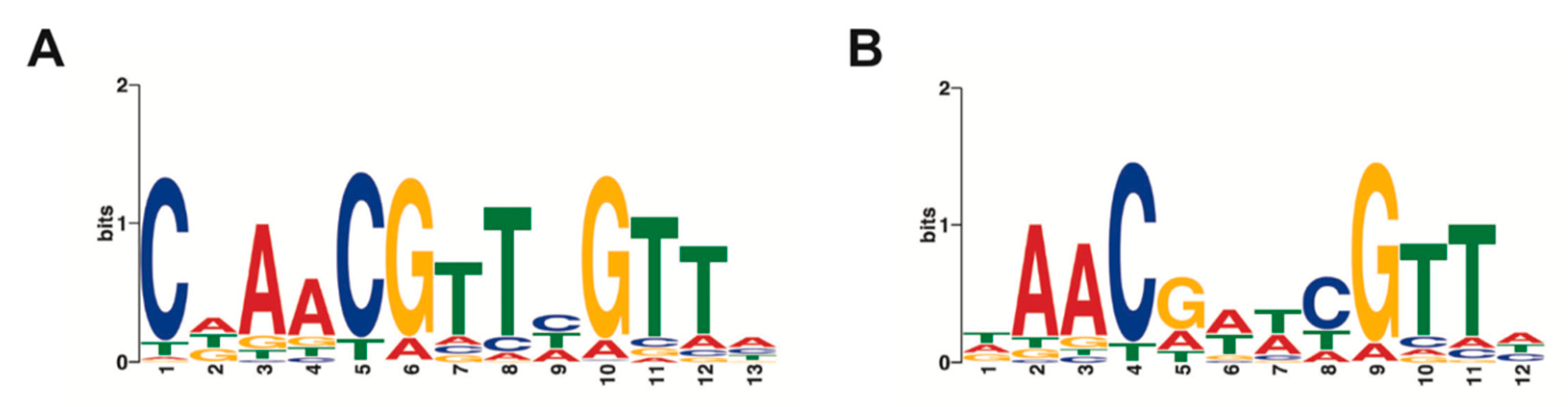
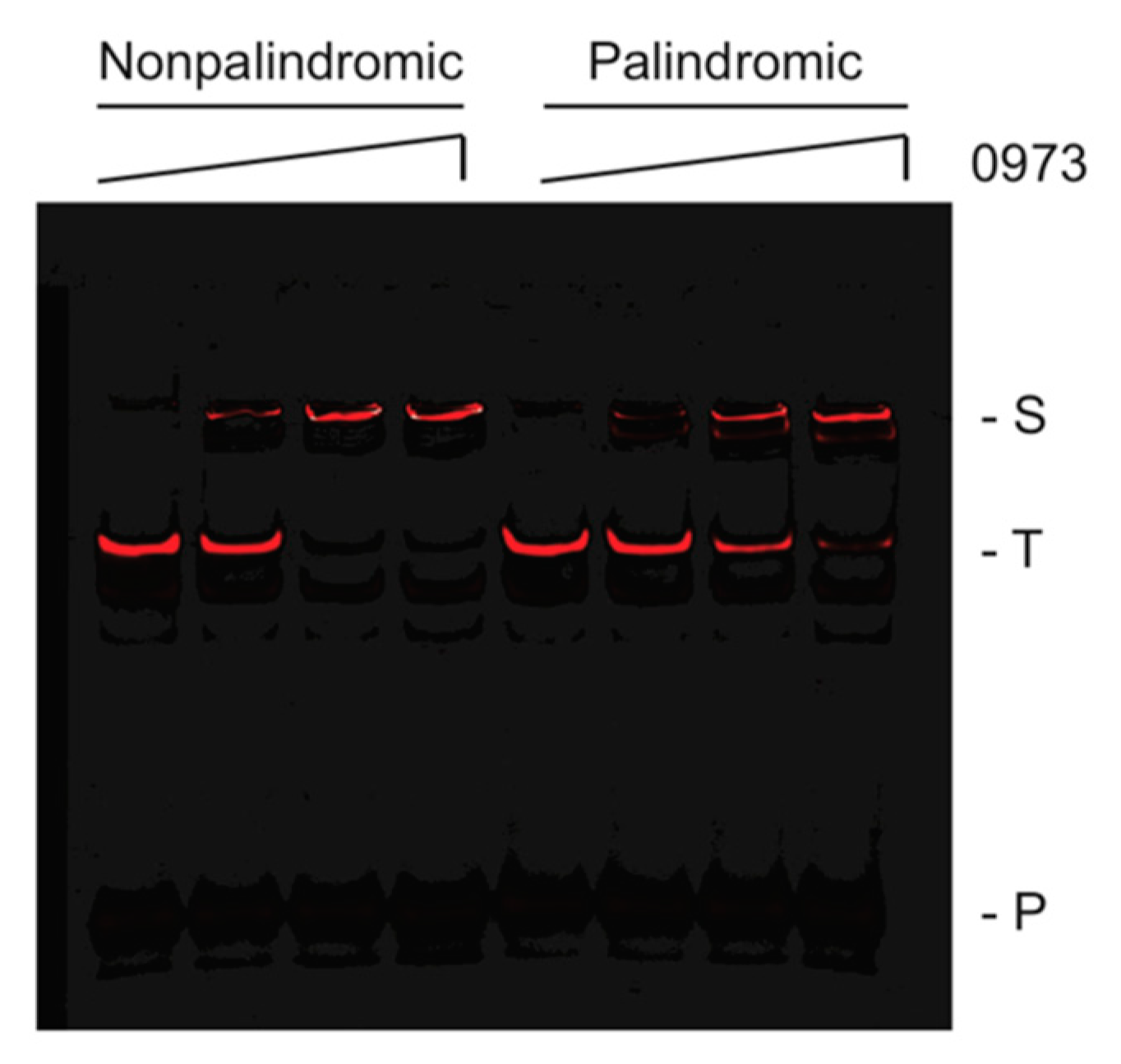
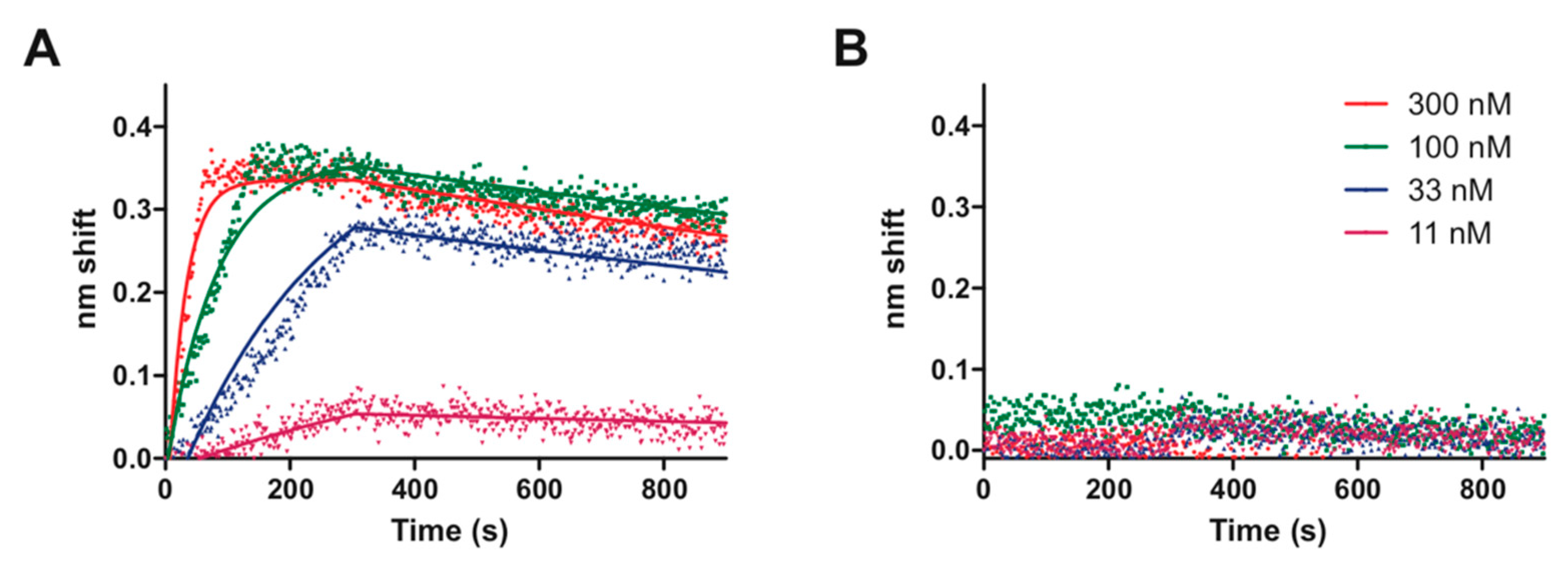

| Name | Sequence | kon (M−1·s−1) | koff (s−1) | KD (M) | R2 |
|---|---|---|---|---|---|
| wt | AACAAACGTTTGTT | 17494 | 9.446 × 10−5 | 5.400 × 10−9 | 0.9716 |
| m1 | tACAAACGTTTGTT | 31516 | 1.422 × 10−4 | 4.512 × 10−9 | 0.9665 |
| m2 | AtCAAACGTTTGTT | 26873 | 1.627 × 10−4 | 6.054 × 10−9 | 0.9681 |
| m3 | AAgAAACGTTTGTT | 24948 | 3.086 × 10−4 | 1.237 × 10−8 | 0.9656 |
| m4 | AACcAACGTTTGTT | 22865 | 1.511 × 10−4 | 6.609 × 10−9 | 0.9803 |
| m5 | AACAcACGTTTGTT | 25404 | 9.445 × 10−5 | 3.718 × 10−9 | 0.9660 |
| m6 | AACAAcCGTTTGTT | 27448 | 1.286 × 10−4 | 4.686 × 10−9 | 0.9440 |
| m7 | AACAAAaGTTTGTT | 26359 | 4.372 × 10−4 | 1.659 × 10−8 | 0.9659 |
| Start | End | P-Value | Q-Value | Sequence | Location | Int? | Gene | Operon |
|---|---|---|---|---|---|---|---|---|
| 229490 | 229503 | 3.65 × 10−6 | 1 | CACCACCGTTTGTT | +1255 | N | TTHA0236 | 5/6 |
| 615296 | 615309 | 3.65 × 10−6 | 1 | ACCTAACGTTCGCT | +733 | N | TTHA0647 | 4/4 |
| 909145 | 909158 | 3.65 × 10−6 | 1 | AACGGCCGTTAGTT | −46 | Y | TTHA0963 | 1/5 |
| 917390 | 917403 | 3.65 × 10−6 | 1 | AACAAACGACCGTT | −3 | Y | TTHA0973 | 1/6 |
