Abstract
Pathogen-induced cell death is closely related to plant disease susceptibility and resistance. The cucumber (Cucumis sativus L.) mildew resistance locus O (CsMLO1) and calmodulin (CsCaM3) genes, as molecular components, are linked to nonhost resistance and hypersensitive cell death. In this study, we demonstrate that CsMLO1 interacts with CsCaM3 via yeast two-hybrid, firefly luciferase (LUC) complementation and bimolecular fluorescence complementation (BiFC) experiments. A subcellular localization analysis of green fluorescent protein (GFP) fusion reveals that CsCaM3 is transferred from the cytoplasm to the plasma membrane in Nicotiana benthamiana, and CsCaM3 green fluorescence is significantly attenuated via the coexpression of CsMLO1 and CsCaM3. CsMLO1 negatively regulates CsCaM3 expression in transiently transformed cucumbers, and hypersensitive cell death is disrupted by CsCaM3 and/or CsMLO1 expression under Corynespora cassiicola infection. Additionally, CsMLO1 silencing significantly enhances the expression of reactive oxygen species (ROS)-related genes (CsPO1, CsRbohD, and CsRbohF), defense marker genes (CsPR1 and CsPR3) and callose deposition-related gene (CsGSL) in infected cucumbers. These results suggest that the interaction of CsMLO1 with CsCaM3 may act as a cell death regulator associated with plant immunity and disease.
1. Introduction
In natural environments, plants are exposed to attack from a variety of microbial pathogens. Therefore, plants have acquired complex innate immune systems to combat long-term biotic threats, thereby reducing the damage caused by pathogen invasion. The evolutionary process of the plant–pathogen interaction is divided into four stages. In the first stage, plant pattern-recognition receptors (PRRs) recognize conserved pathogen-associated molecular patterns (PAMPs) of microorganisms, and PAMP-triggered immunity (PTI) is usually strong enough to prevent the colonization of most microbial pathogens. In the second stage, evolved pathogens secrete virulence factors that inhibit PTI to cause effector-triggered susceptibility (ETS) in plants. In the third stage, plants express a specific R gene to directly or indirectly recognize pathogen-specific effector factors, and then effector-triggered immunity (ETI) accelerates and amplifies PTI to cause disease resistance in plants. In the fourth stage, under the pressure of natural selection, the pathogen is forced to produce new effector factors to inhibit ETI, although plants also produce new R genes to activate ETI to maintain their survival [1,2,3]. Upon recognition of certain effector proteins, host cells undergo rapid programmed cell death, namely, the hypersensitive response (HR), which in turn inhibits pathogen infection [4].
Most plants are resistant to complete species of microbial invaders, a phenomenon that is known as nonhost resistance (NHR) [5]. NHR provides broad-spectrum and strong resistance to nonadapted pathogens in plants [6,7,8]. Currently, the mildew resistance locus O (MLO) gene in nonhost defense plays an important role in the model system of the interaction of barley with Blumeria graminis f. sp. hordei (Bgh) [9,10]. Many reports have found that barley mlo mutant plants are almost completely resistant to powdery mildew pathogens because the host cell wall thickens to prevent the penetration of powdery mildew [6,10,11]. Additionally, the three penetration proteins PEN1, PEN2, and PEN3, which are closely related to the MLO gene defense, are central components of cell wall-based defense against nonadapted powdery mildew [12]. In our laboratory, transcriptome and isobaric tags for relative absolute quantification (iTRAQ) analyses identified two genes, CsMLO1 and CsMLO2, in cucumber that are involved in the response to Corynespora cassiicola. Proteins/genes that are mainly involved in the defense response and oxidative stress and calcium signaling pathways during C. cassiicola infection have been identified in cucumber [13,14]. In addition, our research finds that the functions of CsMLO1 and CsMLO2 in C. cassiicola infection act as negative modulators to enhance the expression of reactive oxygen species (ROS)-related genes and defense-related genes for improved cucumber disease resistance [15]. However, few studies have focused on the resistance mechanism of MLO in cucumbers against C. cassiicola infection.
Hypersensitive cell death leads to the complex activation of defense signaling pathways, as the hallmarks of plant resistance, including the activation of ROS and defense genes, changes in the intercellular calcium levels, and modifications of cell walls and callose deposition [16,17]. The results of genetic epistasis analyses suggest that pathogen-responsive callose deposition has been detected in Atmlo2 mutant plants [18]. Arabidopsis MLO2 is a negative regulator of plant ROS responses to biotic stress [19]. In cucumber transgenic cotyledons, the transcription level of HR-associated genes is negatively regulated by CsMLO1 expression [15]. In addition, reports have shown that a calmodulin-binding domain in the cytoplasmic carboxy-terminal region of the MLO protein interacts with Ca2+-dependent calmodulin (CaM) [20,21]. The function of CaM has been extensively studied as an HR-related gene. For instance, after exposure to Xanthomonas campestris pv. vesicatoria, Agrobacterium spp.-mediated CaCaM1 overexpression activated ROS, nitric oxide (NO) generation, and HR-like cell death in pepper leaves, which exhibited enhanced resistance [22,23,24]. Furthermore, an analysis of a calcium inhibitor demonstrated that the inhibitor obstructed CaCaM1-triggered cell death, thus showing that CaCaM1-induced cell death depended on cytosolic Ca2+ [25,26]. Therefore, we hypothesize that there is a certain regulatory relationship between CsMLO and CsCaM that affects the resistance of cucumber to C. cassiicola infection.
In our laboratory, CsMLO1 (XM_004148737) and CsMLO2 (XM_004142345) of cucumber have been shown to be involved in the response to C. cassiicola. Phenotypic analyses of transgenic cucumber cotyledons inoculated with C. cassiicola revealed that transient overexpression of either CsMLO1 or CsMLO2 in cucumber cotyledons reduced resistance to C. cassiicola, whereas silencing of either CsMLO1 or CsMLO2 enhanced resistance to C. cassiicola. [13,14]. In addition, significant changes of calmodulin involved in calcium signaling were detected after treatment with C. cassiicola by iTRAQ analysis. However, the interaction between MLO and CaM in cucumber has not been precisely demonstrated, and the mechanism underlying the expression regulation between MLO and CaM is poorly understood. In this study, the analysis demonstrated that CsMLO1 stably interacted with CsCaM3 via yeast two-hybrid, firefly luciferase (LUC) complementation and bimolecular fluorescence complementation (BiFC) experiments. The coexpression of CsMLO1 and CsCaM3 in tobacco cells revealed that cytosolic CsCaM3 was transferred to the plasma membrane, where it colocalized with CsMLO1, which resulted in a decrease in the fluorescence of CsCaM3. Additionally, CsMLO1, as a negative modulator, can not only enhance the defense response to cucumber-C. cassiicola interactions, but also negatively regulate the expression of CsCaM3. Furthermore, CsMLO1 and CsCaM3 negatively regulated hypersensitive cell death, including ROS burst, cell death, and defense-related gene expression, under C. cassiicola infection. These observational mechanisms suggested that CsMLO1 interacted with CsCaM3 and negatively regulated CsCaM3 expression, thereby inhibiting plant immune responses.
2. Results
2.1. CsMLO-Mediated Regulation of CsCaM Expression Patterns in Cucumber Cotyledons
CsMLO1 and CsMLO2 were isolated previously and functionally characterized in C. cassiicola-infected cucumber leaves [15]. The sequences of calmodulin-7 (A0A0A0KWT3) were aligned with the cucumber genome database using the service provided by http://cucurbitgenomics.org/BLAST. Finally, we screened three of the most similar cucumber calmodulin genes: CsCaM1 (XM_011655459), CsCaM2 (XM_004144051), and CaM3 (XM_004142130). To further confirm these results, we determined the expression of cucumber calmodulin genes (CaM1, CaM2, and CaM3) in response to exposure to C. cassiicola for different time periods (Figure S1). Reverse-transcription quantitative PCR (RT-qPCR) assays showed that the expression patterns of CaM1, CaM2, and CaM3 were upregulated in the defense response of C. cassiicola, although CsCaM3 expression was rapidly and strongly induced in the early stages of C. cassiicola invasion. These results suggested that CsCaM3 might be a positive regulatory modulator that participates in the immunity of cucumber to C. cassiicola in the early stages of infection.
