Phylogenomics Provides New Insights into Gains and Losses of Selenoproteins among Archaeplastida
Abstract
1. Introduction
2. Results
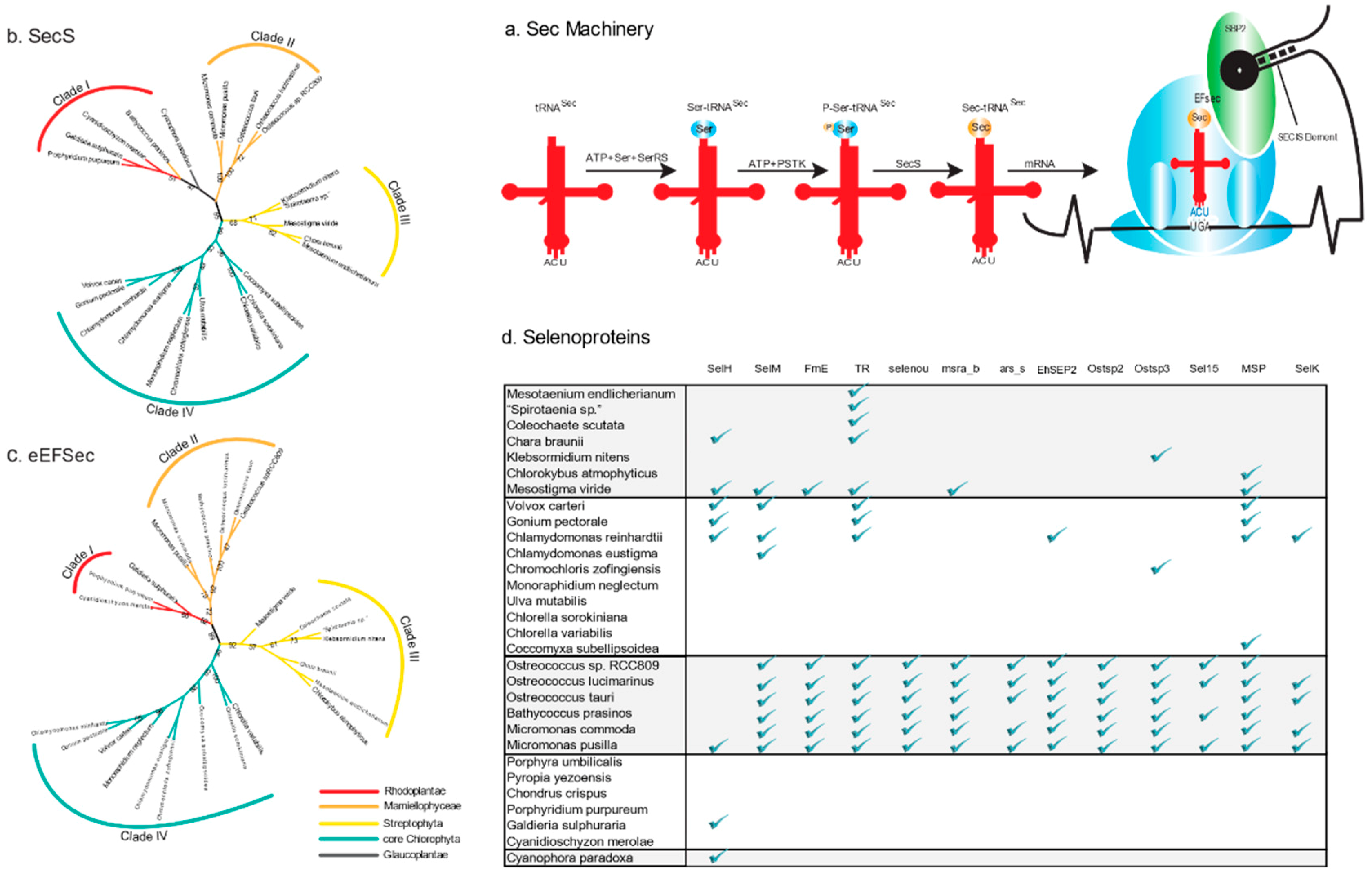
2.1. Sec Machinery in Algae
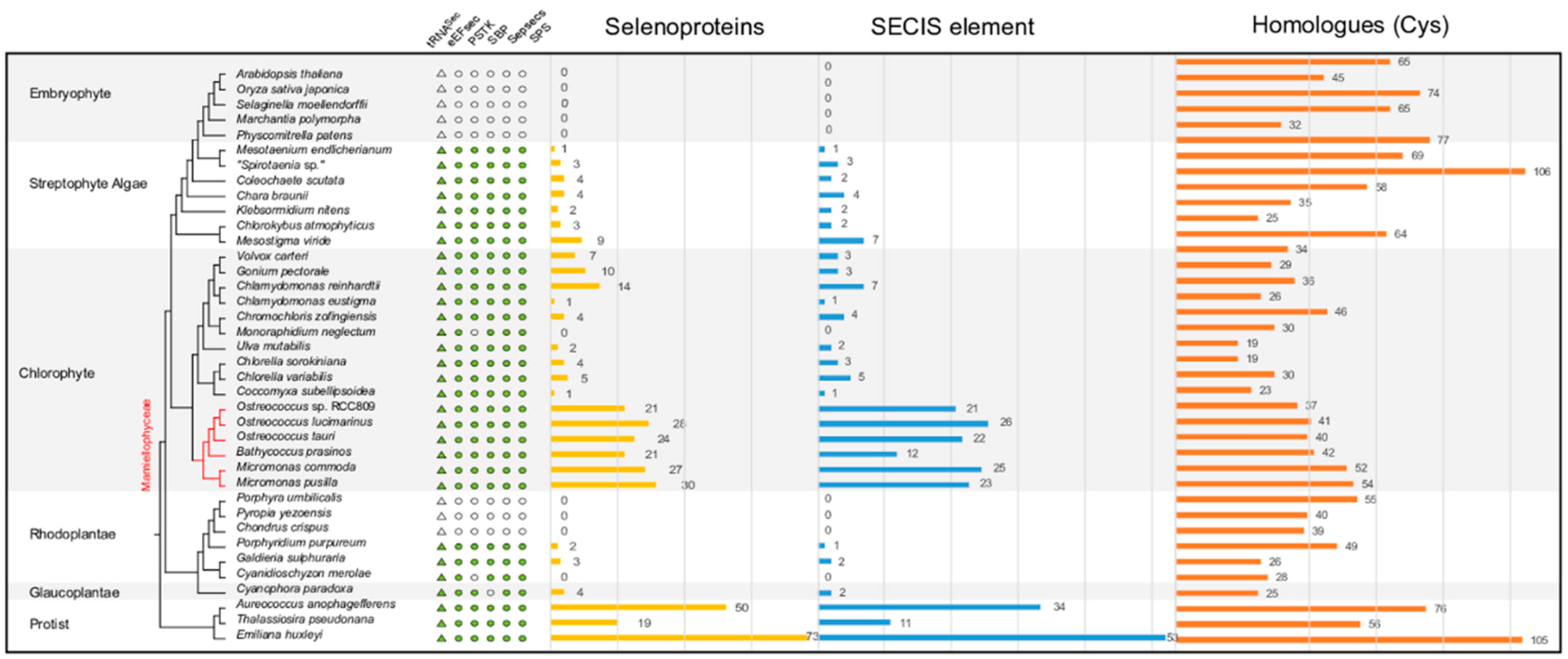
2.2. Sec Incorporation in the Major Algal Lineages
2.3. Variable Number of Selenoproteins Identified in Algae
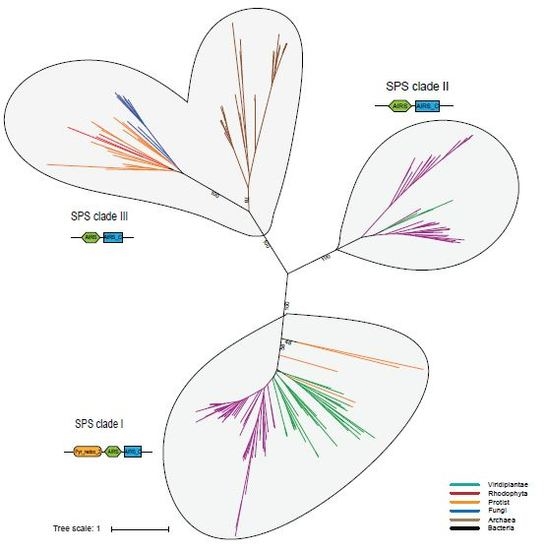
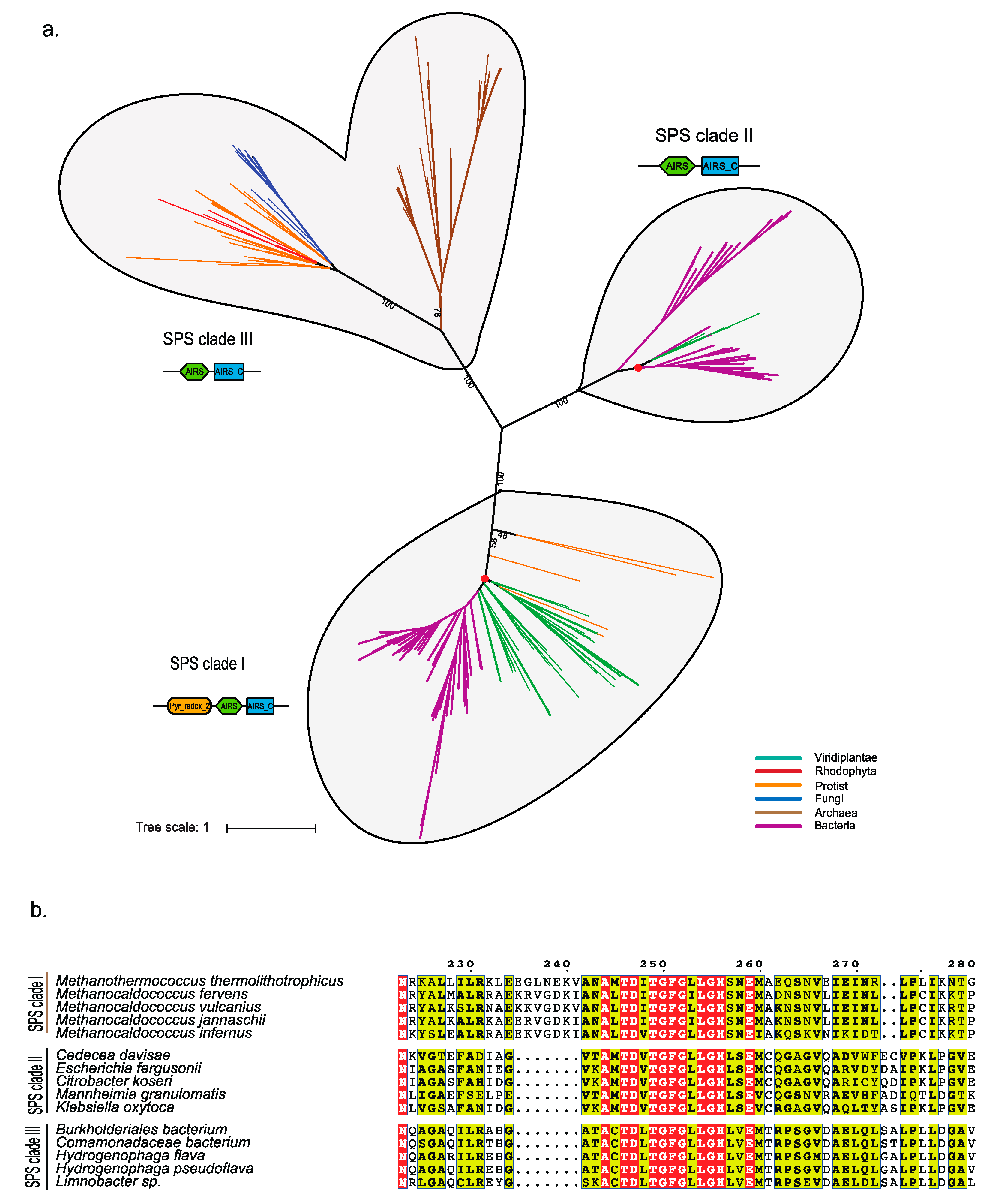
2.4. Phylogenetic Analysis of the Enzymes involved in the Sec Machinery
Phylogenetic Analysis of Eukaryotic SPS Proteins
2.5. Distribution of Types of Selenoproteins among Archaeplastida
3. Discussion
3.1. The Distribution of the Sec Machinery and Selenoproteins in Algae
3.2. Probable Horizontal Gene Transfer of SPS and some Selenoproteins
4. Materials and Methods
4.1. Data Information
4.2. Sec Incorporation Machinery
4.3. Identification of the Selenocysteine tRNA (tRNASec)
4.4. Prediction of Selenoproteins and SECIS Elements
4.5. Phylogenetic Tree Construction
4.6. Identification of Conserved Motifs and Domains.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BV | Bangiophyceae Florideophyceae |
| ROS | Reactive Oxygen Species |
| Sec | Selenocysteine |
| Se | Selenium |
| SECIS | Selenocysteine Insertion Sequence |
| PSTK | O-phosphoseryl-transfer tRNASec kinase |
| SecS | Sec Synthase |
| SPS | Selenophosphate Synthetase 2 |
| SBP2 | SECIS-binding Protein 2 |
| eEFSec | Sec-specific Elongation Factor |
| CTAB | Cetyl Trimethylammonium Bromide |
References
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Avery, J.C.; Hoffmann, P.R. Selenium, Selenoproteins, and Immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef] [PubMed]
