In Vitro Acquisition of Specific Small Interfering RNAs Inhibits the Expression of Some Target Genes in the Plant Ectoparasite Xiphinema index
Abstract
1. Introduction
2. Results
2.1. Optimization of In Vitro Acquisition of siRNA by X. index
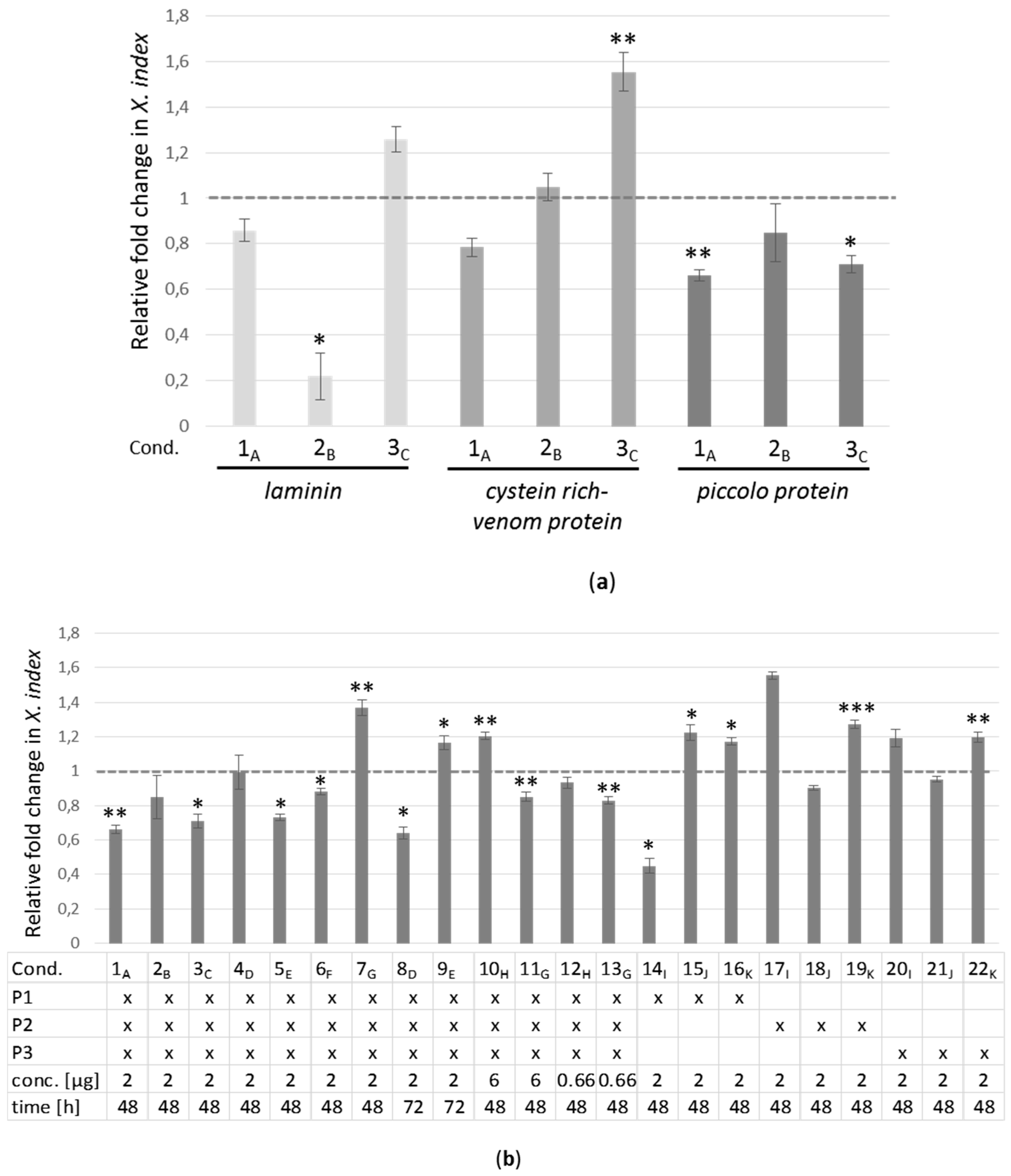
2.2. Selection of X. index Target Genes for RNAi Experiments
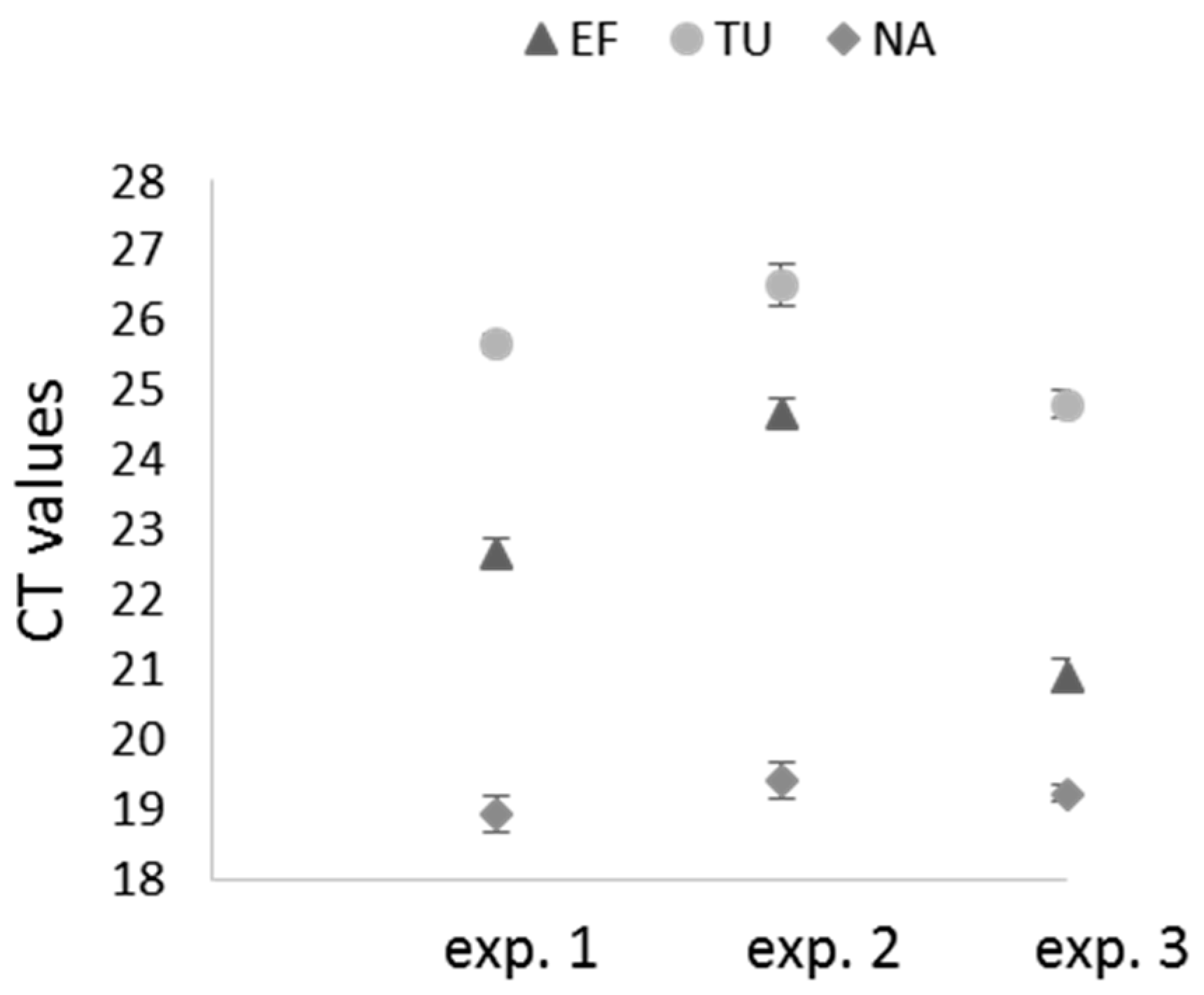
2.3. Selection of Reference Genes for Assessing RNAi Efficiency in X. index
2.4. Real-Time RT-PCR Analyses of Target Genes in X. index after siRNA Acquisition
2.5. Annotation of X. index Putative Proteins Involved in Gene Silencing
3. Discussion
4. Materials and Methods
4.1. Nematode Rearing
4.2. siRNA Design and Synthesis
4.3. Nematode Soaking
4.4. RNA Extraction and Quantitative RT-PCR (RT-qPCR)
4.5. Comparative Functional Annotation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RNAi | RNA interference |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| WAGO | Worm-specific argonaute |
| RdRP | RNA dependent RNA polymerase |
| Cy3 | Cyanine 3 |
References
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-Lopez, R.; Palomares-Rius, J.E.; Wesemael, W.M.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef]
- Nicol, J.M.; Stirling, G.R.; Rose, B.J.; May, P.; Van Heeswijck, R. Impact of nematodes on grapevine growth and productivity: Current knowledge and future directions, with special reference to Australian viticulture. Aust. J. Grape Wine Res. 1999, 5, 109–127. [Google Scholar] [CrossRef]
- Weischer, B.; Wyss, U. Feeding behavior and pathogenicity of Xiphinema index on grapevine roots. Nematologica 1976, 22, 319–325. [Google Scholar] [CrossRef]
- Taylor, C.E.; Brown, D.J.F. Nematode Vectors of Plant Viruses; CAB International: Wallingford, UK, 1997. [Google Scholar]
- Brown, D.J.; Robertson, W.M.; Trudgill, D.L. Transmission of viruses by plant nematodes. Annu. Rev. Phytopathol. 1995, 33, 223–249. [Google Scholar] [CrossRef]
- Wyss, U. Xiphinema Index, Maintenance and Feeding in Monoxenic Cultures; Maramorosch, K., Mahmood, F., Eds.; CRC Press: Boca Raton, USA, 2014; pp. 235–267. [Google Scholar]
- Furlanetto, C.; Cardle, L.; Brown, D.; Jones, J. Analysis of expressed sequence tags from the ectoparasitic nematode Xiphinema index. Nematology 2005, 7, 95–104. [Google Scholar]
