Functional Analysis of M-Locus Protein Kinase Revealed a Novel Regulatory Mechanism of Self-Incompatibility in Brassica napus L.
Abstract
1. Introduction
2. Results
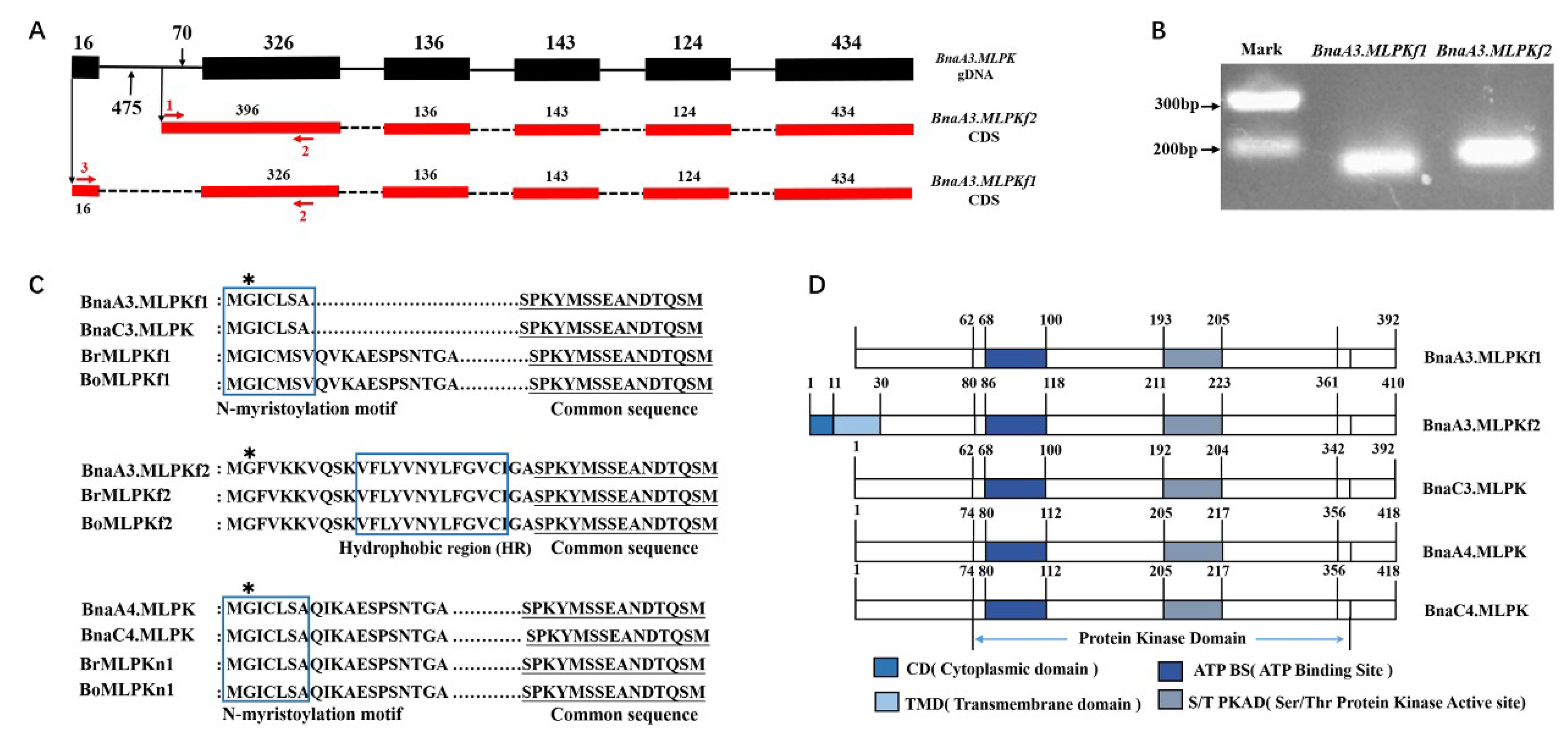
2.1. Cloning and Sequence Analysis of BnaMLPKs from Self-Incompatibile B. napus
2.2. Sequence Analysis of BnaMLPK Transcripts
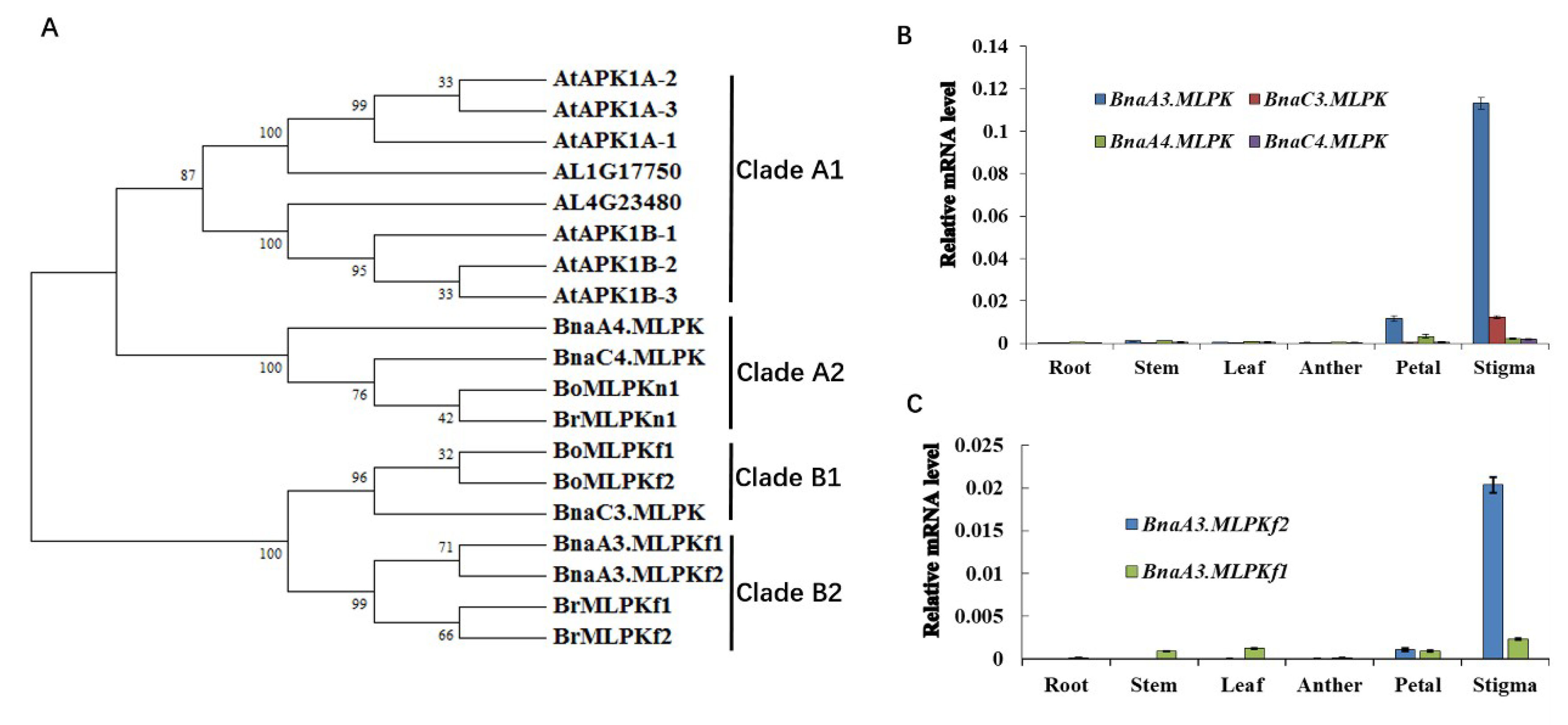
2.3. Phylogenetic and Tissue-Specific Expression Analysis of BnaMLPKs
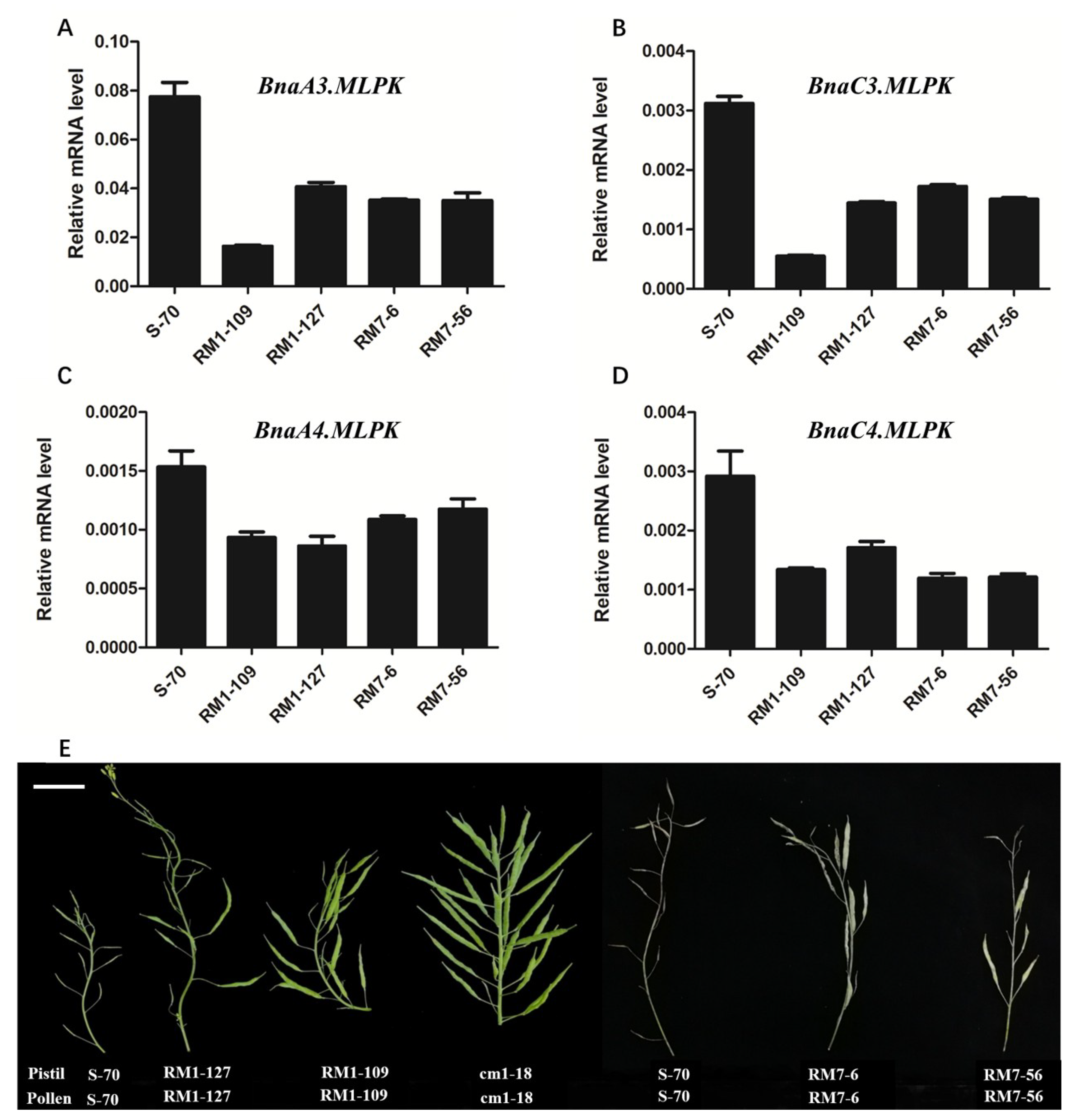
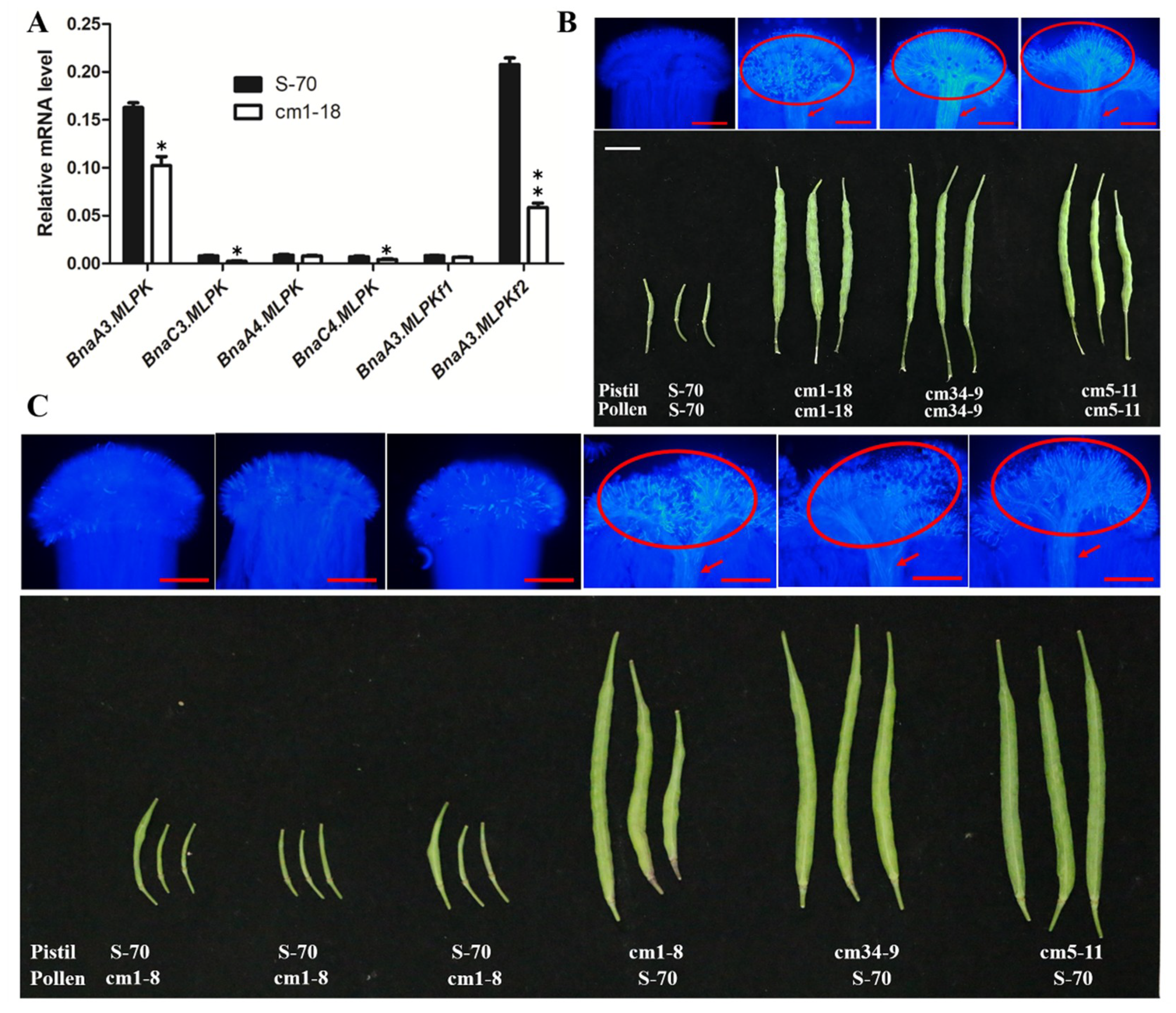
2.4. RNAi Knockdown of BnaMLPKs Partially Suppressed SI Response in B. napus
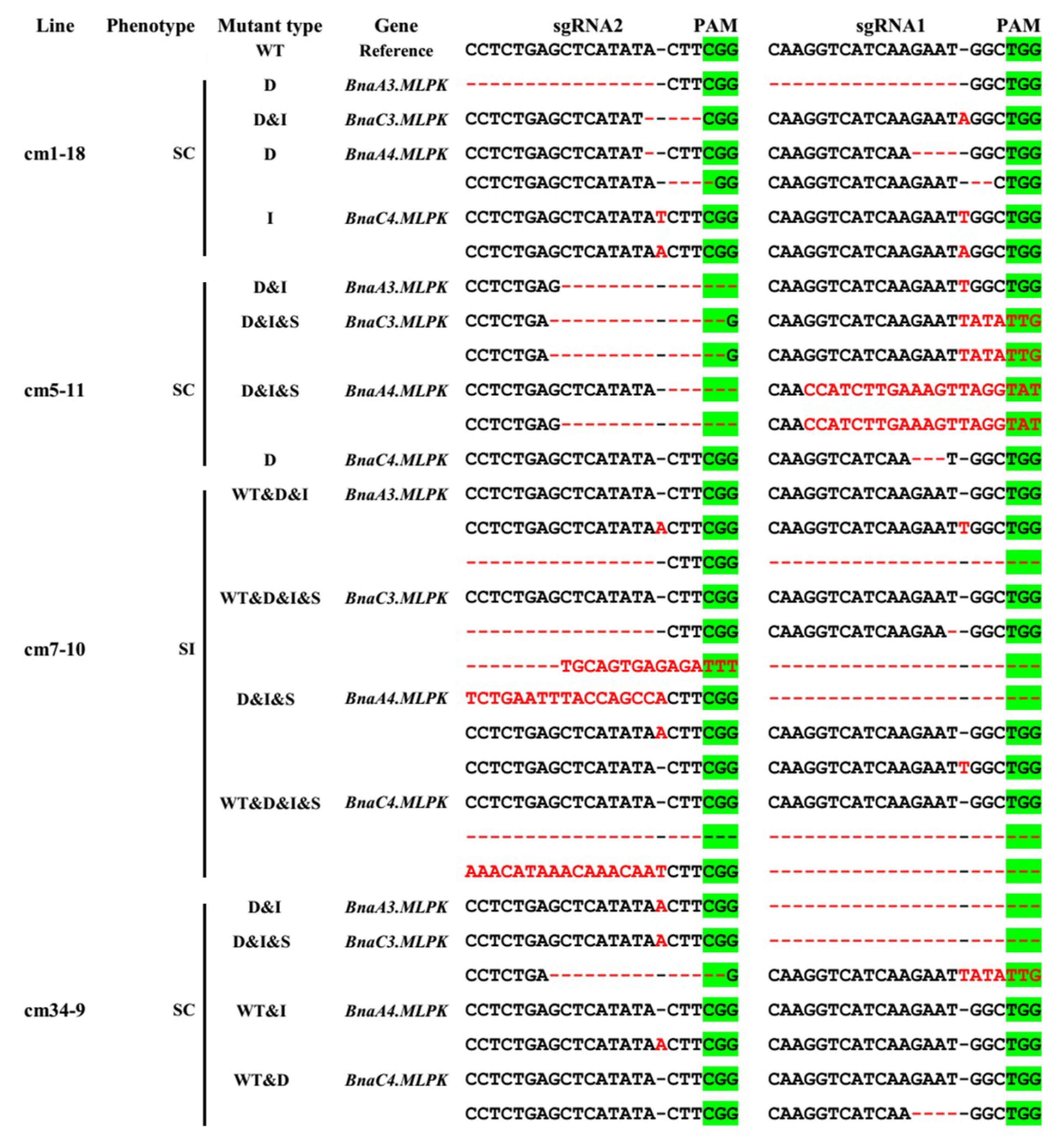
2.5. Knockout Mutant of BnaMLPKs Created Using CRISPR/Cas9 System Completely Breaks Down SI Response in B. napus
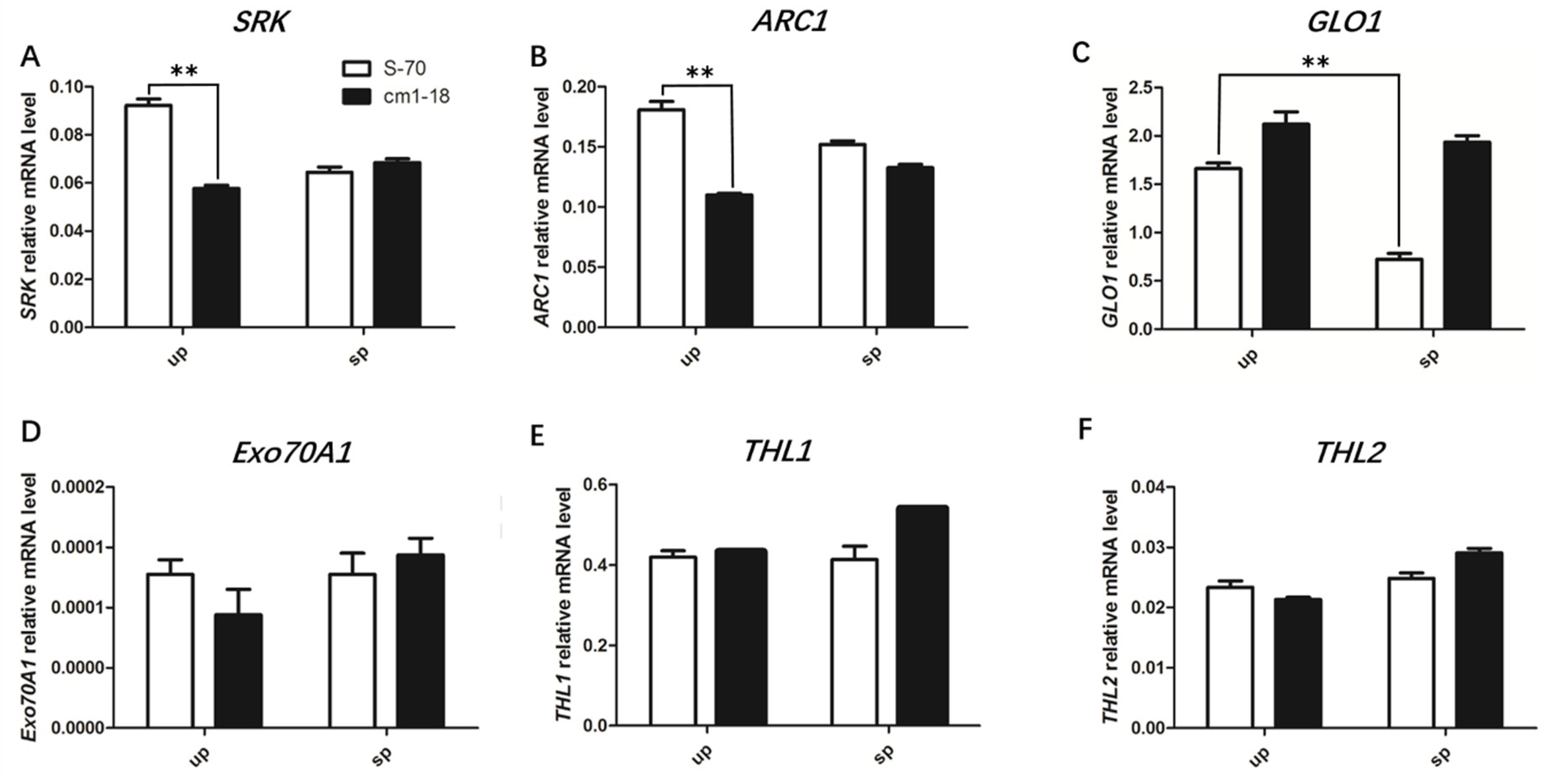
2.6. Expression of SI-Related Genes Changed in bnamlpk Mutant
3. Discussion
3.1. BnaMLPK is a Positive Regulator of SI Response in B. napus
3.2. Alternative Splicing Involved in MLPK-Regulated SI in B. napus
3.3. BnaMLPKs Regulate SI Responses by Influencing the Expression of SCR-SRK Pathway Components
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Sequence Cloning of BnaMLPKs
4.3. Tissue-Specific Expression Analysis of BanMLPKs
4.4. Phylogenetic Analysis
4.5. Plasmid Construction and B. napus Transformation
4.6. Mutant Screening and Validation of Genome Editing
4.7. Aniline Blue Staining Assay
4.8. Self-Incompatibility Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nettancourt, D.D. Incompatibility and Incongruity in Wild and Cultivated Plants, 2nd ed.; Springer: New York, NY, USA, 2001; Volume 3, p. 347. [Google Scholar]