The results showed that the transcriptional levels of CsMLO1, CsMLO2, and CsCaMs changed after C. cassiicola infection, which in turn affected the defense response of cucumber to pathogens. These genes functioned in the defense response to C. cassiicola; thus, whether the CsMLO1 and CsMLO2 genes affected the transcription levels of CsCaM1, CsCaM2, and CsCaM3 was further investigated. In a previous study, an experimental method for the transient agroinfiltration of cucumber cotyledons was established [14,27,28]. Here, a fragment 387 bp in length between the CsMLO1/CsMLO2 and the green fluorescent protein (GFP) fusion genes was detected via PCR analysis in cucumber cotyledons transiently overexpressing CsMLO1/CsMLO2 (Figure 1A). The RT-qPCR assay showed that the expression of the CsMLO1/CsMLO2 genes was higher in the transgenic cucumbers than in the empty-vector control cucumbers (Figure 1B). Furthermore, the GFP imaging assay confirmed the success of the transient overexpression of CsMLO1/CsMLO2 in cucumber protoplasts (Figure 1C). CsMLO1-GFP fluorescence and CsMLO2-GFP fluorescence were located in the plasma membrane, while the GFP control was predominantly located in the cytosol and membrane. In addition, the localization of CsMLO1 and CsMLO2 appeared solely in the plasma membrane in Nicotiana benthamiana leaf cells (Figure S2). The above results further demonstrated that CsMLO1 and CsMLO2 overexpression was successful in transgenic cucumber cotyledons, and they also showed for the first time that the genes were primarily localized in the plasma membrane of cucumber protoplasts.
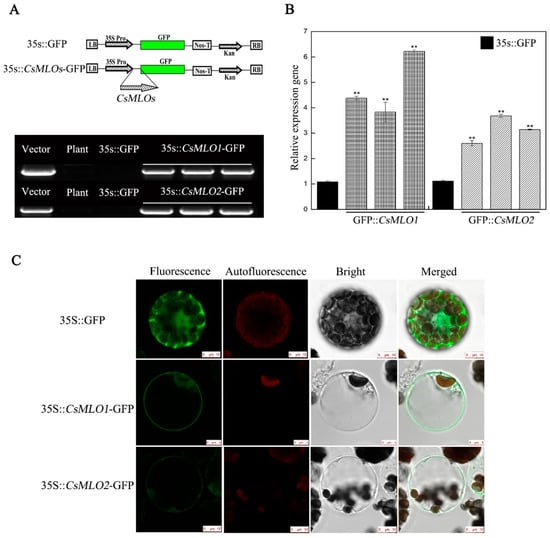
Figure 1.
CsMLO1- and CsMLO2-overexpressing constructs in cucumber cotyledons. (A) Schematic of the CsMLO1-GFP and CsMLO2-GFP constructs. PCR identification of CsMLO1 and CsMLO2 genetically modified of cucumber was successful. Vector, recombinant plasmid; Plant, nontransgenic cucumber; 35S::GFP, empty vector; 35S::CsMLO1-GFP, CsMLO1-overexpressing in cucumbers; 35S::CsMLO2-GFP, CsMLO2-overexpressing in cucumbers; (B) Transgenic plants were identified by RT-qPCR. Data are the means ± standard deviations from three independent experiments, and each column represents a sample containing three cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicate a significant difference (Student’s t test, ** P < 0.01). (C) Green fluorescence protein (GFP) was detected in the cucumber protoplasts of the 35S::GFP, 35S::CsMLO1-GFP and 35S::CsMLO2-GFP constructs. Transient expression in the protoplasts of cucumber cells of 35S::CsMLO1-GFP and 35S::CsMLO2-GFP were primarily localized in the plasma membrane. Fluorescence, chloroplast autofluorescence, bright field and merged images were obtained using a Leica confocal microscope. Scale bars = 10 μm.
Moreover, we used tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) to silence CsMLO1/CsMLO2 transcripts in cucumber cotyledons (Figure 2). After an Agrobacterium suspension (TRV::00, TRV::CsMLO1, and TRV::CsMLO2) was injected into cucumber cotyledons for 10 days, the chlorotic mosaic symptoms of TRV emerged in the cotyledons of transgenic cucumber plants, while no symptoms appeared in the control, suggesting that TRV successfully invaded the cucumber plants (Figure 2A). The efficient silencing of CsMLO1 and CsMLO2 was confirmed by RT-qPCR (Figure 2B). These results indicated that expression was almost abolished in the CsMLO1-silenced and CsMLO2-silenced cucumbers. To explore whether the expression of HR-related genes (CsCaMs) was regulated by CsMLO1/CsMLO2, we analyzed the transcription levels of CsCaM1, CsCaM2 and CsCaM3 in transgenic cucumber cotyledons (Figure 3). In our experiment, the transcript levels of CsCaM1, CsCaM2, and CsCaM3 were upregulated in CsMLO1-silenced cucumber cotyledons compared with those in the TRV::00 plants, although the transcript level of CsCaM3 was more discernibly regulated than CsCaM1 and CsCaM2. CsCaM1 and CsCaM3 expression was reduced while CsCaM2 expression was increased in the CsMLO1-overexpressing plants compared with that in the control plants. However, only the transcript level of CsCaM1 was increased in CsMLO2-silenced cucumber cotyledons but no alteration in gene expression occurred in the other patterns. Based on the above results, the stronger correlation between CsMLO1 and CsCaM3 showed that transient CsMLO1 expression negatively regulated CsCaM3 accumulation in cucumber cotyledons. However, the gene expression of CsCaMs was not significantly regulated by CsMLO2.
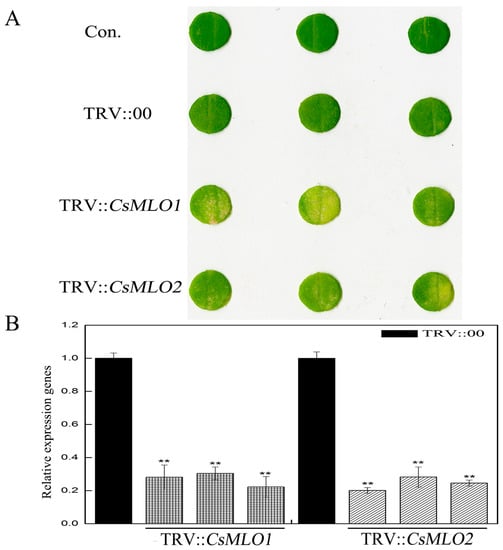
Figure 2.
CsMLO1-silencing and CsMLO2-silencing constructs in cucumber cotyledons. (A) Cucumber cotyledons showed chlorotic mosaic symptoms of TRV in CsMLO1-silencing and CsMLO2-silencing constructs; (B) CsMLO1-silencing and CsMLO2-silencing constructs identified in transgenic plants by RT-qPCR. Black pillars represent the empty vector control; gray grid pillars represent the efficient silencing of CsMLO1 and CsMLO2 by RT-qPCR. Data are the means ± standard deviations from three biological experiments, and each column represents a sample containing three cucumber cotyledons from different plants. Expression analysis of the candidate genes using the 2−ΔΔCt method. The asterisks indicate a significant difference (Student’s t test, ** P < 0.01).
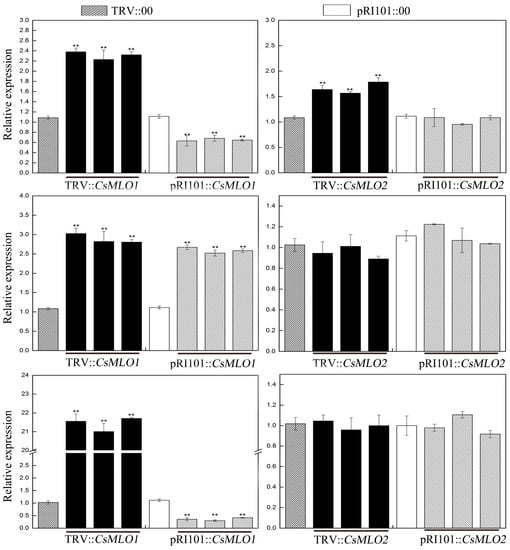
Figure 3.
CsMLO1-mediated expression patterns of CsCaM in cucumber cotyledons via RT-qPCR. Data are the means ± standard deviations from three biological experiments, and each column represents a sample containing three cucumber cotyledons from different plants. Expression analysis of the candidate genes using the 2−ΔΔCt method. The asterisks indicate a significant difference (Student’s t test, ** P < 0.01).