- Andret-Link, P.; Schmitt-Keichinger, C.; Demangeat, G.; Komar, V.; Fuchs, M. The specific transmission of Grapevine fanleaf virus by its nematode vector Xiphinema index is solely determined by the viral coat protein. Virology 2004, 320, 12–22. [Google Scholar] [CrossRef]
- Hewitt, W.B.; Raski, D.J.; Goheen, A.C. Nematode vector of soil-borne fanleaf virus of grapevines. Phytopathology 1958, 48, 586–595. [Google Scholar]
- Raski, D.J.; Goheen, A.C.; Lider, L.A.; Meredith, C.P. Strategies against grapevine fanleaf virus and its nematode vector. Plant Dis. 1983, 67, 335–339. [Google Scholar] [CrossRef]
- Andret-Link, P.; Marmonier, A.; Belval, L.; Hleibieh, K.; Ritzenthaler, C.; Demangeat, G. Nematode vectors for grapevine viruses: Biology, behavior and mechanisms of virus transmission. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Fuchs, M., Golino, D., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 505–529. [Google Scholar]
- Demangeat, G.; Voisin, R.; Minot, J.C.; Bosselut, N.; Fuchs, M.; Esmenjaud, D. Survival of Xiphinema index in Vineyard Soil and Retention of Grapevine fanleaf virus Over Extended Time in the Absence of Host Plants. Phytopathology 2005, 95, 1151–1156. [Google Scholar] [CrossRef]
- Belin, C.; Schmitt, C.; Demangeat, G.; Komar, V.; Pinck, L.; Fuchs, M. Involvement of RNA2-encoded proteins in the specific transmission of Grapevine fanleaf virus by its nematode vector Xiphinema index. Virology 2001, 291, 161–171. [Google Scholar] [CrossRef][Green Version]
- Marmonier, A.; Schellenberger, P.; Esmenjaud, D.; Schmitt-Keichinger, C.; Ritzenthaler, C.; Andret-Link, P.; Lemaire, O.; Fuchs, M.; Demangeat, G. The coat protein determines the specificity of virus transmission by Xiphinema diversicaudatum. J. Plant Pathol. 2010, 92, 275–279. [Google Scholar]
- Schellenberger, P.; Andret-Link, P.; Schmitt-Keichinger, C.; Bergdoll, M.; Marmonier, A.; Vigne, E.; Lemaire, O.; Fuchs, M.; Demangeat, G.; Ritzenthaler, C. A stretch of 11 amino acids in the betaB-betaC loop of the coat protein of grapevine fanleaf virus is essential for transmission by the nematode Xiphinema index. J. Virol. 2010, 84, 7924–7933. [Google Scholar] [CrossRef]
- Schellenberger, P.; Sauter, C.; Lorber, B.; Bron, P.; Trapani, S.; Bergdoll, M.; Marmonier, A.; Schmitt-Keichinger, C.; Lemaire, O.; Demangeat, G.; et al. Structural insights into viral determinants of nematode mediated Grapevine fanleaf virus transmission. PLoS Pathog. 2011, 7, e1002034. [Google Scholar] [CrossRef]