- Bateman, A.J. Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 1955, 9, 52–68. [Google Scholar] [CrossRef]
- Stein, J.C.; Howlett, B.; Boyes, D.C.; Nasrallah, M.E.; Nasrallah, J.B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA. 1991, 88, 8816–8820. [Google Scholar] [CrossRef]
- Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. The Male Determinant of Self-Incompatibility in Brassica. Science 1999, 286, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Kai, N.; Hirose, T.; Fukui, K.; Nishio, T.; Takayama, S.; Isogai, A.; Watanabe, M.; Hinata, K. Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S (9) haplotype of Brassica campestris (syn. rapa). Genetics 1999, 153, 391–403. [Google Scholar] [PubMed]
- Kachroo, A.; Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 2001, 293, 1824–1826. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Shimosato, H.; Shiba, H.; Funato, M.; Che, F.-S.; Watanabe, M.; Iwano, M.; Isogai, A. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature 2001, 413, 534–538. [Google Scholar] [CrossRef]
- Kachroo, A.; Nasrallah, M.E.; Nasrallah, J.B. Self-Incompatibility in the Brassicaceae. Plant Cell 2002, 14 (Suppl. 1), S227–S238. [Google Scholar] [CrossRef]
- Haffani, Y.; Gaude, T.; Cock, J.; Goring, D. Antisense suppression of thioredoxin h mRNA in Brassica napus cv Westar pistils causes a low level constitutive pollen rejection response. Plant Mol. Biol. 2004, 55, 619–630. [Google Scholar] [CrossRef]