2.2. CsMLO1 Interacts with CsCaM3 in Yeast and Plants
Next, we examined the interaction between CsMLO1 or CsMLO2 and CsCaM1, CsCaM2, and CsCaM3 (Figure 4). Two cDNAs from the CaM-binding domain of CsMLO1 and CsMLO2 (CaMBD2 and CaMBD1, respectively) were constructed into pGBKT7 (BD/CsMLO1 and BD/CsMLO2). Three cDNAs from CsCaM1, CsCaM2, and CsCaM3 were constructed into pGADT7 (AD/CaM1, AD/CaM2, AD/CaM3, respectively). All yeast strains cotransformed with plasmids were grown on synthetic dropout (SD) medium lacking Trp-Leu-His-Ade. The results showed that the yeast combinations BD/CsMLO1+AD/CsCaM3 grew well on SD/-Trp-Leu-His-Ade plates while other strains cotransformed with plasmids did not grow normally (Figure 4A). Collectively, these results indicated that CaMBD1 of CsMLO1 specifically interacted with CsCaM3 in the yeast two-hybrid system.
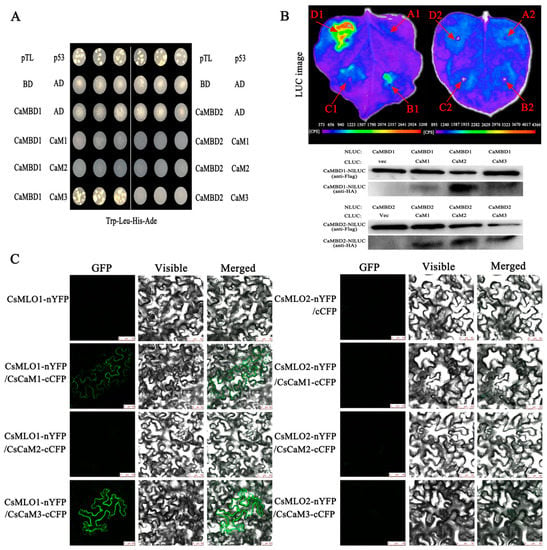
Figure 4.
CsMLO interacts with CsCaM in yeast and N. benthamiana epidermal cells. (A) BD/CsMLO1 or BD/CsMLO2 interacts with AD/CaM1, AD/CaM2, and AD/CaM3 on synthetic dropout medium lacking Trp-Leu-His-Ade in the yeast two-hybrid system. BD, bait; AD, prey; and p53/pTL, positive control. (B) Firefly luciferase complementation (LUC) imaging assay and western blot analysis of the interaction between CsMLO and CsCaM in N. benthamiana leaves. Arrows indicate leaf panels that were infiltrated with Agrobacterium containing the indicated constructs. A1: cLUC+CaMBD1-nLUC; B1: cLUC-CaM1+CaMBD1-nLUC; C1: cLUC-CaM2+CaMBD1-nLUC; D1: cLUC-CaM3+CaMBD1-nLUC; A2: cLUC+CaMBD2-nLUC, B2: cLUC-CaM1+CaMBD2-nLUC, C2: cLUC-CaM2+CaMBD2-nLUC; and D2: cLUC-CaM3+CaMBD2-nLUC. The western blot below shows the expression levels of cLUC- and nLUC-fusion proteins. (C) Bimolecular fluorescence complementation (BiFC) analysis of the interaction with CsMLO1/CsCaM1, CsMLO1/CsCaM2, CsMLO1/CsCaM3, CsMLO2/CsCaM1, CsMLO2/CsCaM2, and CsMLO2/CsCaM3 in N. benthamiana leaves infiltrated with Agrobacterium. (Left) Reconstitution of YFP-derived fluorescence. (Middle) Bright field images. (Right) Merged fluorescence images. (Scale bar: 50 μm.).
The LUC assay is a convenient system and provides a rapid analysis of protein functions in N. benthamiana plants presenting Agrobacterium-mediated transient expression of genes [29,30]. Therefore, we tested the LUC activity in leaves coexpressing different constructs by Agrobacterium-mediated transient expression. Figure 4B shows that nLUC-MLO1 (CaMBD1) and the empty cLUC vector did not display LUC complementation while coinfiltration of Agrobacterium containing nLUC-MLO1 (CaMBD1) and cLUC-CaM1/cLUC-CaM2/cLUC-CaM3 resulted in LUC complementation. However, strong fluorescent signaling was detected at nLUC-MLO1 (CaMBD1) and cLUC-CaM3. We also detected LUC complementation following the coexpression of nLUC-MLO2 (CaMBD2) and cLUC-CaM1/cLUC-CaM2/cLUC-CaM3, which showed a very low background as the negative control construct containing nLUC-MLO2 and the empty cLUC vector. Western blotting showed that the corresponding proteins were coexpressed in tobacco leaves, indicating that the LUC fluorescence of tobacco leaves was due to interprotein interactions.
The interaction in plants was also examined via a BiFC analysis [31]. The NH2-proximal half of yellow fluorescent protein (YFP) (nYFP) was fused to CsMLO1 and CsMLO2; and the C-proximal half of CFP (cCFP) was fused to CsCaM1, CsCaM2, and CsCaM3. Six combinations of fusion proteins, CsMLO1-nYFP+CsCaM1-cCFP, CsMLO1-nYFP+CsCaM2-cCFP, CsMLO1-nYFP+CsCaM3-cCFP, CsMLO2-nYFP+CsCaM1-cCFP, CsMLO2-nYFP+CsCaM2-cCFP, and CsMLO2-nYFP+CsCaM3-cCFP, were transiently co-overexpressed in N. benthamiana leaves (Figure 4C). The results showed that the green fluorescence was increased in leaves transiently co-overexpressing CsMLO1-nYFP+CsCaM1-cCFP, CsMLO1-nYFP+CsCaM2-cCFP, and CsMLO1-nYFP+CsCaM3-cCFP compared with the leaves of the control (CsMLO1-nYFP+cCFP), indicating that CsMLO1 interacted with CsCaM1, CsCaM2, and CsCaM3. Interestingly, the green fluorescence of the CsMLO1-nYFP+CsCaM3-cCFP fusion proteins was brighter than that of the other fusion proteins (CsMLO1-nYFP+CsCaM1-cCFP and CsMLO1-nYFP+CsCaM2-cCFP). However, no or weaker green fluorescence was detected in the fusion proteins (CsMLO2-nYFP+CsCaM1-cCFP, CsMLO2-nYFP+CsCaM2-cCFP, and CsMLO2-nYFP+CsCaM3-cCFP). The above experiments indicate that the interaction between CsMLO1 and CsCaM3 was highly stable. Collectively, our results supported the hypothesis that CsMLO1 interacted with CsCaM3 in plants.
2.3. Subcellular Colocalization of CsMLO1 and CsCaM3 in Plants
To investigate whether CsCaM3 localization was altered via the coexpression with CsMLO1, transient coexpression of CaCaM3 and CaMLO1 was observed in N. benthamiana leaves and protoplasts (Figure 5; Figure 6). CsCaM3-GFP fluorescence was predominantly located in the cytosol and plasma membrane, which was the same as in the GFP control, while CsMLO1-GFP was mainly localized in the plasma membrane. The coexpression of CsCaM3-GFP and CsMLO1-GFP transferred the fluorescence of CsCaM3-GFP from the cytoplasm to the plasma membrane, thereby resulting in a significant reduction in the fluorescence emission of CsCaM3-GFP compared to that in cells expressing CsCaM3-GFP alone. However, the coexpression of the empty vector and CsMLO3-GFP did not reveal this phenomenon of the fluorescence transfer of CsMLO3-GFP (Figure S3). This result further confirmed the interaction between CsCaM3-GFP and CsMLO1-GFP in the tobacco epidermis and protoplasts. Additionally, CsMLO1-GFP blocked the accumulation of CsCaM3-GFP in tobacco cells. Here, the result of CsMLO1 negatively regulating CsCaM3 expression was further confirmed.
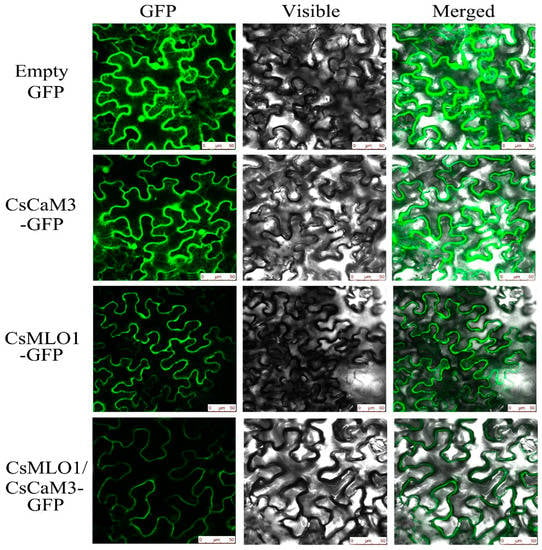
Figure 5.