- Danchin, E.G.J.; Perfus-Barbeoch, L.; Rancurel, C.; Thorpe, P.; Da Rocha, M.; Bajew, S.; Neilson, R.; Guzeeva, E.S.; Da Silva, C.; Guy, J.; et al. The Transcriptomes of Xiphinema index and Longidorus elongatus Suggest Independent Acquisition of Some Plant Parasitism Genes by Horizontal Gene Transfer in Early-Branching Nematodes. Genes 2017, 8, 287. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Britton, C.; Samarasinghe, B.; Knox, D.P. Ups and downs of RNA interference in parasitic nematodes. Exp. Parasitol. 2012, 132, 56–61. [Google Scholar] [CrossRef]
- Kimber, M.J.; McKinney, S.; McMaster, S.; Day, T.A.; Fleming, C.C.; Maule, A.G. flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J. 2007, 21, 1233–1243. [Google Scholar] [CrossRef]
- Urwin, P.E.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef]
- Bakhetia, M.; Charlton, W.; Atkinson, H.J.; McPherson, M.J. RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2005, 18, 1099–1106. [Google Scholar] [CrossRef]
- Rosso, M.N.; Dubrana, M.P.; Cimbolini, N.; Jaubert, S.; Abad, P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microbe Interact. 2005, 18, 615–620. [Google Scholar] [CrossRef]
- Shingles, J.; Lilley, C.J.; Atkinson, H.J.; Urwin, P.E. Meloidogyne incognita: Molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp. Parasitol. 2007, 115, 114–120. [Google Scholar] [CrossRef]
- Arguel, M.J.; Jaouannet, M.; Magliano, M.; Abad, P.; Rosso, M.N. siRNAs Trigger Efficient Silencing of a Parasitism Gene in Plant Parasitic Root-Knot Nematodes. Genes 2012, 3, 391–408. [Google Scholar] [CrossRef]
- Dalzell, J.J.; McMaster, S.; Fleming, C.C.; Maule, A.G. Short interfering RNA-mediated gene silencing in Globodera pallida and Meloidogyne incognita infective stage juveniles. Int. J. Parasitol. 2010, 40, 91–100. [Google Scholar] [CrossRef]
- Dalzell, J.J.; Warnock, N.D.; Stevenson, M.A.; Mousley, A.; Fleming, C.C.; Maule, A.G. Short interfering RNA-mediated knockdown of drosha and pasha in undifferentiated Meloidogyne incognita eggs leads to irregular growth and embryonic lethality. Int. J. Parasitol. 2010, 40, 1303–1310. [Google Scholar] [CrossRef]
- Issa, Z.; Grant, W.N.; Stasiuk, S.; Shoemaker, C.B. Development of methods for RNA interference in the sheep gastrointestinal parasite, Trichostrongylus colubriformis. Int. J. Parasitol. 2005, 35, 935–940. [Google Scholar] [CrossRef]