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, A.; Takayama, S. Two Distinct Forms of M-Locus Protein Kinase Localize to the Plasma Membrane and Interact Directly with S-Locus Receptor Kinase to Transduce Self-Incompatibility Signaling in Brassica rapa. Plant Cell 2007, 19, 3961–3973. [Google Scholar] [CrossRef]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 Is an E3 Ubiquitin Ligase and Promotes the Ubiquitination of Proteins during the Rejection of Self-Incompatible Brassica Pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef]
- Samuel, M.A.; Chong, Y.T.; Haasen, K.E.; Aldea-Brydges, M.G.; Stone, S.L.; Goring, D.R. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 2009, 21, 2655–2671. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen Acceptance or Rejection: A Tale of Two Pathways. Trends Plant Sci. 2016, 21, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 2015, 1, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. The glyoxalase system: New developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990, 269, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, J.B.; Nasrallah, M.E. Robust Self-Incompatibility in the Absence of a Functional ARC1 Gene in Arabidopsis thaliana. Plant Cell 2014, 26, 3838–3841. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goring, D.R.; Indriolo, E.; Samuel, M.A. The ARC1 E3 Ligase Promotes a Strong and Stable Self-Incompatibility Response in Arabidopsis Species: Response to the Nasrallah and Nasrallah Commentary. Plant Cell 2014, 26, 3842–3846. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nasrallah, J.B. In planta assessment of the role of thioredoxin h proteins in the regulation of S-locus receptor kinase signaling in transgenic Arabidopsis thaliana. Plant Physiol. 2013, 163, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Shiba, H.; Iwano, M.; Che, F.-S.; Watanabe, M.; Isogai, A.; Takayama, S. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 2004, 303, 1516–1519. [Google Scholar] [CrossRef]
- Kitashiba, H.; Liu, P.; Nishio, T.; Nasrallah, J.B.; Nasrallah, M.E. Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 18173–18178. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Gao, Q.; Shi, S.; Liu, Y.; Pu, Q.; Liu, X.; Zhang, Y.; Zhu, L. Identification of a novel MLPK homologous gene MLPKn1 and its expression analysis in Brassica oleracea. Plant Reprod. 2016, 29, 239–250. [Google Scholar] [CrossRef]
- Hanks, S.K.; Quinn, A.M. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991, 200, 38–62. [Google Scholar]
- Boisson, B.; Giglione, C.; Meinnel, T. Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 2003, 278, 43418–43429. [Google Scholar] [CrossRef]
- Safavian, D.; Zayed, Y.; Indriolo, E.; Chapman, L.; Ahmed, A.; Goring, D. RNA silencing of exocyst genes in the stigma impairs the acceptance of compatible pollen in Arabidopsis. Plant Physiol. 2015, 169, 2526–2538. [Google Scholar] [CrossRef]
- Bower, M.S.; Matias, D.D.; Fernandes-Carvalho, E.; Mazzurco, M.; Gu, T.; Rothstein, S.J.; Goring, D.R. Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 1996, 8, 1641–1650. [Google Scholar]