Subcellular localization analyses of CsCaM3-GFP, CsMLO1-GFP, and CsCaM3-GFP+CsMLO1-GFP in transiently transformed N. benthamiana leaves. Confocal images of green fluorescent protein (GFP), CsCaM3-GFP, CsMLO1-GFP, and CsCaM3-GFP+ CsMLO1-GFP in N. benthamiana epidermal cells 3 days after Agroinfiltration. Bars = 50 μm.
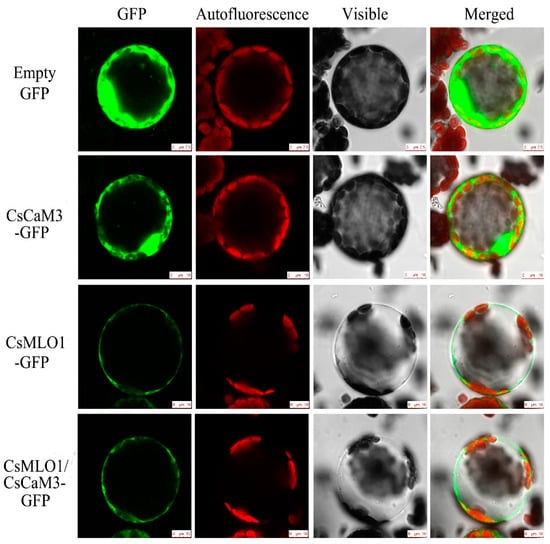
Figure 6.
Subcellular localization analyses of CsCaM3-GFP, CsMLO1-GFP, and CsCaM3-GFP+CsMLO1-GFP in transiently transformed N. benthamiana protoplasts. Confocal images of green fluorescent protein (GFP), CsCaM3-GFP, CsMLO1-GFP, and CsCaM3-GFP+CsMLO1-GFP in N. benthamiana protoplasts 3 days after Agroinfiltration. Chloroplast autofluorescence (red). Bars = 50 μm.
2.4. Inhibition of CsCaM3-Triggered Defense Response in CsMLO1-Overexpressing Cucumber Cotyledons
To examine the function of coexpressing CsMLO1 and CsCaM3 in the defense response of cucumber plants to C. cassiicola infection, Agrobacterium carrying the 35S::00, 35S::CsMLO1, 35S::CsCaM3, and 35S::CsMLO1/35S::CsCaM3 constructs was infiltrated into the cucumber cotyledons (Figure 7). Compared with the control cucumbers, cucumbers transiently overexpressing CsMLO1 had strongly induced necrotic lesions in the cotyledons at 5 dpi of C. cassiicola invasion. However, transient overexpression of CsCaM3 reduced necrotic lesions in cucumber cotyledons, whereas coexpression of CsMLO1 and CsCaM3 in cucumber cotyledons enhanced necrotic lesions compared with the overexpression of CsCaM3 alone. In addition, we also investigated disease status via a disease index analysis (Table 1). CsMLO1-overexpressing cucumber cotyledons were susceptible and presented an infection index of 73.61%, and CsCaM3-overexpressing cucumber cotyledons showed high resistance and presented a low infection index of 21.48%. However, the disease index (49.46%) of CsMLO1/CsCaM3-overexpressing cucumber cotyledons significantly increased or decreased compared with that of CsCaM3- or CsMLO1-overexpressing cucumber cotyledons, respectively.
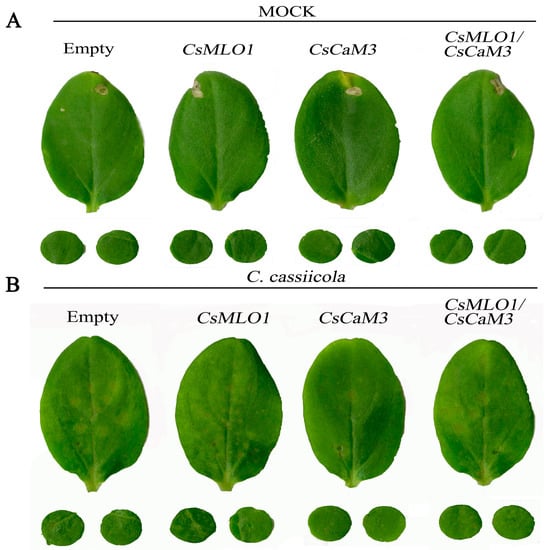
Figure 7.
Identification of disease-resistance of empty vector control, CsCaM3-overexpressing, CsMLO1-overexpressing, and CsCaM3-GFP+CsMLO1-GFP-overexpressing constructs in cucumber cotyledons. (A) Phenotype of uninfected cucumber cotyledons. (B) Phenotype of cucumber cotyledons after C. cassiicola challenge.

Table 1.
Disease index of different cucumber cotyledons to C. cassiicola.
To define the defense response of cucumber plants at the cellular level, we performed various histochemical analyses on cucumber cotyledons infected with C. cassiicola for 3 days (Figure 8). The induction of HR-like cell death and callose deposition was significantly increased in CsCaM3-overexpressing cucumber cotyledons at 5 days after Agrobacterium transformation by trypan blue and aniline staining analysis, although the coexpression of CsMLO1 and CsCaM3 significantly suppressed cell death in these tissues (Figure 8A). When challenged with C. cassiicola, the cucumber cotyledons overexpressing CsCaM3 demonstrated the highest rate of cell death, while the cucumber cotyledons overexpressing CsMLO1 demonstrated the lowest rate of cell death. Cell death and callose deposition were reduced in plant tissues coexpressing CsMLO1 and CsCaM3 compared to plant tissues overexpressing CsCaM3, and this reduction might be mediated by CsMLO1 after 3 days of C. cassiicola infection (Figure 8B). Similarly, the CsCaM3-overexpressing cucumber cotyledons showed large brown spots compared with the empty-vector control, and this phenomenon showed that the cotyledons accumulated high levels of H2O2 in the CsCaM3-overexpressing cucumber cotyledons when detected by 3,3′-diaminobenzidine (DAB, a histochemical reagent for H2O2) staining. Interestingly, coexpression of CsMLO1 and CsCaM3 in cucumber cotyledons strongly reduced H2O2 accumulation compared to that in the CsCaM3-overexpressing cucumber cotyledons. Furthermore, the infection of cucumber cotyledons also triggered H2O2 accumulation (Figure 8A). The accumulation of H2O2 was evidently increased in the CsCaM3-overexpressing cucumber cotyledons after inoculation with C. cassiicola, although this accumulation was also inhibited in the CsMLO1 and CsCaM3-coexpressing cucumber cotyledons based on DAB staining (Figure 8B).
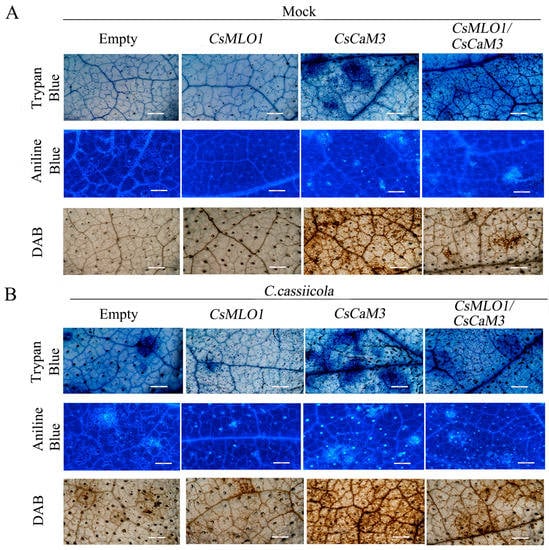
Figure 8.
Effects of agroexpression of CsCaM3 and CsMLO1 on cell death and defense responses of cucumber leaves. (A) Trypan blue, aniline blue, and DAB staining of cucumber cotyledons 5 d after agroexpression with the indicated transgenes. (B) Trypan blue, aniline blue, and DAB staining of cucumber cotyledons 3 d after C. cassiicola challenge with transgenes. Bars = 500 μm.