- Danchin, E.G.; Arguel, M.J.; Campan-Fournier, A.; Perfus-Barbeoch, L.; Magliano, M.; Rosso, M.N.; Da Rocha, M.; Da Silva, C.; Nottet, N.; Labadie, K.; et al. Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PLoS Pathog. 2013, 9, e1003745. [Google Scholar] [CrossRef]
- Dubreuil, G.; Magliano, M.; Dubrana, M.P.; Lozano, J.; Lecomte, P.; Favery, B.; Abad, P.; Rosso, M.N. Tobacco rattle virus mediates gene silencing in a plant parasitic root-knot nematode. J. Exp. Bot. 2009, 60, 4041–4050. [Google Scholar] [CrossRef]
- Fairbairn, D.J.; Cavallaro, A.S.; Bernard, M.; Mahalinga-Iyer, J.; Graham, M.W.; Botella, J.R. Host-delivered RNAi: An effective strategy to silence genes in plant parasitic nematodes. Planta 2007, 226, 1525–1533. [Google Scholar] [CrossRef]
- Huang, G.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef]
- Nguyen, C.N.; Perfus-Barbeoch, L.; Quentin, M.; Zhao, J.; Magliano, M.; Marteu, N.; Da Rocha, M.; Nottet, N.; Abad, P.; Favery, B. A root-knot nematode small glycine and cysteine-rich secreted effector, MiSGCR1, is involved in plant parasitism. New Phytol. 2018, 217, 687–699. [Google Scholar] [CrossRef]
- Yadav, B.C.; Veluthambi, K.; Subramaniam, K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 2006, 148, 219–222. [Google Scholar] [CrossRef]
- Van Megen, H.; Van den Elsen, S.; Holterman, M.; Karssen, G.; Mooyman, P.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 2009, 11, 927–950. [Google Scholar]
- Truong, N.M.; Nguyen, C.-N.; Abad, P.; Quentin, M.; Favery, B. Chapter Twelve—Function of Root-Knot Nematode Effectors and Their Targets in Plant Parasitism. In Advances in Botanical Research; Escobar, C., Fenoll, C., Eds.; Academic Press: Oxford, UK, 2015; Volume 73, pp. 293–324. [Google Scholar]
- Huang, G.; Gao, B.; Maier, T.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. A profile of putative parasitism genes expressed in the esophageal gland cells of the root-knot nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2003, 16, 376–381. [Google Scholar] [CrossRef]
- Jaouannet, M.; Magliano, M.; Arguel, M.J.; Gourgues, M.; Evangelisti, E.; Abad, P.; Rosso, M.N. The Root-Knot Nematode Calreticulin Mi-CRT Is a Key Effector in Plant Defense Suppression. Mol. Plant Microbe Interact. 2012, 26, 97–105. [Google Scholar] [CrossRef]
- Dinh, P.T.; Brown, C.R.; Elling, A.A. RNA Interference of Effector Gene Mc16D10L Confers Resistance Against Meloidogyne chitwoodi in Arabidopsis and Potato. Phytopathology 2014, 104, 1098–1106. [Google Scholar] [CrossRef]
