QTL Analysis of Resistance to High-Intensity UV-B Irradiation in Soybean (Glycine max [L.] Merr.)
Abstract
1. Introduction
2. Results
2.1. Phenotypic Evaluation of UV-B Stress Resistance in a RIL Population of Cheongja 3 × Buseok
2.2. Genotyping-by-Sequencing Analysis and Genetic Linkage Map Construction
2.3. Identification of QTLs for UV-B Resistance
2.4. SNPs and Gene Expression Differences in the QTLs in the Mapping Parents
2.5. Comparisons of UV-B Stress Resistance QTLs with Known Stress-Related QTLs Based on Synteny
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. UV-B Treatment and Phenotypic Evaluation
4.3. Genotyping-by-Sequencing
4.4. Sequence Analysis, Genetic Map Construction, and QTL Analysis
4.5. SNP Survey and qRT-PCR Analysis of Genes in the QTLs for UV-B Stress Resistance
4.6. Co-Localization of Stress-Related QTLs with UV-B Resistance QTLs
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| UV-B | Ultraviolet-B |
| QTL | Quantitative trait locus |
| RIL | Recombinant inbred line |
| LOD | Logarithm of odds |
| GBS | Genotyping-by-sequencing |
| SNP | Single nucleotide polymorphism |
| WGD | Whole genome duplication |
| SCN | Soybean cyst nematode |
| SPTBN | Spectrin beta chain, brain |
| bZIP TF21/TGA9 | bZIP transcription factor21/TGACC motif-binding 9 |
| LRR | Leucine-rich repeat |
References
- World Meteorological Organization, 2008. WMO Statement on the Status of the Global Climate in 2007. WMO-No. 1031. Available online: https://library.wmo.int/doc_num.php?explnum_id=3457 (accessed on 20 February 2019).
- Petrescu, R.V.; Aversa, R.; Apicella, A.; Petrescu, F.I. NASA sees first in 2018 the direct proof of ozone hole recovery. J. Aircr. Spacecr. Technolog. 2018, 2, 53–64. [Google Scholar] [CrossRef]