Transcription level measurements supported significantly attenuated or strengthened cell death and H2O2 burst in CsMLO1- or CsCaM3-overexpressing cucumber cotyledons, respectively (Figure 9; Figure 10). CsCaM3 overexpression strongly induced H2O2 signaling-related genes, including ascorbate peroxidase (CsPO1) and NADPH oxidase homolog (CsRbohD and CsRbohF), including defense-related genes (CsPR1 and CsPR3) and callose deposition-related gene (CsGSL). However, these related genes were not induced in CsMLO1-overexpressing cells (Figure 9). Notably, the extent of the increase in the transcription levels of these genes was significantly inhibited after coexpression of CsMLO1 and CsCaM3. To examine whether the transcription level of related defense genes in transgenic cucumbers was changed under the stress of C. cassiicola, we also detected the expression of the above related genes (Figure 10). The data showed that changes in the expression of these genes were consistent with the trend of the above analyzed defense-related genes. Interestingly, coexpression of both CsMLO1 and CsCaM3 in cucumber cotyledons strongly inhibited the expression of CsPR3 compared to that in the CsCaM3-overexpressing cotyledons. Collectively, these results suggested that CsMLO1 overexpression suppressed CsCaM3-regulated cell death and defense responses to C. cassiicola, including defense gene expression and ROS burst. The above experiments demonstrated that CsMLO1 can negatively regulate the expression of CsCaM3, thereby changing the HR effect of cucumber resistance on C. cassiicola.
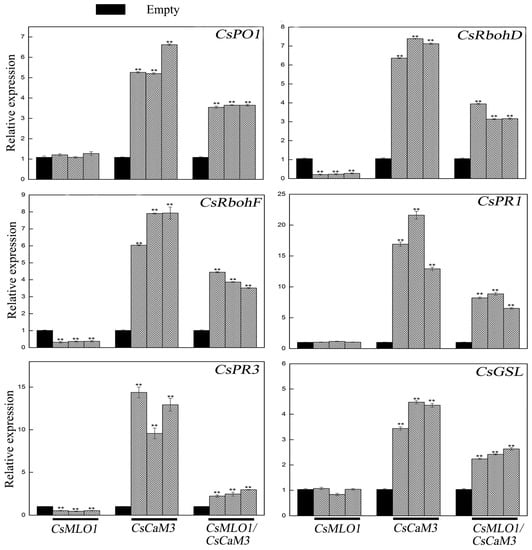
Figure 9.
Expression analysis of the CsRbohD (cucumber NADPH oxidase homolog D), CsRbohF (cucumber NADPH oxidase homolog F), CsPO1 (cucumber ascorbate peroxidase), CsPR1 (cucumber defense marker gene), CsPR3 (cucumber defense marker gene), and CsGSL (cucumber callose deposition-related gene) genes were determined by RT-qPCR in cucumber cotyledons 5 d after agroexpression. Data are the means ± standard deviations from three biological experiments, and each column represents a sample containing three cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicate a significant difference (Student’s t test, ** P < 0.01).
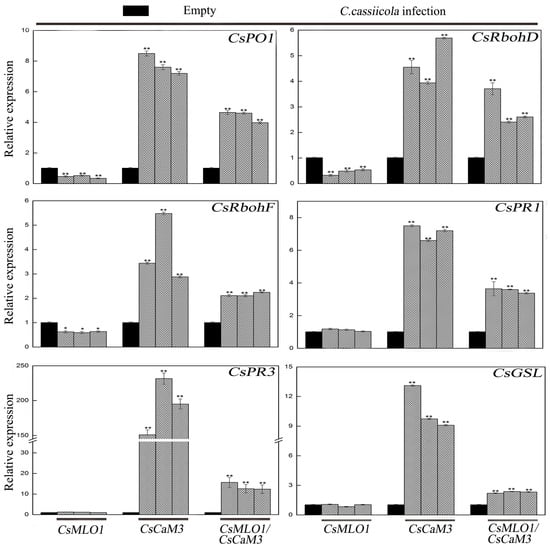
Figure 10.
Expression analysis of the CsRbohD, CsRbohF, CsPO1, CsPR1, CsPR3 and CsGSL genes were determined by RT-qPCR in cucumber cotyledons 5 d after C. cassiicola inoculation. Data are the means ± standard deviations from three biological experiments, and each column represents a sample containing three of cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicate a significant difference (Student’s t test, * P < 0.05 or ** P < 0.01).
2.5. CsMLO1 Silencing Enhances Cucumber Resistance to C. cassiicola Infection
TRV-based VIGS technology has been effectively used in cucumber reverse genetics research and can detect the functions of defense-related genes in cucumber plants [14,27,28]. The defense resistance of CsMLO1 silencing was evaluated following C. cassiicola inoculation for 5 days in transgenic cucumbers (Figure 11). The CsMLO1-silenced cucumbers exhibited enhanced disease resistance as demonstrated by milder symptoms than those of the controls (Figure 11A). To determine whether CsMLO1 silencing regulated the expression of genes involved in ROS signaling and cell death under C. cassiicola infection, we analyzed the transcript levels of these defense-related genes by RT-qPCR (Figure 11B). After 5 days of infection, CsMLO1 silencing distinctly induced the expression of genes, including CsPO1, CsRhobD, CsRhobF, CsPR1, CsPR3, CsGSL, and CsCaM3, compared with the empty-vector control leaves. In particular, the transcript levels of CsPR3 were more significantly multiplied in CsMLO1-silenced cucumber cotyledons. These results indicated that a high level of defense-related genes is required for the CsMLO1-mediated resistance response to C. cassiicola.
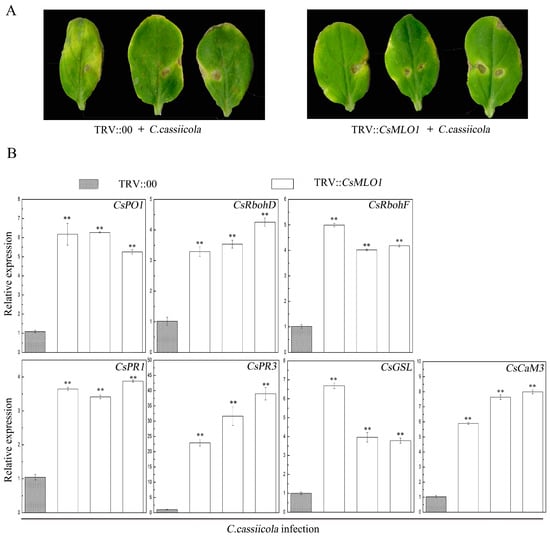
Figure 11.
Identification of disease resistance in the empty vector control and CsCsMLO1 silencing in cucumber cotyledons. (A) Phenotype of the cucumber cotyledons after C. cassiicola challenge. (B) Expression analysis of the CsRbohD, CsRbohF, CsPO1, CsPR1, CsPR3, CsGSL and CsCaM3 (cucumber Calmodulin) genes were determined via RT-qPCR in CsMLO1-silencing cucumber cotyledons 5 d after C. cassiicola inoculation. Data are the means ± standard deviations from three biological experiments, and each column represents a sample containing three cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicate a significant difference (Student’s t test, ** P < 0.01).
3. Discussion
Traditionally, MLO functions have been associated with susceptibility and resistance to powdery mildew disease [32]. Powdery mildew, as an ascomycete pathogen, can infiltrate into epidermal host cells and trigger various plant defense-related responses, including transcriptional activation of PR genes, callose deposition of papillae, and biosynthesis of antimicrobial biomolecules [33]. In Arabidopsis, the callose deposition in papillae acts as a physical barrier against powdery mildew invasion to improve resistance [18,34]. Callose is a polymeric (1→3)-β-D-glucan that is synthesized by plasma membrane-resident glucan synthase-like (GSL) proteins. In an Arabidopsis mlo mutant genetic screen, GSL5-generated callose is mainly deposited in the wound site and pathogen-triggered papillae [18]. The Arabidopsis mlo2 mutation confers partial resistance to Golovinomyces orontii, while the mlo2 mlo6 mlo12 mutation results in complete immunity. These phenomena are mainly due to the prevention of fungal onset before the fungus successfully penetrates the host cell wall [18]. Papilla formation is usually accompanied by a spatial and temporal burst of ROS, and ROS act as a signal transduction intermediate to activate the relevant signaling pathways [35,36]. Studies in barley are consistent with those in Arabidopsis because the mlo mutants also enhanced callose deposition, ROS bursts and the pathogen-induced expression of PR genes in resistance to Blumeria graminis f. sp. hordei (Bgh) [34]. Therefore, MLO can negatively regulate the resistance of powdery mildew in plants.