- Iberkleid, I.; Vieira, P.; de Almeida Engler, J.; Firester, K.; Spiegel, Y.; Horowitz, S.B. Fatty acid-and retinol-binding protein, Mj-FAR-1 induces tomato host susceptibility to root-knot nematodes. PLoS ONE 2013, 8, e64586. [Google Scholar] [CrossRef]
- Hoogstrate, S.W.; Volkers, R.J.; Sterken, M.G.; Kammenga, J.E.; Snoek, L.B. Nematode endogenous small RNA pathways. Worm 2014, 3, e28234. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Samarasinghe, B.; Knox, D.P.; Britton, C. Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. Int. J. Parasitol. 2011, 41, 51–59. [Google Scholar] [CrossRef]
- Stefanic, S.; Dvorak, J.; Horn, M.; Braschi, S.; Sojka, D.; Ruelas, D.S.; Suzuki, B.; Lim, K.C.; Hopkins, S.D.; McKerrow, J.H.; et al. RNA interference in Schistosoma mansoni schistosomula: Selectivity, sensitivity and operation for larger-scale screening. PLoS Negl. Trop. Dis. 2010, 4, e850. [Google Scholar] [CrossRef]
- Visser, A.; Geldhof, P.; de Maere, V.; Knox, D.P.; Vercruysse, J.; Claerebout, E. Efficacy and specificity of RNA interference in larval life-stages of Ostertagia ostertagi. Parasitology 2006, 133, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Blaxter, M. Functional diversification of Argonautes in nematodes: An expanding universe. Biochem. Soc. Trans. 2013, 41, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Sijen, T.; Fleenor, J.; Simmer, F.; Thijssen, K.L.; Parrish, S.; Timmons, L.; Plasterk, R.H.; Fire, A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 2001, 107, 465–476. [Google Scholar] [CrossRef]
- Zhang, C.; Ruvkun, G. New insights into siRNA amplification and RNAi. RNA Biol. 2012, 9, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Winston, W.M.; Molodowitch, C.; Hunter, C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 2002, 295, 2456–2459. [Google Scholar] [CrossRef] [PubMed]
- Winston, W.M.; Sutherlin, M.; Wright, A.J.; Feinberg, E.H.; Hunter, C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 2007, 104, 10565–10570. [Google Scholar] [CrossRef] [PubMed]
- Geldhof, P.; Visser, A.; Clark, D.; Saunders, G.; Britton, C.; Gilleard, J.; Berriman, M.; Knox, D. RNA interference in parasitic helminths: Current situation, potential pitfalls and future prospects. Parasitology 2007, 134, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Lendner, M.; Doligalska, M.; Lucius, R.; Hartmann, S. Attempts to establish RNA interference in the parasitic nematode Heligmosomoides polygyrus. Mol. Biochem. Parasitol. 