| 220957 | 220970 | 5.86 × 10−6 | 1 | AAGAAAGGTTAGAT | +175 | ~ | TTHB214 | 2/2 |
| 584275 | 584288 | 1.02 × 10−5 | 1 | AACTAAGGATTGGT | −1 | Y | TTHA0615 | 2/3 |
| 615792 | 615805 | 1.02 × 10−5 | 1 | ACCTAGCGTTACTT | +237 | N | TTHA0647 | 4/4 |
| 59241 | 59254 | 1.21 × 10−5 | 1 | AACCAGCCTTCCTT | +88 | N | TTHB067 | N |
| 143843 | 143856 | 1.21 × 10−5 | 1 | AAAGAACCTTCGCT | +131 | N | TTHB153 | N |
| 260398 | 260411 | 1.21 × 10−5 | 1 | AATGACCCTTGGTT | −41 | Y | TTHA0272 | 1/2 |
| Gene | Sequence | kon (M−1·s−1) | koff (s−1) | KD (M) | R2 |
|---|---|---|---|---|---|
| TTHA0236 | CACCACCGTTTGTT | 100803 | 8.463 × 10−4 | 8.396 × 10−9 | 0.9631 |
| TTHA0647 | ACCTAACGTTCGCT | 102574 | 4.044 × 10−4 | 3.943 × 10−9 | 0.9588 |
| TTHA0963 | AACGGCCGTTAGTT | 135763 | 5.176 × 10−4 | 3.813 × 10−9 | 0.9569 |
| TTHA0973 | AACAAACGACCGTT | 140260 | 4.286 × 10−4 | 3.056 × 10−9 | 0.9632 |
| TTHB214 | AAGAAAGGTTAGAT | nab | |||
| TTHA0615 | AACTAAGGATTGGT | 60936 | 2.300 × 10−4 | 3.774 × 10−8 | 0.9586 |
| TTHA0647b | ACCTAGCGTTACTT | 38264 | 3.807 × 10−4 | 9.950 × 10−8 | 0.9741 |
| TTHB067 | AACCAGCCTTCCTT | Nab | |||
| TTHB153 | AAAGAACCTTCGCT | 95001 | 2.833 × 10−4 | 2.982 × 10−7 | 0.9018 |
| TTHA0272 | AATGACCCTTGGTT | nab |
| Gene | LogFC | Adj. P-Value | P-Value | t | B |
|---|---|---|---|---|---|