- Rozema, J.; van de Staaij, J.; Björn, L.O.; Caldwell, M. UV-B as an environmental factor in plant life: Stress and regulation. Trends Ecol. Evolut. 1997, 12, 22–28. [Google Scholar] [CrossRef]
- Frohnmeyer, H.; Staiger, D. Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 2003, 133, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- UNEP. 2010 Annual Report. 2011. Available online: https://wedocs.unep.org/handle/20.500.11822/7915 (accessed on 20 February 2019).
- Murali, N.; Teramura, A.H.; Randall, S.K. Response differences between two soybean cultivars with contrasting UV-B radiation sensitivities. Photochem. Photobiol. 1988, 48, 653–657. [Google Scholar] [CrossRef]
- Teramura, A.H.; Sullivan, J.H.; Lydon, J. Effects of UV-B radiation on soybean yield and seed quality: A 6-year field study. Physiol. Plantarum. 1990, 80, 5–11. [Google Scholar] [CrossRef]
- Baroniya, S.S.; Kataria, S.; Pandey, G.P.; Guruprasad, K.N. Intraspecific variation in sensitivity to ambient ultraviolet-B radiation in growth and yield characteristics of eight soybean cultivars grown under field conditions. Braz. J. Plant Physiol. 2011, 23, 197–202. [Google Scholar] [CrossRef]
- Shim, H.C.; Ha, B.K.; Yoo, M.; Kang, S.T. Detection of quantitative trait loci controlling UV-B resistance in soybean. Euphytica 2015, 202, 109–118. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, S.; Ha, B.K.; Kang, S.T. Positional mapping and identification of novel quantitative trait locus responsible for UV-B radiation tolerance in soybean (Glycine max [L.] Merr.). Mol. Breed. 2016, 36, 1–10. [Google Scholar] [CrossRef]