2008, 161, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Dalzell, J.J.; McVeigh, P.; Warnock, N.D.; Mitreva, M.; Bird, D.M.; Abad, P.; Fleming, C.C.; Day, T.A.; Mousley, A.; Marks, N.J.; et al. RNAi effector diversity in nematodes. PLoS Negl. Trop. Dis. 2011, 5, e1176. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Qian, Z.; Shen, S.; Min, T.; Tan, C.; Xu, J.; Zhao, Y.; Huang, W. High doses of siRNAs induce eri-1 and adar-1 gene expression and reduce the efficiency of RNA interference in the mouse. Biochem. J. 2005, 390, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Geldhof, P.; Murray, L.; Couthier, A.; Gilleard, J.S.; McLauchlan, G.; Knox, D.P.; Britton, C. Testing the efficacy of RNA interference in Haemonchus contortus. Int. J. Parasitol. 2006, 36, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Bakhetia, M.; Urwin, P.E.; Atkinson, H.J. QPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Mol. Plant Microbe Interact. 2007, 20, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Bakhetia, M.; Urwin, P.E.; Atkinson, H.J. Characterisation by RNAi of pioneer genes expressed in the dorsal pharyngeal gland cell of Heterodera glycines and the effects of combinatorial RNAi. Int. J. Parasitol. 2008, 38, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, A.S.; Maier, T.R.; Mitchum, M.G.; Hussey, R.S.; Davis, E.L.; Baum, T.J. Effective and specific in planta RNAi in cyst nematodes: Expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 2009, 60, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Steeves, R.M.; Todd, T.C.; Essig, J.S.; Trick, H.N. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct. Plant Biol. 2006, 33, 991–999. [Google Scholar] [CrossRef]
- Dinh, P.T.; Zhang, L.; Mojtahedi, H.; Brown, C.R.; Elling, A.A. Broad Meloidogyne Resistance in Potato Based on RNA Interference of Effector Gene 16D10. J. Nematol. 2015, 47, 71–78. [Google Scholar] [PubMed]
- Yang, Y.; Jittayasothorn, Y.; Chronis, D.; Wang, X.; Cousins, P.; Zhong, G.Y. Molecular characteristics and efficacy of 16D10 siRNAs in inhibiting root-knot nematode infection in transgenic grape hairy roots. PLoS ONE 2013, 8, e69463. [Google Scholar] [CrossRef]
- Adams, S.; Pathak, P.; Shao, H.; Lok, J.B.; Pires-daSilva, A. Liposome-based transfection enhances RNAi and CRISPR-mediated mutagenesis in non-model nematode systems. Sci. Rep. 2019, 9, 483. [Google Scholar] [CrossRef]