- Novoselov, S.V.; Rao, M.; Onoshko, N.V.; Zhi, H.; Kryukov, G.V.; Xiang, Y.; Weeks, D.P.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002, 21, 3681–3693. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, M.; Salinas, G.; Gabaldon, T.; Gladyshev, V.N. Utilization of selenocysteine in early-branching fungal phyla. Nat. Microbiol. 2019, 4, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Araie, H.; Suzuki, I.; Shiraiwa, Y. Identification and characterization of a selenoprotein, thioredoxin reductase, in a unicellular marine haptophyte alga, Emiliania huxleyi. J. Biol. Chem. 2008, 283, 35329–35336. [Google Scholar] [CrossRef]
- Papp, L.V.; Holmgren, A.; Khanna, K.K. Selenium and selenoproteins in health and disease. Antioxid. Redox. Signal 2010, 12, 793–795. [Google Scholar] [CrossRef]
- Lobanov, A.V.; Fomenko, D.E.; Zhang, Y.; Sengupta, A.; Hatfield, D.L.; Gladyshev, V.N. Evolutionary dynamics of eukaryotic selenoproteomes: Large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007, 8, 1–16. [Google Scholar] [CrossRef]
- Bulteau, A.L.; Chavatte, L. Update on selenoprotein biosynthesis. Antioxid. Redox Signal. 2015, 23, 775–794. [Google Scholar] [CrossRef]
- Schiavon, M.; Pilon-Smits, E.A. The fascinating facets of plant selenium accumulation—Biochemistry, physiology, evolution and ecology. New Phytol. 2017, 213, 1582–1596. [Google Scholar] [CrossRef]
- Böck, A.; Forchhammer, K.; Heider, J.; Barion, C. Selenoprotein synthesis: An expansion of the genetic code. Trends Biochem. Sci. 1991, 16, 463–467. [Google Scholar] [CrossRef]
- Berry, M.J.; Banu, L.; Chen, Y.; Mandel, S.J.; Kieffer, J.D.; Harney, J.W.; Larsen, P.R. Recognition of UGA as a selenocysteine codon in Type I deiodinase requires sequences in the 3′ untranslated region. Nature 1991, 353, 273–276. [Google Scholar] [CrossRef]
- Low, S.C.; Berry, M.J. Knowing when not to stop: Selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 1996, 21, 203–208. [Google Scholar] [CrossRef]