- Kim, K.D.; Yun, M.Y.; Shin, J.H.; Kang, Y.J.; Kim, M.Y.; Lee, S.H. Underlying genetic variation in the response of cultivated and wild soybean to enhanced ultraviolet-B radiation. Euphytica 2015, 202, 207–217. [Google Scholar] [CrossRef]
- Yoon, M.Y.; Kim, M.Y.; Shim, S.; Kim, K.D.; Ha, J.; Shin, J.H.; Lee, S.H. Transcriptomic profiling of soybean in response to high-intensity UV-B irradiation reveals stress defense signaling. Front. Plant Sci. 2016, 7, 1917. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Knight, H.; Knight, M.R. Abiotic stress signaling pathways: Specificity and cross-talk. Trends Plant Sci. 2001, 6, 262–267. [Google Scholar] [CrossRef]
- Jordan, B.R. Review: Molecular response of plant cells to UV-B stress. Funct. Plant Biol. 2002, 29, 909–916. [Google Scholar] [CrossRef]
- Paul, N.D.; Gwynn-Jones, D. Ecological roles of solar UV radiation: Towards an integrated approach. Trends Ecol. Evolut. 2003, 18, 48–55. [Google Scholar] [CrossRef]
- Jenkins, G.I. Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 2009, 60, 407–431. [Google Scholar] [CrossRef]
- Heijde, M.; Ulm, R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl. Acad. Sci. USA 2013, 110, 1113–1118. [Google Scholar] [CrossRef]
- Müller-Xing, R.; Xing, Q.; Goodrich, J. Footprints of the sun: Memory of UV and light stress in plants. Front. Plant Sci. 2014, 5, 474. [Google Scholar] [CrossRef]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Changing scenario in plant UV-B research: UV-B from a generic stressor to a specific regulator. J. Photochem. Photobiol. B Biol. 2015, 153, 334–343. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.Y.; Kim, M.Y.; Lee, J.; Lee, T.; Kim, K.H.; Ha, J.; Kim, Y.H.; Lee, S.H. Transcriptomic profiling of soybean in response to UV-B and Xanthomonas axonopodis treatment reveals shared gene components in stress defense pathways. Genes Genom. 2017, 39, 225–236. [Google Scholar] [CrossRef]
- Machnicka, B.; Czogalla, A.; Hryniewicz-Jankowska, A.; Bogusławska, D.M.; Grochowalska, R.; Heger, E.; Sikorski, A.F. Spectrins: A structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim. Biophys. Acta Biomembr 2014, 1838, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Bennett, V.; Baines, A.J. Spectrin and ankyrin-based pathways: Metazoan inventions for integrating cells into tissues. Physiol. Rev. 2001, 81, 1353–1392. [Google Scholar] [CrossRef] [PubMed]
- Drøbak, B.K.; Franklin-Tong, V.E.; Staiger, C.J. The role of the actin cytoskeleton in plant cell signaling. New Phytol. 2004, 163, 13–30. [Google Scholar] [CrossRef]
- Young, K.G.; Kothary, R. Spectrin repeat proteins in the nucleus. BioEssays 2005, 2 7, 144–152. [Google Scholar] [CrossRef]
- Holzinger, A.; De Ruijter, N.; Emons, A.M.; LüTz-Meindl, U. Spectrin-like proteins in green algae (Desmidiaceae). Cell Biol. Int. 1999, 23, 335–344. [Google Scholar] [CrossRef]
- Braun, M. Association of spectrin-like proteins with the actin-organized aggregate of endoplasmic reticulum in the spitzenkörper of gravitropically tip-growing plant cells. Plant Physiol. 2001, 125, 1611–1619. [Google Scholar] [CrossRef][Green Version]
- Pérez-Munive, C.; Moreno Diaz de la Espina, S. Nuclear spectrin-like proteins are structural actin-binding proteins in plants. Biol. Cell 2011, 103, 145–157. [Google Scholar] [CrossRef]