Additionally, barley and Arabidopsis mlo resistance requires three resistance components, i.e., the PEN genes, which were initially discovered to defend against nonadaptive powdery mildew Blumeria graminis f. sp. hordei and Erysiphe pisi [9,12]. Inducible defense responses in nonhost plants include the accumulation of ROS, activation of pathogenesis-related genes, localized reinforcement of the plant cell walls and HR [6,37]. Currently, information regarding the defense mechanisms of the cucumber–C. cassiicola interaction is limited. In our previous study, an iTRAQ analysis suggest that the occurrence of dynamic changes in the early stages of C. cassiicola stress. The accumulation of NADPH oxidases (RBOHs) and level of PR proteins in cucumber leaves under C. cassiicola infection at 24 h post-infection show that cucumber plants increase the synthesis of ROS and defense-related genes to prevent the invasion of pathogenic fungi [13]. Wang has shown that secondary metabolism and ROS accumulation play important roles in disease resistance during cucumber–C. cassiicola interactions [16]. Moreover, we first found that CsMLO1 is a negative modulator that regulates the defense response of cucumbers to C. cassiicola [15]. Next, the relationships of CsPR and ROS-associated genes with the overexpression and silencing of CsMLO1 in noninfiltrated cucumber plants was investigated, and the results show that CsMLO1 acts as a negative regulator to activate/inhibit the expression of related genes [15]. Thus, we propose that these genes, including ROS-related genes and defense marker genes, are important in cucumber immunity to C. cassiicola infection. The function of the HR-related gene CaM has been fully studied in recent years. For example, the transient expression of CaCaM1 increased the ROS burst and hypersensitive cell death to improve the defense responses in pepper [23]. In this study, CsMLO1 and CsCaM3 are linked to the defense response to C. cassiicola, and we hypothesized that the resistance mechanism of CsMLO1 and CsCaM3 in the regulation of cucumbers to C. cassiicola was related to NHR.
We adopted the yeast two-hybrid, LUC and BiFC methods to test the interaction and role of CsMLO and CsCaM in plant defense responses. The results showed that CsMLO1 stably interacted with CsCaM3. In barley, a CaM-binding domain (CaMBD) in MLO interacts with Ca2+-dependent calmodulin proteins, and this loss of binding halves the ability of MLO to negatively regulate powdery mildew in vivo [20]. Pathogen-triggered Ca2+ fluxes can enhance the oxidative burst to activate resistance responses [16,17]. However, increasing Ca2+ ions also promotes the affinity of CaM for its target protein, MLO, to suppress resistance in barley [20]. At present, the function of the CaM–MLO complex is rarely studied in cucumber resistance or susceptibility to C. cassiicola. In the Agrobacterium-mediated transient transformation, CsCaM3 was transferred from the cytoplasm to the plasma membrane with a reduction in CsCaM3 accumulation when CsMLO1 and CsCaM3 were coexpressed in tobacco cells. Furthermore, CsMLO1 in transiently transformed transgenic cucumbers can negatively regulate the expression of CsCaM3 by RT-qPCR. In contrast, CsMLO1 silencing in cucumber plants significantly enhances the expression of the cell death marker genes CsCaM3 and CsPO1, ROS-related genes, defense marker genes, and callose deposition-related genes during C. cassiicola infection. In pepper, the pathogen-response genes CaCaM1 and CaPO2 can activate ROS signaling and HRs to improve defense resistance in pepper leaves [23]. CaMLO2 silencing enhanced resistance against virulent Xcv infection, which is related to ROS bursts and PR gene expression, suggesting that CaMLO2 plays a role in abolishing stress-induced cell death and H2O2 burst at the infection site [38]. Collectively, these findings support the hypothesis that CsMLO1 can negatively regulate CsCaM3 expression to affect the HR of cucumbers to C. cassiicola.
Further studies are required to define the molecular mechanisms underlying CsCaM3–CsMLO1 complex-induced cell death signaling in the cucumber-C. cassiicola interaction. Agrobacterium-mediated transient CsCaM3 overexpression induced cell death and enhanced defense against C. cassiicola, suggesting that CsCaM3 might act as a positive regulator of cell death and disease resistance in cucumber plants. As expected, cell death, callose deposition, and ROS burst were rapidly induced in CsCaM3-overexpressing leaves under C. cassiicola infection. In pepper, early defense indicators have been detected, such as callose deposition and ROS generation during infection, suggesting that transient CaCaM1 overexpression induces localized cell death and enhances locally acquired resistance against virulent Xcv in pepper leaves [38,39]. Furthermore, coexpression of CaCaM1 and CaMLO2 significantly suppresses AvrB-triggered cell death and defense responses [40]. In our study, coexpression of CsMLO1 with CsCaM3 also significantly reduced HR-associated cell death in response to C. cassiicola infection. Increasing evidence indicates that plant responses to pathogen invasions via the regulation of the accumulation of ROS, such as H2O2 and O2·-, are related to AtrbohD and AtrbohF [41,42,43,44,45,46]. ROS generation and cell death induction are also regulated by the expression of the peroxidase gene CaPO2 in pepper leaves [47]. Under C. cassiicola infection, the mRNA levels of CsPO1, CsRbohD, and CsRbohF were significantly increased in CsCaM3-overexpressing cotyledons, although their transcription levels were inhibited via the coexpression of CsCaM3 and CsMLO1 in cucumber. Furthermore, triggering HR-like cell death is accompanied by callose deposition and defense marker gene induction, including pathogenesis-related protein 1-1a (PR1-1a) and chitinase (PR3), which are upregulated to promote resistance to pathogens [18,39,48,49]. Similar to these findings, the strong induction of defense marker genes (CsPR1 and CsPR3) and callose deposition-related genes (CsGSL) was observed in CsCaM3-overexpressing cotyledons, although the coexpression of CsCaM3 with CsMLO1 compromised the accumulation of the transcription levels of CsPR1 and CsPR3 and CsGSL. Collectively, our findings combined with these data support the hypothesis that the coexpression of CsMLO1 with CsCaM3 negatively regulates an early step in C. cassiicola-triggered cell death and immunity in cucumber leaves. In our study, we did not detect the interaction of the CsCaM protein with CsMLO2. We speculate that there may be other proteins that interact with CsMLO2, such as the membrane protein ROP (Rho-related GTPase of plant) [13,50]. In addition, the role of CsMLO2 as a negative regulator in cucumber disease resistance remains unknown. These functional speculations about CsMLO2 will be the focus of further studies.
In conclusion, CsMLO1 is a negative modulator that enhances the defense response of cucumbers and stably interacts with CsCaM3 and transfers CsCaM3 in the cytoplasm to the plasma membrane, thereby blocking the accumulation of CsCaM3. Furthermore, the coexpression of CsCaM3 and CsMLO1 significantly inhibited hypersensitive cell death after C. cassiicola infection, suggesting that CsMLO1 negatively regulates CsCaM3 expression, which results in the inhibition of defense-related gene activation.
4. Materials and methods
4.1. Plant Materials and Pathogen Inoculation
Cucumber (C. sativus L. cv. Xintaimici) and tobacco (N. benthamiana) plants were sown in pots with peat:vermiculite (1:2, v/v) under greenhouse conditions (25 °C, 16 h light/8 h dark cycle). At the second-third leaf stage, the plants were sprayed with a C. cassiicola spore suspension, for which the spores had been harvested in sterile water and adjusted to a final concentration of 105 microconidia/mL. Cucumber leaves were incubated at 25 °C after C. cassiicola inoculation, and then the second leaves were collected from three plants at 3 h, 6 h, 12 h, 24 h, 48 h, 72 h, and 144 h. The control plants were inoculated with distilled water. Infected leaves were sampled at various time points for the fungal growth assay, RNA isolation, and the histochemical assay.
The disease progression of corynespora leaf spot was estimated on the basis of the severity of leaf scabs as follows: Grade 0: no scabs observed; Grade 1: less than 1/10 of leaves infected; Grade 3: 1/10–1/4 of leaves infected; Grade 5: 1/4–1/2 of leaves infected; Grade 7: 1/2–3/4 of leaves infected; and Grade 9: more than 3/4 of leaves infected. Disease index = 100 × ∑(no. of diseased leaves of each grade × disease grade) / (total no. leaves × 9).
4.2. RNA Extraction, cDNA Synthesis, and Reverse-Transcription Quantitative PCR
Total RNA extraction and cDNA synthesis were performed as previously described [15]. The primers for candidate genes were designed using QuantPrime—a flexible primer design tool for high-throughput qPCR by http://quantprime.mpimp-golm.mpg.de/. RT-qPCR was performed on a SYBR Green I 96-I system (Roche fluorescence quantitative PCR instrument, Basel). The cucumber actin gene was used as the internal reference [51]. The relative expression of the target genes was calculated using the 2−ΔΔCT method [52]. Leaves/cotyledons sampled at each sampling point were divided into three groups (three leaves per group) and then evenly mixed for RNA extraction. RNA extracted from each group was used as one biological replicate for RT-qPCR. A total of three biological replicates were performed. All primers used to detect gene expression are listed in Table S1.