- Haasen, K.E.; Goring, D.R. The recognition and rejection of self-incompatible pollen in the Brassicaceae. Bot. Stud. 2010, 51, 1–6. [Google Scholar]
- Ivanov, R.; Fobis-Loisy, I.; Gaude, T. When no means no: Guide to Brassicaceae self-incompatibility. Trends Plant Sci. 2010, 15, 387–394. [Google Scholar] [CrossRef]
- Tantikanjana, T.; Nasrallah, M.E.; Nasrallah, J.B. Complex networks of self-incompatibility signaling in the Brassicaceae. Curr. Opin. Plant Biol. 2010, 13, 520–526. [Google Scholar] [CrossRef]
- Li, Y.; Dai, C.; Hu, C.; Liu, Z.; Kang, C. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 2017, 90, 164. [Google Scholar] [CrossRef]
- Zavolan, M.; Van, N.E.; Gaasterland, T. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 2002, 12, 1377–1385. [Google Scholar] [CrossRef]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef]
- Nasrallah, J.B. Self-incompatibility in the Brassicaceae: Regulation and mechanism of self-recognition. Curr. Top. Dev. Biol. 2019, 131, 435–452. [Google Scholar] [CrossRef]
- Silva, N.F.; Stone, S.L.; Christie, L.N.; Sulaman, W.; Nazarian, K.A.; Burnett, L.A.; Arnoldo, M.A.; Rothstein, S.J.; Goring, D.R. Expression of the S receptor kinase in self-compatible Brassica napus cv. Westar leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Mol. Genet. Genom. MGG 2001, 265, 552–559. [Google Scholar]