- Michaud, D.; Guillet, G.; Roger, P.A.; Charest, P.M. Identification of a 220-kDa membrane-associated plant cell protein immunologically related to human β-spectrin. FEBS Lett. 1991, 294, 77–80. [Google Scholar] [CrossRef]
- De Ruijter, N.C.A.; Emons, A.M.C. Immunodetection of spectrin in plant cells. Cell Biol. Int. 1993, 17, 169–182. [Google Scholar] [CrossRef]
- Khan, A.A.; Hanada, T.; Mohseni, M.; Jeong, J.J.; Zeng, L.; Gaetani, M.; Li, D.; Reed, B.C.; Speicher, D.W.; Chishti, A.H. Dematin and adducing provide a novel link between the spectrin cytoskeleton and human erythrocyte membrane by directly interacting with glucose transporter-1. J. Biol. Chem. 2008, 283, 14600–14609. [Google Scholar] [CrossRef] [PubMed]
- Salomao, M.; Zhang, X.; Yang, Y.; Lee, S.; Hartwig, J.H.; Chasis, J.A.; Mohandas, N.; An, X. Protein 4.1R-dependent multiprotein complex: New insights into the structural organization of the red blood cell membrane. Proc. Natl. Acad. Sci. USA 2008, 105, 8026–8031. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, B.; Grochowalska, R.; Bogusławska, D.M.; Sikorski, A.F.; Lecomte, M.C. Spectrin-based skeleton as an actor in cell signaling. Cell Mol. Life Sci. 2012, 69, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Haslam, R.J.; Koide, H.B.; Hemmings, B.A. Pleckstrin domain homology. Nature 1993, 363, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Gibson, T.; Rice, P.; Thompson, J.; Saraste, M. The PH domain: A common piece in the structural patchwork of signaling proteins. Trends Biochem. Sci. 1993, 18, 343–348. [Google Scholar] [CrossRef]
- Macias, M.J.; Musacchio, A.; Ponstingl, H.; Nilges, M.; Saraste, M.; Oschkinat, H. Structure of the pleckstrin homology domain from beta-spectrin. Nature 1994, 369, 675–677. [Google Scholar] [CrossRef] [PubMed]