4.3. Yeast Two-Hybrid Assay
The PCR-amplified full-length coding sequence (CDS) regions of CsCaM1, CsCaM2, and CsCaM3 were ligated into the pGADT7 vector using the EcoRI and BamHI restriction sites. CaMBD1 (CsMLO1) and CaMBD2 (CsMLO2) were ligated into the pGBKT7 vector using the EcoRI and BamHI restriction sites. The recombinant plasmids BD/CsMLO1, BD/CsMLO2, AD/CsCaM1, AD/CsCaM2, and AD/CsCaM3 were generated. The primers used are shown in Table S2. BD and AD vectors were cotransformed into the Y2HGold yeast strain. Transformants were arrayed on interaction-selection medium (SD medium lacking adenine, histidine, leucine, and tryptophan, supplemented with 10 mmol·L−1 CaCl2), and growth was scored as an indicator of the interactions between two proteins.
4.4. Firefly Luciferase Complementation Imaging Assay and Western Blot
The CaM-binding domains CaMBD1 (CsMLO1) and CaMBD2 (CsMLO2) were ligated into the pCAMBIA1300-NLUC vector using the KpnI and SalI restriction sites. The CDS regions of CsCaM1, CsCaM2 and CsCaM3 were ligated into the pCAMBIA1300-CLUC vector using the KpnI and SalI restriction sites. After sequencing, 35S::CaMBD1-nLUC, 35S::CaMBD2-nLUC, 35S::cLUC-CaM1, 35S::cLUC-CaM2, and 35S::cLUC-CaM3 fusion constructs were transformed into A. tumefaciens strain EHA105. Agrobacterium suspensions harboring the 35S::cLUC vector/35S::CaMBD1-nLUC, 35S::cLUC-CaM1/35S::CaMBD1-nLUC, 35S::cLUC-CaM2/35S::CaMBD1-nLUC, 35S::cLUC-CaM3/35S::CaMBD1-nLUC, 35S::cLUC vector/35S::CaMBD2-nLUC, 35S::cLUC-CaM1/35S::CaMBD2-nLUC, 35S::cLUC-CaM2/35S::CaMBD2-nLUC, and 35S::cLUC-CaM3/35S::CaMBD2-nLUC were infiltrated into N. benthamiana leaves using a needleless syringe. Luciferase activity was detected 3 days after infiltration using the 699 NightSHADE LB 985 imaging system (Berthold Technologies). Then, 0.2 mmol·L−1 luciferin (Promega; Madison, WI, USA) was sprayed on the surface of the tobacco leaves, and the LUC fluorescence intensity was measured after 5 min.
Total proteins were extracted from infiltrated leaves by grinding 0.3 g of leaf tissue in 3 mL buffer (0.0625 mol·L−1 Tris-HCl, pH 6.8, 0.5% SDS, 5% glycerin, 3% β-mercaptoethanol and 100% Triton X-100) at 4 °C for 1 h. Then, the solution was centrifuged at 10,000× g for 20 min at 4 °C. The resulting supernatant was the total protein extract fraction. The supernatant was supplemented with 10 mmol·L−1 CaCl2 and four volumes of acetone solution at -20 °C overnight. The next day, the crude extract was clarified by centrifugation at 10,000× g for 20 min at 4 °C. Twenty-four milligrams of protein dry powder was added to 50 µL of protein sample treatment solution (0.125 mol·L−1 Tris-HCl, pH 8.0, 2% SDS, 5% β-mercaptoethanol, 10% glycerin and 0.02% bromophenol blue) and boiling water for 3–5 min and then stored at −20 °C. The fractionated proteins were subjected to SDS-PAGE, and immunoblotting was performed using standard methods. Immunoprecipitates were electrophoretically separated by SDS-PAGE and transferred to a nitrocellulose membrane (Amersham, Shanghai, China). Related proteins (Flag-tagged CsCaMBD1, Flag-tagged CsCaMBD2, HA-tagged CsCaM1, HA-tagged CsCaM2 and HA-tagged CsCaM3 using Flag and HA antibodies) were detected by immunoblot using antibodies anti-Flag (Beyotime, Shanghai, China) and anti-HA (Beyotime, Shanghai, China). Anti-Flag and anti-HA were diluted 1:1000 with TBST buffer. The secondary antibody (anti-mouse, Cell Signaling Technology, Boston, MA, USA) was diluted 1:1000 with TBST buffer. The primers used are shown in Table S2.
4.5. Bimolecular Fluorescence Complementation Analysis
A BiFC analysis was performed to identify the interactions between CsMLO1/CsCaM1, CsMLO1/CsCaM2, CsMLO1/CsCaM3, CsMLO2/CsCaM1, CsMLO2/CsCaM2, and CsMLO2/CsCaM3. The BiFC constructs with the full-length CDS regions CsMLO1, CsMLO2, CsCaM1, CsCaM2, and CsCaM3 (without termination codons) were ligated into intermediate vectors pDONR221 by the BP reaction. Finally, the target gene fragments were individually constructed into the corresponding functional vectors PXNGW (nYFP) and PXCGW (cCFP) by the LR reaction. A. tumefaciens strain GV3101 containing BiFC constructs was coinfiltrated into N. benthamiana leaves. Leaves were visualized 2–3 days after agroinfiltration using a confocal laser scanning microscope (Leica TCS SP8, Solms, Germany) to detect YFP. All assays were repeated independently at least three times with comparable results. The primers used are shown in Table S3.
4.6. Construction of Fusion Proteins, Plant Transformation, and Fluorescence Microscopy
The CDSs of CsMLO1 and CsCaM3 were inserted between the CaMV 35S promoter and the soluble-modified GFP (eGFP) in the pRI101-GFP vector to generate the eGFP-fusion proteins CsMLO1-GFP and CsCaM3-GFP, respectively. To determine CsCaM3 localization in the presence of CsMLO1, the CsCaM3-GFP construct was cobombarded with pRI10135S::CsMLO1 lacking the C-terminal GFP. Recombinant plasmids were transformed into A. tumefaciens strain EHA 105 and then centrifuged after overnight culture. The precipitate was cultured in induction medium (10 mmol·L−1 ethanesulfonic acid (pH 5.7), 10 mmol·L−1 MgCl2 and 200 mmol·L−1 acetosyringone), harvested, diluted to OD600 = 0.6, and then injected into N. benthamiana leaves. Two days after infiltration, the epidermal cells and protoplasts were observed using a confocal laser scanning microscope (Leica TCS SP8, Solms, Germany). The filter sets BP505-530 (excitation 488 nm, emission 505 to 530 nm) were used to detect GFP. Protoplasts were extracted from transformed tobacco leaves, which were cut into 0.5–1 mm thin strips, quickly transferred to the enzymatic hydrolysate (20 mmol·L−1 MES, 1.5% Cellulase R-10, 0.4% Macerozyme R-10, 20 mmol·L−1 KCL, 0.4 mol·L−1 mannitol, 10 mmol·L−1 CaCl2, and 0.1% BSA, pH 5.7), subjected to 30 min of vacuum infiltration, and then shaken at 50 rpm for 6 h. The enzymatic hydrolysate was diluted with an equal amount of W5 (0.2 mol·L−1 MES, 1.54 mol·L−1 NaCl, 1 mol·L−1 CaCl2, and 0.2 mol·L−1 KCl, pH 5.7) and then filtered through a 200-m nylon membrane. The collected filtrate was centrifuged at 100× g for 2 min. The supernatant was slowly removed, and the remaining green liquid contained protoplasts. An equal amount of precooled MGG was placed in the protoplasts, which were placed on ice for observation using a confocal laser scanning microscope. The primers used are shown in Table S3.