- Carlson, B.A.; Xu, X.M.; Kryukov, G.V.; Rao, M.; Berry, M.J.; Gladyshev, V.N.; Hatfield, D.L. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 12848–12853. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Carlson, B.A.; Irons, R.; Mix, H.; Zhong, N.; Gladyshev, V.N.; Hatfield, D.L. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem. J. 2007, 404, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.E.; Copeland, P.R.; Driscoll, D.M.; Krol, A. The selenocysteine incorporation machinery: Interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA 2001, 7, 1442–1453. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Gobler, C.J.; Lobanov, A.V.; Tang, Y.Z.; Turanov, A.A.; Zhang, Y.; Doblin, M.; Taylor, G.T.; Sanudo-Wilhelmy, S.A.; Grigoriev, I.V.; Gladyshev, V.N. The central role of selenium in the biochemistry and ecology of the harmful pelagophyte, Aureococcus anophagefferens. ISME J. 2013, 7, 1333–1343. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef]
- Palenik, B.; Grimwood, J.; Aerts, A.; Salamov, A.; Putnam, N.H.; Dupont, C.L.; Jorgensen, R.A.; Rombauts, S.; Zhou, K.; Otillar, R.; et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef]
- Araie, H.; Shiraiwa, Y. Selenium utilization strategy by microalgae. Molecules 2009, 14, 4880–4891. [Google Scholar] [CrossRef]
- Mariotti, M.; Guigo, R. Selenoprofiles: Profile-based scanning of eukaryotic genome sequences for selenoprotein genes. BMC Bioinform. 2010, 26, 2656–2663. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Nelson, W.; Linstrom, S.C.; Boo, S.M.; Pueschel, C.; Qiu, H.; Bhattacharya, D. Rhodophyta. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 367–406. [Google Scholar]
- Parte, S.; Sirisha, V.L.; D’Souza, J.S. Biotechnological applications of marine enzymes from algae, bacteria, fungi, and sponges. Adv. Food Nutr. Res. 2017, 80, 75–106. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Yoon, H.S.; Bhattacharya, D. Red algal phylogenomics provides a robust framework for inferring evolution of key metabolic pathways. PLoS Curr. 2016, 8. [Google Scholar] [CrossRef]