- DeWald, D.B.; Torabinejad, J.; Jones, C.A.; Shope, J.C.; Cangelosi, A.R.; Thompson, J.E.; Prestwich, G.D.; Hama, H. Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 2001, 126, 759–769. [Google Scholar] [CrossRef]
- Ulm, R.; Baumann, A.; Oravecz, A.; Máté, Z.; Ádám, É.; Oakeley, E.J.; Nagy, F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1397–1402. [Google Scholar] [CrossRef]
- Favory, J.J.; Stec, A.; Gruber, H.; Rizzini, L.; Oravecz, A.; Funk, M.; Seidlitz, H.K. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009, 28, 591–601. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Murmu, J.; Bush, M.J.; DeLong, C.; Li, S.; Xu, M.; Khan, M.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Shigeoka, S.; Maruta, T. Cellular redox regulation, signaling, and stress response in plants. Biosci. Biotechnol. Biochem. 2014, 78, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Noshi, M.; Mori, D.; Tanabe, N.; Maruta, T.; Shigeoka, S. Arabidopsis clade IV TGA transcription factors, TGA10 and TGA9, are involved in ROS-mediated responses to bacterial PAMP flg22. Plant Sci. 2016, 252, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Gunawardena, H.P.; Huang, Y.; Kenjale, R.; Wang, H.; Xie, L.; Chen, X. Unambiguous characterization of site-specific phosphorylation of leucine-rich repeat Fli-I-interacting protein 2 (LRRFIP2) in toll-like receptor 4 (TLR4)-mediated signaling. J. Biol. Chem. 2011, 286, 10897–10910. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef]