- Goring, D.R.; Glavin, T.L.; Schafer, U.; Rothstein, S.J. An S receptor kinase gene in self-compatible Brassica napus has a 1-bp deletion. Plant Cell 1993, 5, 531–539. [Google Scholar] [CrossRef]
- Gao, C.; Ma, C.; Zhang, X.; Li, F.; Zhang, J.; Zhai, W.; Wang, Y.; Tu, J.; Shen, J.; Fu, T. The genetic characterization of self-incompatibility in a Brassica napus line with promising breeding potential. Mol. Breed. 2013, 31, 485–493. [Google Scholar] [CrossRef]
- Sambrock, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 3, pp. 639–646. [Google Scholar]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Gao, X.; Yan, P.; Shen, W.; Li, X.; Zhou, P.; Li, Y. Modular construction of plasmids by parallel assembly of linear vector components. Anal. Biochem. 2013, 437, 172–177. [Google Scholar] [CrossRef]
- Franklin, T.M.; Oldknow, J.; Trick, M. SLR1 function is dispensable for both self-incompatible rejection and self-compatible pollination processes in Brassica. Sex. Plant Reprod. 1996, 9, 203–208. [Google Scholar] [CrossRef]
- Yang, H.; Wu, J.J.; Tang, T.; Liu, K.D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Z.; Zhang, T.; Zhou, G.; Duan, Z.; Li, B.; Dou, S.; Liang, X.; Tu, J.; Shen, J.; et al. Mechanism of Salt-Induced Self-Compatibility Dissected by Comparative Proteomic Analysis in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 1652. [Google Scholar] [CrossRef]
| Gene | BnaA3.MLPK | BnaC3.MLPK | BnaA4.MLPK | BnaC4.MLPK |
|---|---|---|---|---|
| BnaA3.MLPK | - | 98.47% | 74.88% | 74.88% |
| BnaC3.MLPK | 98.56% | - | 75.60% | 75.60% |
| BnaA4.MLPK | 84.73% | 84.82% | - | 99.76% |