- Price, N.M.; Harrison, P.J. Specific selenium-containing macromolecules in the marine diatom Thalassiosira pseudonana. Plant Physiol. 1988, 86, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Marin, B. Nested in the Chlorellales or independent class? Phylogeny and classification of the Pedinophyceae (Viridiplantae) revealed by molecular phylogenetic analyses of complete nuclear and plastid-encoded rRNA operons. Protist 2012, 163, 778–805. [Google Scholar] [CrossRef] [PubMed]
- Acton, E. Coccomyxa subellipsoidea, a new member of the palmellaceae. Annals Bot. 1909, 23, 573–577. [Google Scholar] [CrossRef]
- Mariotti, M.; Lobanov, A.V.; Guigo, R.; Gladyshev, V.N. SECISearch3 and Seblastian: New tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res. 2013, 41, e149. [Google Scholar] [CrossRef]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Gann, E.R.; Wilhelm, S.W. Infection by a giant virus (AaV) induces widespread physiological reprogramming in Aureococcus anophagefferens CCMP1984—A harmful bloom algae. Front Microbiol. 2018, 9, 752. [Google Scholar] [CrossRef]
- Weynberg, K.D.; Allen, M.J.; Wilson, W.H. Marine prasinoviruses and their tiny plankton hosts. Viruses 2017, 9, 43. [Google Scholar] [CrossRef]
- Melkonian, M. Virus-like particles in the scaly green flagellate Mesostigma viride. Br. Phycol. J. 1982, 17, 63–68. [Google Scholar] [CrossRef]
- Surek, B.; Melkonian, M. The filose amoeba Vampyrellidium perforans nov. sp. (Vampyrellidae, Aconchulinida): Axenic culture, feeding behaviour and host range specificity. Arch. Protistenkd. 1980, 123, 166–191. [Google Scholar] [CrossRef]
- Hess, S. Hunting for agile prey: Trophic specialisation in leptophryid amoebae (Vampyrellida, Rhizaria) revealed by two novel predators of planktonic algae. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed]