- Au, V.; Li-Leger, E.; Raymant, G.; Flibotte, S.; Chen, G.; Martin, K.; Fernando, L.; Doell, C.; Rosell, F.I.; Wang, S.; et al. CRISPR/Cas9 Methodology for the Generation of Knockout Deletions in Caenorhabditis elegans. G3 2019, 9, 135–144. [Google Scholar] [CrossRef]
- Flegg, J.J.M. Extraction of Xiphinema and Longidorus species from soil by a modification of Cobb’s decanting and sieving technique. Ann. Appl. Biol. 1967, 60, 420–437. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Cottret, L.; Rancurel, C.; Briand, M.; Carrere, S. Family-companion: Analyse, visualise, browse, query and share your homology clusters. bioRxiv 2018. [Google Scholar] [CrossRef]
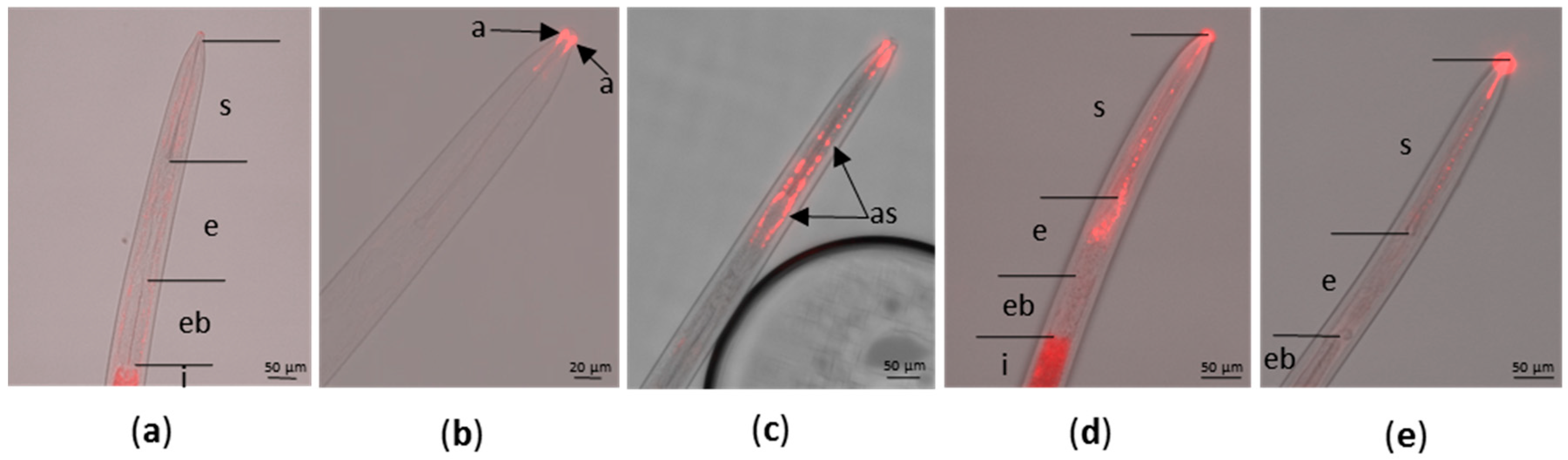

| Cy3-siRNA Solution | Acquisition Period | % of Viable Nematodes | Labeling of Amphid Sheath 1 | Labeling of Alimentary Tract 1 |
|---|---|---|---|---|
| Mineral water | 1 h | 100% | 9/20 (45%) | 3/20 (15%) |
| 24 h | 100% | 16/30 (53%) | 6/30 (20%) | |
| 48 h | 100% | 16/30 (53%) | 7/30 (23%) | |
| Mineral water + Cellfectin II | 1 h | 100% | 0/26 (0%) | 0/26 (0%) |
| 24 h | 100% | 0/29 (0%) | 0/29 (0%) | |
| 48 h | 50% | 0/26 (0%) | 0/26 (0%) | |
| Mineral water + TransIT-insect transfection reagent | 1 h | 100% | 0/23 (0%) | 0/23 (0%) |
| 24 h | 10% | 0/20 (0%) | 0/20 (0%) | |
| 48 h | 0% | nd 2 | nd |
| Reference Genes | Number of EST in X. index | % of Sequence Identity with Orthologous Sequences in Root-Knot Nematodes |
|---|---|---|
| tubulin | 19 | 70 to 75% 1 |
| elongation factor | 2 | 81% 1 |
| 18S | 10 | 70 to 76% 1 |
| NADH dehydrogenase | 40 | nd 2 |
| Name of Family or Complex Involved in C. Elegans Gene Silencing | Name 1 | C. elegans Wormbase Accession Number 2 | Presence/Absence of Putative X. index Proteins in Ortholog Group 3 |
|---|---|---|---|
| AGO | ALG-1 | WBGene00000105 | Presence of AGO ortholog |
| ALG-2 | WBGene00000106 | ||
| HPO-24/ALG-5 | WBGene00011945 | ||
| ALG-3 | WBGene00011910 | ||
| ALG-4 | WBGene00006449 | ||
| RDE-1 | WBGene00004323 | ||
| ZK218.8 | WBGene00013942 | ||
| PIWI | ERGO-1 | WBGene00019971 | No X. index putative prot. |
| PRG-1 | WBGene00004178 | Presence of PRG-1 ortholog | |
| WAGO | WAGO-1 | WBGene00011061 | No X. index putative prot. |
| WAGO-2 | WBGene00018862 | ||
| WAGO-4 | WBGene00010263 | ||
| WAGO-5 | WBGene00022877 | ||
| SAGO-1 | WBGene00019666 | ||
| SAGO-2 | WBGene00018921 | ||
| PPW-1 | WBGene00004093 | ||
| PPW-2 | WBGene00004094 | ||
| HRDE-1 | WBGene00007624 | ||