| BnaC4.MLPK | 84.82% | 84.90% | 98.09% | - |
| Transcripts | BrMLPKf1 | BrMLPKf2 | BrMLPKn | BoMLPKf1 | BoMLPKf2 | BoMLPKn |
|---|---|---|---|---|---|---|
| BnaA3.MLPK | 99.83% | 99.32% | 81.34% | 98.64% | 93.29% | 84.82% |
| BnaC3.MLPK | 98.56% | 98.05% | 83.55% | 98.90% | 94.14% | 84.90% |
| BnaA4.MLPK | 85.93% | 84.02% | 91.77% | 86.01% | 80.03% | 98.09% |
| BnaC4.MLPK | 86.01% | 84.10% | 91.69% | 86.09% | 80.03% | 100.00% |
| Target Gene | sgRNA | Number of Plants Examined | Number of Plants with Mutations | Mutation Rate (%) | Homozygous Mutations | |
|---|---|---|---|---|---|---|
| Number | Rate (%) | |||||
| BnaA3.MLPK | sgRNA1 | 6 | 6 | 100.0 | 1 | 0.17 |
| sgRNA2 | 6 | 100.0 | 0 | 0 | ||
| BnaC3.MLPK | sgRNA1 | 6 | 6 | 100.0 | 0 | 0 |
| sgRNA2 | 6 | 100.0 | 0 | 0 | ||
| BnaA4.MLPK | sgRNA1 | 6 | 5 | 83.3 | 0 | 0 |
| sgRNA2 | 5 | 83.3 | 0 | 0 | ||
| BnaC4.MLPK | sgRNA1 | 6 | 6 | 100.0 | 0 | 0 |
| sgRNA2 | 4 | 66.7 | 0 | 0 | ||
| Target Gene | Sites | Number of Examined Plants | Genotype | ||||
|---|---|---|---|---|---|---|---|
| Homozygote | Heterozygote | Bi-allele | Chimera | WT | |||
| BnaA3.MLPK | sgRNA1 | 6 | 1 (16.7%) | 3 (50.0%) | 2 (33.3%) | ||
| sgRNA2 | 6 | 3 (50.0%) | 2 (33.3%) | ||||
| BnaC3.MLPK | sgRNA1 | 6 | 2 (33.3%) | 4 (66.7%) | |||
| sgRNA2 | 6 | 4 (66.7%) | 2 (33.3%) | ||||
| BnaA4.MLPK | sgRNA1 | 6 | 1 (16.7%) | 1 (16.7%) | 1 (16.7%) | 2 (33.3%) | 1 (16.7%) |
| sgRNA2 | 6 | 1 (16.7%) | 1 (16.7%) | 3 (50.0%) | 1 (16.7%) | ||
| BnaC4.MLPK | sgRNA1 | 6 | 1 (16.7%) | 2 (16.7%) | 3 (50.0%) | ||