- Seto, K.; Degawa, Y. Collimyces mutans gen. et sp. nov. (Rhizophydiales, Collimycetaceae fam. nov.), a new chytrid parasite of Microglena (Volvocales, clade Monadinia). Protist 2018, 169, 507–520. [Google Scholar] [CrossRef]
- Matsuzaki, M.; Misumi, O.; Shin-I, T.; Ma ruyama, S.; Takahara, M.; Miyagishima, S.Y.; Mori, T.; Nishida, K.; Yagisawa, F.; Nishida, K.; et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae. Nature 2004, 428, 653–657. [Google Scholar] [CrossRef]
- Schiavon, M.; Ertani, A.; Parrasia, S.; Vecchia, F.D. Selenium accumulation and metabolism in algae. Aquat. Toxicol. 2017, 189, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gojkovic, Ž.; Garbayo, I.; Ariza, J.L.G.; Márová, I.; Vílchez, C. Selenium bioaccumulation and toxicity in cultures of green microalgae. Algal Res. 2015, 7, 106–116. [Google Scholar] [CrossRef]
- Kim, J.W.; Brawley, S.H.; Prochnik, S.; Chovatia, M.; Grimwood, J.; Jenkins, J.; LaButti, K.; Mavromatis, K.; Nolan, M.; Zane, M.; et al. Genome analysis of Planctomycetes inhabiting blades of the red alga Porphyra umbilicalis. PLoS ONE 2016, 11, e0151883. [Google Scholar] [CrossRef]
- Maruyama, S.; Misumi, O.; Ishii, Y.; Asakawa, S.; Shimizu, A.; Sasaki, T.; Matsuzaki, M.; Shin-i, T.; Nozaki, H.; Kohara, Y.; et al. The minimal eukaryotic ribosomal DNA units in the primitive red alga Cyanidioschyzon merolae. DNA Res. 2004, 11, 83–91. [Google Scholar] [CrossRef]
- Raven, J.A.; Giordano, M. Algae. Curr. Biol. 2014, 24, R590–R595. [Google Scholar] [CrossRef]
- Zhang, Y.; Romero, H.; Salinas, G.; Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006, 7, R94. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Lin, J.; Xu, Y.Z.; Zhang, Y. Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. ISME J. 2016, 10, 2048–2059. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.T.; Carpenter, E.J.; Tian, Z.; Bruskiewich, R.; Burris, J.N.; Carrigan, C.T.; Chase, M.W.; Clarke, N.D.; Covshoff, S.; Depamphilis, C.W.; et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE 2012, 7, e50226. [Google Scholar] [CrossRef] [PubMed]