| TTHA0236 | −0.807 | 0.132 | 4.84 × 10−2 | −2.42 | −4.48 |
| TTHA0647 | −0.386 | 0.213 | 1.01 × 10−1 | −1.90 | −5.21 |
| TTHA0963 | 2.09 | 0.0247 | 7.84 × 10−4 | 5.85 | −0.137 |
| TTHA0973 | 6.68 | 0.00594 | 1.57 × 10−5 | 11.2 | 3.80 |
| TTHB214 | −0.335 | 0.725 | 6.05 × 10−1 | −0.543 | −6.68 |
| TTHA0615 | 0.851 | 0.0539 | 8.65 × 10−3 | 3.69 | −2.68 |
| TTHB067 | −0.666 | 0.0695 | 1.58 × 10−2 | −3.22 | −3.32 |
| TTHB153 | 0.304 | 0.501 | 3.55 × 10−1 | 0.994 | −6.33 |
| TTHA0272 | −0.0794 | 0.870 | 7.97 × 10−1 | −0.269 | −6.81 |
| Promoter | Operon | Gene | Role | LogFC | Adj P-Value |
|---|---|---|---|---|---|
| ~ | 1 | TTHA0963 | Pseudogene | 2.09 | 7.84 × 10−4 |
| 2 | TTHA0965 | phenylacetic acid degradation protein PaaI | 2.62 | 1.00 × 10−3 | |
| 3 | TTHA0966 | phenylacetyl-CoA ligase | 2.41 | 8.56 × 10−4 | |
| 4 | TTHA0967 | hypothetical protein | 2.58 | 2.19 × 10−2 | |
| Y | 1 | TTHA0973 | TetR family transcriptional regulator PaaR | 6.68 | 1.57 × 10−5 |
| 2 | TTHA0972 | phenylacetate-CoA oxygenase subunit PaaA | 3.49 | 1.09 × 10−5 | |
| 3 | TTHA0971 | phenylacetic acid degradation protein PaaB | 3.98 | 8.23 × 10−7 | |
| 4 | TTHA0970 | phenylacetic acid degradation protein PaaC | 4.11 | 1.81 × 10−7 | |