- Fu, S.F.; Chen, P.Y.; Nguyen, Q.T.T.; Huang, L.Y.; Zeng, G.R.; Huang, T.L.; Lin, C.Y.; Huang, H.J. Transcriptome profiling of genes and pathways associated with arsenic toxicity and tolerance in Arabidopsis. BMC Plant Biol. 2014, 14, 94. [Google Scholar] [CrossRef]
- Chang, G.J.; Hwang, S.G.; Yong, C.P.; Hyeon, M.P.; Dong, S.K.; Duck, H.P.; Cheol, S.J. Molecular characterization of the cold- and heat-induced Arabidopsis PXL1 gene and its potential role in transduction pathways under temperature fluctuations. J. Plant Physiol. 2015, 176, 138–146. [Google Scholar] [CrossRef]
- Ma, X.L.; Cui, W.N.; Zhao, Q.; Zhao, J.; Hou, X.N.; Li, D.Y.; Chen, Z.L.; Shen, Y.Z.; Huang, Z.J. Functional study of a salt-inducible TaSR gene in Triticum aestivum. Physiol. Plant. 2015, 156, 40–53. [Google Scholar] [CrossRef]
- Wu, F.; Sheng, P.; Tan, J.; Chen, X.L.; Lu, G.W.; Ma, W.W.; Heng, Y.Q.; Lin, Q.B.; Zhu, S.S.; Wang, J.L.; et al. Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the drought and salt tolerance transcription factor to regulate drought sensitivity in rice. J. Exp. Bot. 2015, 66, 271–281. [Google Scholar] [CrossRef]
- Caldwell, M.M. Solar UV irradiation and the growth and development of higher plants. Photophysiology 1971, 6, 131–177. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchellet, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010, 38, D843–D846. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Hui, G.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
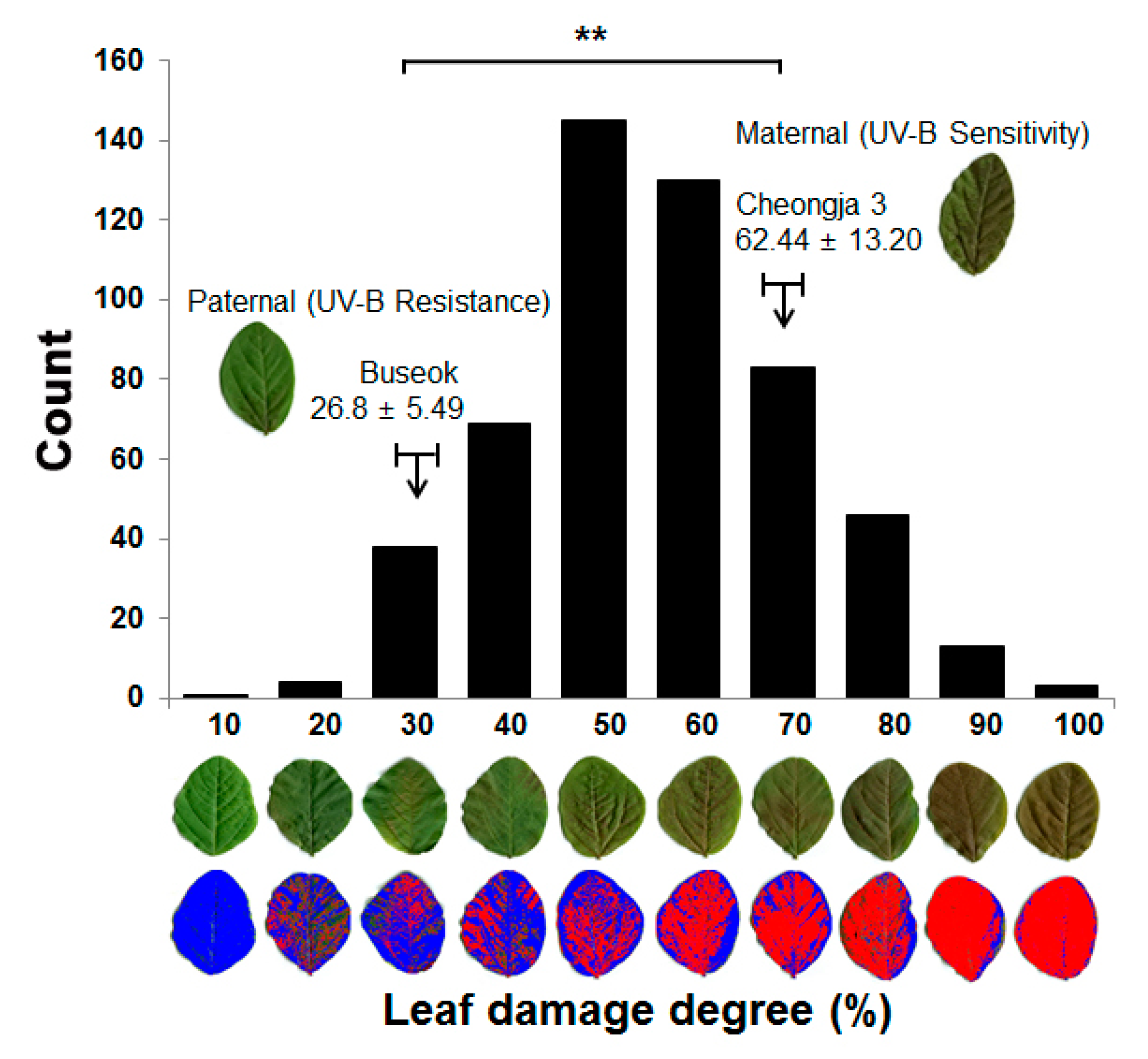
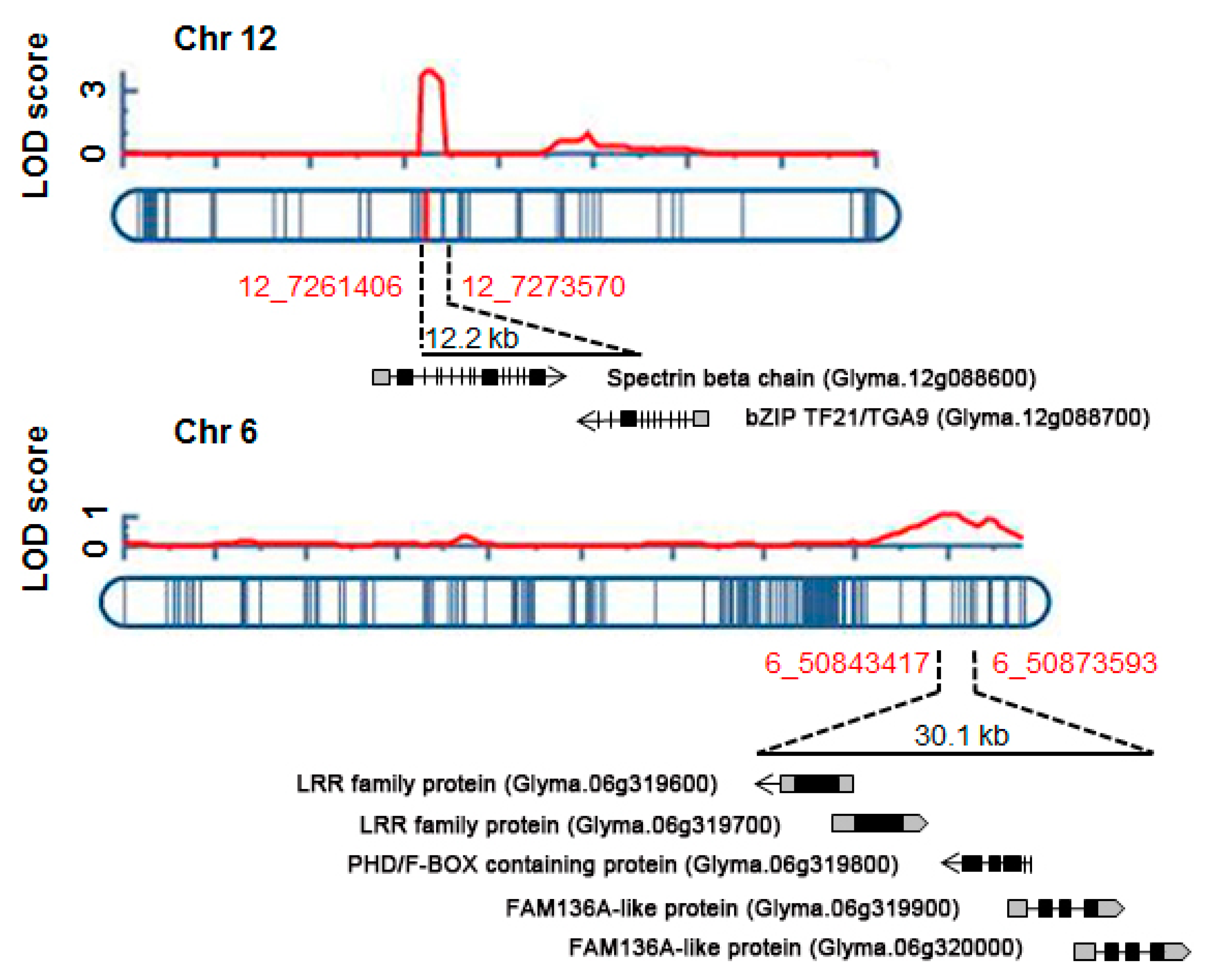
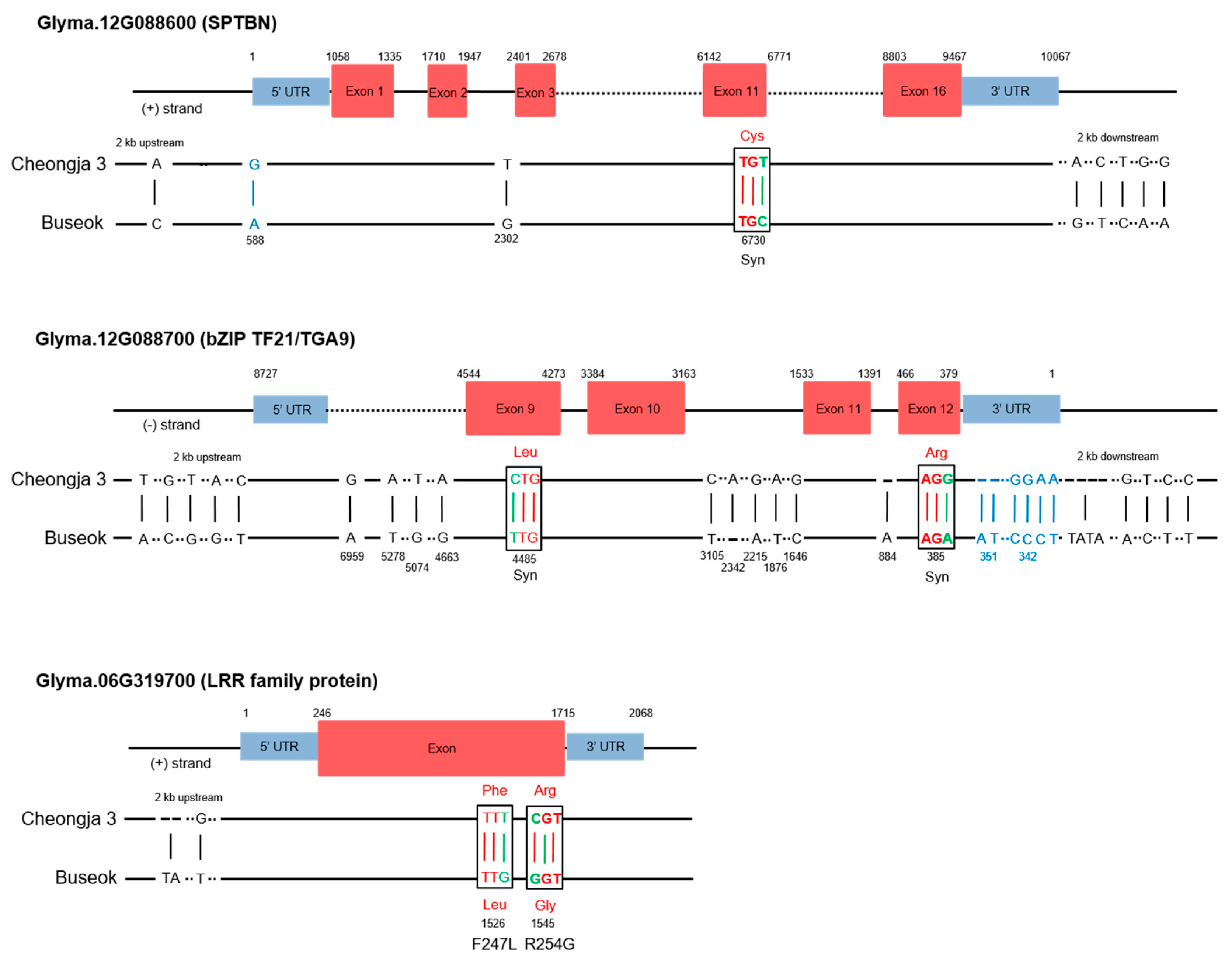
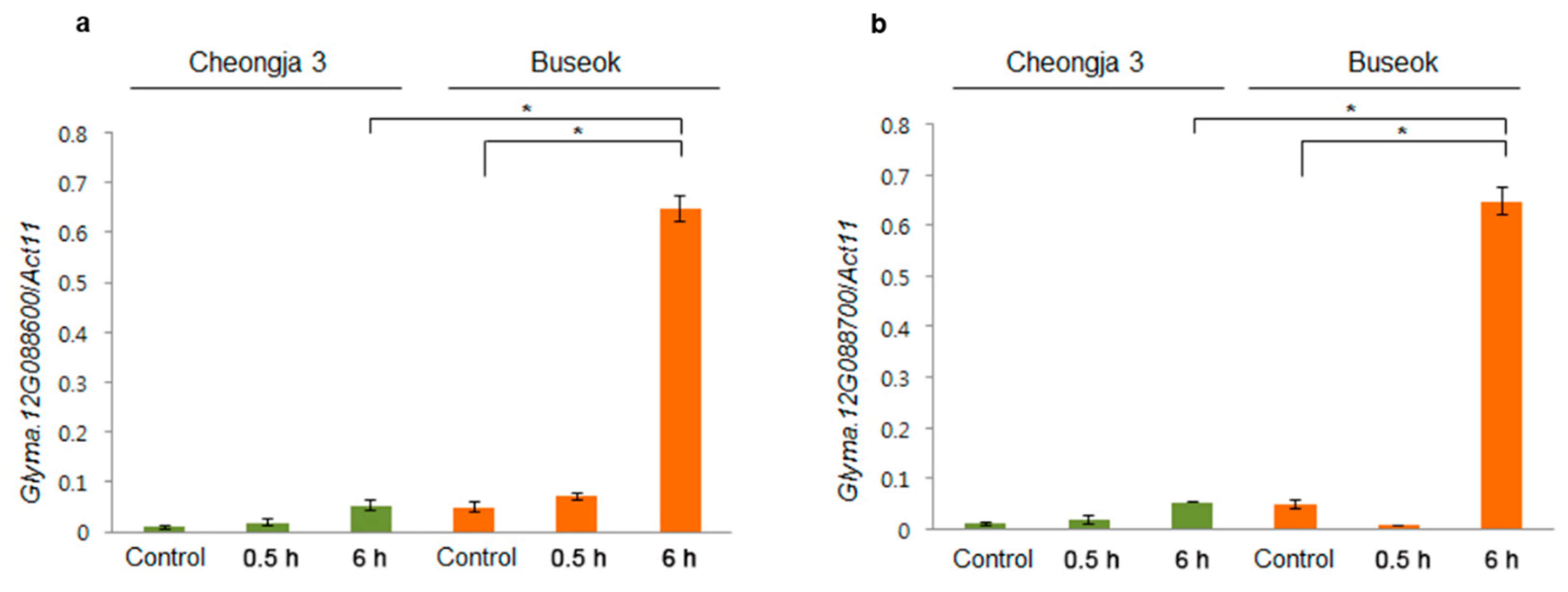
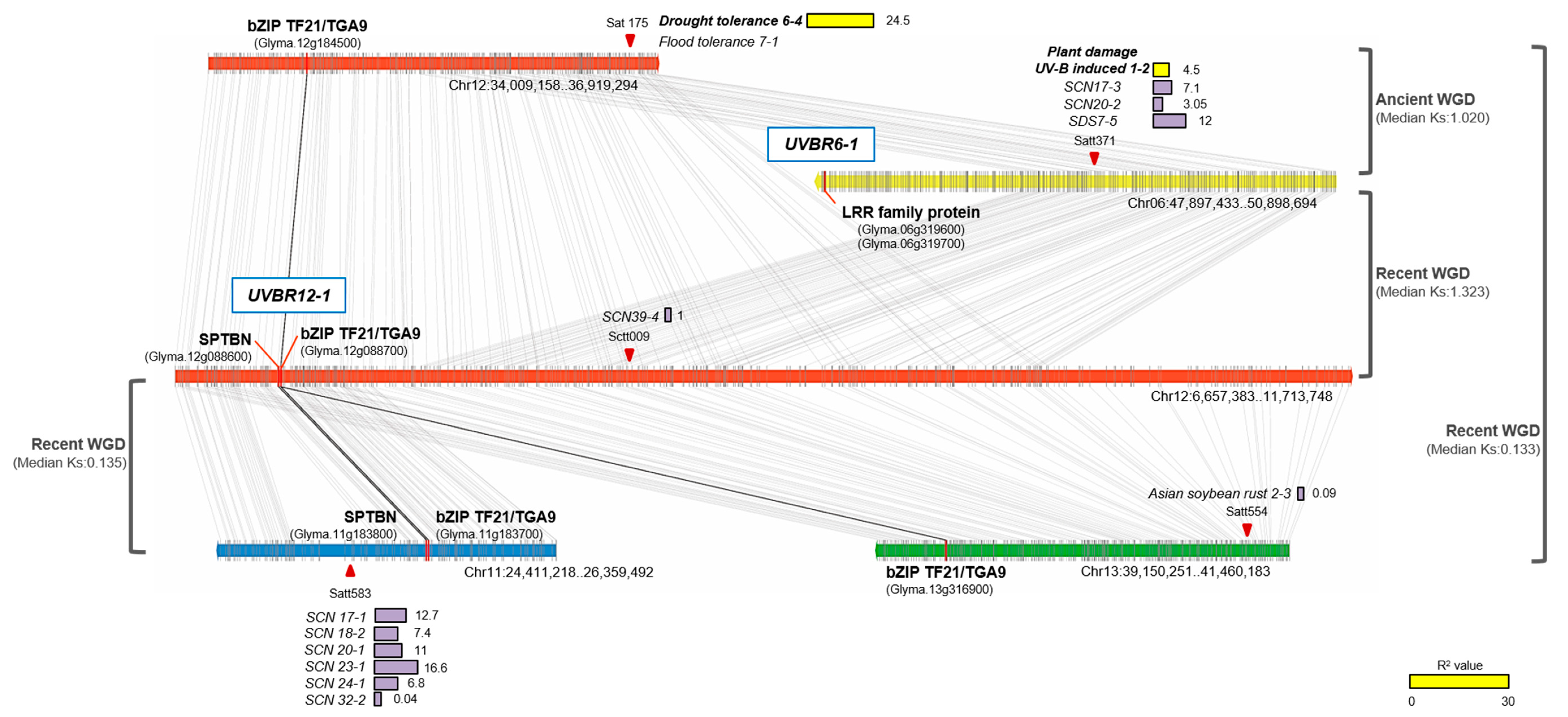
| Locus | Left Marker | Right Marker | Position a (cM) | LOD b | Add c | R2 d (%) | No. of Genes e | Known Stress-Related QTLs f |