- Rogers SO, B.A. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant. Plant Mol. Biol. 1985, 5, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Thangaraj, M.; Kathiresan, K. DNA Extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol. Biol. 2012, 2012, 205049. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. BMC Bioinform. 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Santesmasses, D.; Mariotti, M.; Guigo, R. Selenoprofiles: A computational pipeline for annotation of selenoproteins. Methods Mol. Biol. 2018, 1661, 17–28. [Google Scholar] [CrossRef]
- Santesmasses, D.; Mariotti, M.; Guigó, R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput. Biol. 2017, 13, e1005383. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. BMC Bioinform. 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. BMC Bioinform. 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
| 1KP Group | Sec Machinery | Selenoproteins (Sec) & Homologues | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group (513) | Clade/Order | Species Number | eEFSec | PSTK | SBP2 | SecS | SPS | Sec | Cys | Other |
| Vascular (175) | Conifers | 76 | 0 | 0 | 0 | 0 | 0 | 0 | 5256 | 2574 |
| Lycophytes | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 1473 | 678 | |
| Eusporangiate Monilo-phytes | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 640 | 280 | |
| Leptosporangiate Monilophytes | 68 | 0 | 0 | 0 | 0 | 0 | 0 | 4999 | 2184 | |
| Non-Vascular (70) | Hornworts | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 245 | 120 |
| Mosses | 39 | 0 | 1 | 5 | 17 | 0 | 1 | 3297 | 1485 | |
| Liverworts | 25 | 1 | 0 | 5 | 0 | 1 | 2 | 2184 | 1044 | |