| 5 | TTHA0969 | phenylacetic acid degradation protein PaaD | 3.74 | 7.58 × 10−7 | |
| 6 | TTHA0968 | bifunctional aldehyde dehydrogenase/enoyl-CoA hydratase | 4.07 | 5.78 × 10−7 | |
| Y | 1 | TTHA0615 | ATP-dependent Clp protease, protease subunit | 0.85 | 8.65 × 10−3 |
| 2 | TTHA0616 | ATP-dependent protease ATP-binding subunit ClpX | −0.032 | 8.74 × 10−1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cox, J.S.; Moncja, K.; Mckinnes, M.; Van Dyke, M.W. Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973. Int. J. Mol. Sci. 2019, 20, 3336. https://doi.org/10.3390/ijms20133336
Cox JS, Moncja K, Mckinnes M, Van Dyke MW. Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973. International Journal of Molecular Sciences. 2019; 20(13):3336. https://doi.org/10.3390/ijms20133336
Chicago/Turabian StyleCox, James Shell, Kristi Moncja, Mykala Mckinnes, and Michael W. Van Dyke. 2019. "Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973" International Journal of Molecular Sciences 20, no. 13: 3336. https://doi.org/10.3390/ijms20133336
APA StyleCox, J. S., Moncja, K., Mckinnes, M., & Van Dyke, M. W. (2019). Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973. International Journal of Molecular Sciences, 20(13), 3336. https://doi.org/10.3390/ijms20133336