|---|---|---|---|---|---|---|---|---|
| UVBR12-1 | Chr12:7261406 | Chr12:7273570 | 74 | 3.8 | -0.4 | 9.5 | 2 | Drought tolerance 6-4 SCN 39-4 |
| UVBR14-1 | Chr14:46720930 | Chr14:47368499 | 189 | 2.2 | 0.3 | 5.3 | 60 | Fe-effect 3-2, 9-2, 10-3 |
| UVBR10-1 | Chr10:41185273 | Chr10:42517624 | 160 | 1.1 | -0.2 | 2.7 | 142 | Flood tolerance 4-7 Drought tolerance 6-3 Sclero 2-23, 4-10, 3-18 Phytoph 5-3 |
| UVBR6-1 | Chr06:50843417 | Chr06:50873593 | 38 | 1.1 | 0.2 | 2.6 | 5 | Plant damage, UV-B induced 1-2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, M.Y.; Kim, M.Y.; Ha, J.; Lee, T.; Kim, K.D.; Lee, S.-H. QTL Analysis of Resistance to High-Intensity UV-B Irradiation in Soybean (Glycine max [L.] Merr.). Int. J. Mol. Sci. 2019, 20, 3287. https://doi.org/10.3390/ijms20133287
Yoon MY, Kim MY, Ha J, Lee T, Kim KD, Lee S-H. QTL Analysis of Resistance to High-Intensity UV-B Irradiation in Soybean (Glycine max [L.] Merr.). International Journal of Molecular Sciences. 2019; 20(13):3287. https://doi.org/10.3390/ijms20133287
Chicago/Turabian StyleYoon, Min Young, Moon Young Kim, Jungmin Ha, Taeyoung Lee, Kyung Do Kim, and Suk-Ha Lee. 2019. "QTL Analysis of Resistance to High-Intensity UV-B Irradiation in Soybean (Glycine max [L.] Merr.)" International Journal of Molecular Sciences 20, no. 13: 3287. https://doi.org/10.3390/ijms20133287
APA StyleYoon, M. Y., Kim, M. Y., Ha, J., Lee, T., Kim, K. D., & Lee, S.-H. (2019). QTL Analysis of Resistance to High-Intensity UV-B Irradiation in Soybean (Glycine max [L.] Merr.). International Journal of Molecular Sciences, 20(13), 3287. https://doi.org/10.3390/ijms20133287