| Algae (268) | Zygnematophyceae | 40 | 51 | 7 | 27 | 34 | 25 | 15 | 2426 | 1298 |
| Coleochaetophyceae | 4 | 3 | 2 | 1 | 1 | 1 | 1 | 217 | 120 | |
| Charophyceae | 2 | 1 | 2 | 1 | 1 | 2 | 4 | 103 | 65 | |
| Klebsormidiophyceae | 5 | 10 | 6 | 4 | 5 | 4 | 5 | 295 | 131 | |
| Mesostigmatophyceae | 4 | 4 | 2 | 4 | 4 | 1 | 12 | 199 | 124 | |
| Chlorophyta | 137 | 174 | 50 | 83 | 112 | 75 | 222 | 7518 | 4489 | |
| Glaucoplantae) | 6 | 6 | 4 | 5 | 2 | 3 | 4 | 281 | 177 | |
| Rhodoplantae | 35 | 6 | 4 | 4 | 9 | 5 | 5 | 1296 | 736 | |
| Chromista (algae) | 35 | 45 | 1 | 24 | 32 | 20 | 4 | 2100 | 1127 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, H.; Wei, T.; Xu, Y.; Li, L.; Kumar Sahu, S.; Wang, H.; Li, H.; Fu, X.; Zhang, G.; Melkonian, M.; et al. Phylogenomics Provides New Insights into Gains and Losses of Selenoproteins among Archaeplastida. Int. J. Mol. Sci. 2019, 20, 3020. https://doi.org/10.3390/ijms20123020
Liang H, Wei T, Xu Y, Li L, Kumar Sahu S, Wang H, Li H, Fu X, Zhang G, Melkonian M, et al. Phylogenomics Provides New Insights into Gains and Losses of Selenoproteins among Archaeplastida. International Journal of Molecular Sciences. 2019; 20(12):3020. https://doi.org/10.3390/ijms20123020
Chicago/Turabian StyleLiang, Hongping, Tong Wei, Yan Xu, Linzhou Li, Sunil Kumar Sahu, Hongli Wang, Haoyuan Li, Xian Fu, Gengyun Zhang, Michael Melkonian, and et al. 2019. "Phylogenomics Provides New Insights into Gains and Losses of Selenoproteins among Archaeplastida" International Journal of Molecular Sciences 20, no. 12: 3020. https://doi.org/10.3390/ijms20123020
APA StyleLiang, H., Wei, T., Xu, Y., Li, L., Kumar Sahu, S., Wang, H., Li, H., Fu, X., Zhang, G., Melkonian, M., Liu, X., Wang, S., & Liu, H. (2019). Phylogenomics Provides New Insights into Gains and Losses of Selenoproteins among Archaeplastida. International Journal of Molecular Sciences, 20(12), 3020. https://doi.org/10.3390/ijms20123020