| sgRNA2 | 6 | 1 (16.7%) | 1 (16.7%) | 2 (33.3%) | 2 (33.3%) | ||
| Total | 48 | 5 (10.4%) | 5 (10.4%) | 21 (43.8%) | 12 (25.0%) | 4 (8.3%) | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Yang, Y.; Li, B.; Liu, Z.; Khan, F.; Zhang, T.; Zhou, G.; Tu, J.; Shen, J.; Yi, B.; et al. Functional Analysis of M-Locus Protein Kinase Revealed a Novel Regulatory Mechanism of Self-Incompatibility in Brassica napus L. Int. J. Mol. Sci. 2019, 20, 3303. https://doi.org/10.3390/ijms20133303
Chen F, Yang Y, Li B, Liu Z, Khan F, Zhang T, Zhou G, Tu J, Shen J, Yi B, et al. Functional Analysis of M-Locus Protein Kinase Revealed a Novel Regulatory Mechanism of Self-Incompatibility in Brassica napus L. International Journal of Molecular Sciences. 2019; 20(13):3303. https://doi.org/10.3390/ijms20133303
Chicago/Turabian StyleChen, Fang, Yong Yang, Bing Li, Zhiquan Liu, Fawad Khan, Tong Zhang, Guilong Zhou, Jinxing Tu, Jinxiong Shen, Bin Yi, and et al. 2019. "Functional Analysis of M-Locus Protein Kinase Revealed a Novel Regulatory Mechanism of Self-Incompatibility in Brassica napus L." International Journal of Molecular Sciences 20, no. 13: 3303. https://doi.org/10.3390/ijms20133303
APA StyleChen, F., Yang, Y., Li, B., Liu, Z., Khan, F., Zhang, T., Zhou, G., Tu, J., Shen, J., Yi, B., Fu, T., Dai, C., & Ma, C. (2019). Functional Analysis of M-Locus Protein Kinase Revealed a Novel Regulatory Mechanism of Self-Incompatibility in Brassica napus L. International Journal of Molecular Sciences, 20(13), 3303. https://doi.org/10.3390/ijms20133303