| HRDE-2 | WBGene00011324 | ||
| NRDE-3 | WBGene00019862 | ||
| WAGO-10 | WBGene00020707 | ||
| WAGO-11 | WBGene00021711 | ||
| C14B1.7 | WBGene00007578 | ||
| CSR-1 | WBGene00017641 | ||
| C04F12.1 | WBGene00007297 | ||
| ERI/DICER complex | ERI-1 | WBGene00001332 | Presence of ERI ortholog |
| ERI-3 | WBGene00021103 | ||
| ERI-5 | WBGene00021419 | ||
| DCR-1 (Dicer) | WBGene00000939 | Presence of DCR-1 ortholog | |
| DICER related complex | PASH1 | WBGene00011908 | Presence of PASH1 ortholog |
| DRH1 | WBGene00001090 | No X. index putative prot. | |
| DRH3 | WBGene00008400 | ||
| DRSH1 (Drosha) | WBGene00009163 | ||
| RdRP | RRF-3 | WBGene00004510 | No X. index putative prot. |
| EGO-1 | WBGene00001214 | ||
| RRF-1 | WBGene00004508 | ||
| RRF-2 | WBGene00004509 | ||
| Systemic RNA interference defective | Sid-1 | WBGene00004795 | No X. index putative prot. |
| Sid-2 | WBGene00004796 | ||
| ribonuclease | Xrn-2 | WBGene00006964 | Presence of Xrn-2 ortholog |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marmonier, A.; Perfus-Barbeoch, L.; Rancurel, C.; Boissinot, S.; Favery, B.; Demangeat, G.; Brault, V. In Vitro Acquisition of Specific Small Interfering RNAs Inhibits the Expression of Some Target Genes in the Plant Ectoparasite Xiphinema index. Int. J. Mol. Sci. 2019, 20, 3266. https://doi.org/10.3390/ijms20133266
Marmonier A, Perfus-Barbeoch L, Rancurel C, Boissinot S, Favery B, Demangeat G, Brault V. In Vitro Acquisition of Specific Small Interfering RNAs Inhibits the Expression of Some Target Genes in the Plant Ectoparasite Xiphinema index. International Journal of Molecular Sciences. 2019; 20(13):3266. https://doi.org/10.3390/ijms20133266
Chicago/Turabian StyleMarmonier, Aurélie, Laetitia Perfus-Barbeoch, Corinne Rancurel, Sylvaine Boissinot, Bruno Favery, Gérard Demangeat, and Véronique Brault. 2019. "In Vitro Acquisition of Specific Small Interfering RNAs Inhibits the Expression of Some Target Genes in the Plant Ectoparasite Xiphinema index" International Journal of Molecular Sciences 20, no. 13: 3266. https://doi.org/10.3390/ijms20133266
APA StyleMarmonier, A., Perfus-Barbeoch, L., Rancurel, C., Boissinot, S., Favery, B., Demangeat, G., & Brault, V. (2019). In Vitro Acquisition of Specific Small Interfering RNAs Inhibits the Expression of Some Target Genes in the Plant Ectoparasite Xiphinema index. International Journal of Molecular Sciences, 20(13), 3266. https://doi.org/10.3390/ijms20133266