The CDSs of CsMLO1 and CsCaM3 were inserted between the CaMV 35S promoter and the soluble-modified green fluorescent protein (GFP) (eGFP) in the pRI101-GFP vector to generate the eGFP-fusion proteins CsMLO1-GFP and CsCaM3-GFP, respectively. To determine CsCaM3 localization in the presence of CsMLO1, the CsCaM3-GFP construct was cobombarded with pRI10135S::CsMLO1 lacking the C-terminal GFP. Recombinant plasmids were transformed into A. tumefaciens strain EHA 105, which were centrifuged after overnight culture. The precipitate was cultured in induction medium (10 mmol·L−1 ethanesulfonic acid, pH 5.7, 10 mmol·L−1 MgCl2 and 200 mmol·L−1 acetosyringone), harvested and diluted to OD600 = 0.6, and then injected into N. benthamiana leaves. Two days after infiltration, the epidermal cells and protoplasts were observed using a confocal laser scanning microscope (Leica TCS SP8, Solms, Germany). The filter sets BP505-530 (excitation 488 nm, emission 505 to 530 nm) were used to detect GFP. Protoplasts were extracted from transformed tobacco leaves. The transformed tobacco leaves were cut into 0.5–1 mm thin strips, quickly transferred to the enzymatic hydrolysate (20 mmol·L−1 MES, 1.5% Cellulase R-10, 0.4% Macerozyme R-10, 20 mmol·L−1 KCL, 0.4 mol·L−1 mannitol, 10 mmol·L−1 CaCl2, and 0.1% BSA, pH 5.7), and after 30 min of vacuum infiltration, shaken at 50 rpm for 6 h. The enzymatic hydrolysate was diluted with an equal amount of W5 (0.2 mol·L−1 MES, 1.54 mol·L−1 NaCl, 1 mol·L−1 CaCl2, and 0.2 mol·L−1 KCl, pH 5.7) and then filtered through a 100 μm nylon membrane. The collected filtrate was centrifuged at 100× g for 2 min. The supernatant was slowly removed, and the remaining green liquid contained protoplasts. An equal amount of precooled MGG was placed in the protoplasts and placed on ice for observation using a confocal laser scanning microscope. The primers used are shown in Table S3.
4.7. Histochemical Analysis
For the visualization of H2O2 accumulation with 3,3’-diaminobenzidine (DAB; Sigma, St. Louis, MO, USA) [53], cucumber cotyledons transiently expressing CsMLO1 and/or CsCaM3 were assessed by DAB staining. The infected cucumber cotyledons were soaked in 1 mg·mL−1 DAB for 8 h, boiled for 20 min in 3:1:1 ethanol/lactic acid/glycerol and then transferred to 95% ethanol at 4 °C for storage. Three independent replicates were performed for each assay.
We detected the cell death of cucumber cotyledons by trypan blue staining [54]. The treated cucumber cotyledons were immersed in trypan blue solution, boiled for 10 min, and placed at 37 °C overnight. Then, the dyed leaves were repeatedly subjected to decolorizing and photographed.
We detected callose deposition in cucumber cotyledons transiently expressing CsMLO1 and/or CsCaM3 by aniline blue staining [55,56]. The cucumber cotyledons were stored in FAA fixative (50% ethanol, 5% glacial acetic acid, 10% formaldehyde and 35% H2O) for 24 h. The cotyledons were carefully washed with 100% ethanol, and 500 μL of 50% ethanol was added and incubated for 30 min on the bench after removing 100% ethanol. After ethanol was removed, 500 μL 67 mM K2HPO4 (pH 12) was added, and the leaves were incubated for 30 min. Finally, 500 μL staining solution (0.01% aniline blue in 67 mmol·L−1 K2HPO4) was added, and the cotyledons were incubated for 1 h in the dark to perform UV microscopy. Three independent replicates were performed for each assay.
4.8. Cucumber Cotyledon Transformation
The CDS regions of CsMLO1 and CsMLO2 were ligated into the pRI101-eGFP vector using the SalI and BamHI restriction sites as previously described. Then, recombinant plasmids were generated to transiently overexpress the CsMLO1 gene and CsMLO2 gene in cucumber cotyledons. pTRV-based VIGS was performed to knock down the CsMLO1 gene and CsMLO2 gene in cucumber cotyledons. A 402 bp fragment within the 3′ region of CsMLO1 cDNA (nucleotides 1348–1749) and a 387 bp fragment within the 3′ region of CsMLO2 cDNA (nucleotides 1347–1725) were cloned into the pTRV vector (TRV:CsMLO1 and TRV:CsMLO2, respectively) as previously described [16]. All recombinant plasmids were transformed into A. tumefaciens strain EHA105. Agrobacterium-mediated transformations with the 35s::GFP, GFP::CsMLO1, GFP::CsMLO2, pTRV, pTRV-CsMLO1, and pTRV-CsMLO2 genes were carried out. The samples were supplemented with 10 mmol·L−1 MES, 10 mmol·L−1 MgCl2, and 200 μmol·L−1 AS and then infiltrated into fully expanded cotyledons of cucumber plants (OD600 = 0.4 for each construct). The plants were then placed in a growth room at 22 °C with a 16 h light and 8 h dark photoperiod for growth.
4.9. Statistical Analysis
Primer design and sequence alignment were conducted using Primer 5 software. Data are the mean ± standard deviation from three biological replicates per cultivar. Standard errors of deviation were assessed by Excel. Statistical significance was analyzed by Student’s t-test (P < 0.05 or P < 0.01) using SPSS software.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/12/2995/s1.
Author Contributions
G.Y. contributed to the experimental design, cucumber planting and sampling, tobacco planting and experimenting, data processing, results analysis and writing. X.W. and Q.C. provide assistance during the experiment. H.F., N.C. and Y.Y. defined the work objectives and technical approach, results analysis and writing. All authors read and approved the final manuscript.
Funding
This study was financially supported by the National Key Research Program of China (2016YFD0201004), Key Projects of Basic Scientific Research Projects in Liaoning Province Colleges and Universities (2017004) and Shenyang Young and Middle-aged Science and Technology Innovation Talents Project (RC170439).
Acknowledgments
We thank Zhihong Zhang and Yuanhu Xuan for donating the carrier.
Conflicts of Interest
The authors declare that this research is conducted in the absence of any financial or commercial relationships that could be interpreted as a potential conflict of interest.
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Göhre, V.; Robatzek, S. Breaking the Barriers: Microbial Effector Molecules Subvert Plant Immunity. Annu. Rev. Phytopathol. 2008, 46, 189–215. [Google Scholar] [CrossRef] [PubMed]
- Bent, A.F.; Mackey, D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 2007, 45, 399–436. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Nürnberger, T.; Lipka, V. Non-host resistance in plants: New insights into an old phenomenon. Mol. Plant Pathol. 2005, 6, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Thordal-Christensen, H. Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 2003, 6, 351–357. [Google Scholar] [CrossRef]
- Jaouannet, M.; Rodriguez, P.A.; Thorpe, P.; Lenoir, C.J.; MacLeod, R.; Escudero-Martinez, C.; Bos, J.I. Plant immunity in plant-aphid interactions. Front. Plant Sci. 2014, 5, 663. [Google Scholar] [CrossRef] [PubMed]
- Strugala, R.; Delventhal, R.; Schaffrath, U. An organ-specific view on non-host resistance. Front. Plant Sci. 2015, 6, 526. [Google Scholar] [CrossRef] [PubMed]
- Humphry, M.; Consonni, C.; Panstruga, R. Mlo-based powdery mildew immunity: Silver bullet or simply non-host resistance? Mol. Plant Pathol. 2006, 7, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Lipka, U.; Fuchs, R.; Lipka, V. Arabidopsis non-host resistance to powdery mildews. Curr. Opin. Plant Biol. 2008, 11, 404–411. [Google Scholar] [CrossRef]
- Collins, N.C.; Thordal-Christensen, H.; Lipka, V.; Bau, S.; Kombrink, E.; Qiu, J.L.; Hückelhoven, R.; Stein, M.; Freialdenhoven, A.; Somerville, S.C.; et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 2003, 425, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Johansson, O.N.; Fantozzi, E.; Fahlberg, P.; Nilsson, A.K.; Buhot, N.; Tör, M.; Andersson, M.X. Role of the penetration resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race specific resistance in Arabidopsis thaliana. Plant J. 2014, 79, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.C.; Yu, Y.; Fan, H.; Zhang, D.; Cui, N.; Wang, X.; Jia, S.; Yang, Y.; Zhao, J. Analysis of protein regulation in the leaves of resistant cucumber after inoculation with Corynespora cassiicola: A proteomic approach. Biochem. (Mosc.), Accepted but not yet published.
- Wang, X.; Zhang, D.; Cui, N.; Yu, Y.; Yu, G.; Fan, H. Transcriptome and miRNA analyses of the response to Corynespora cassiicolain cucumber. Sci. Rep. 2018, 8, 7798. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.C.; Fan, H.Y.; Cui, N.; Wang, X.Y.; Meng, X.N. Mildew resistance locus O genes CsMLO1 and CsMLO2 are negative modulators of the Cucumis sativus defense response to Corynespora cassiicola. Unpublished.
- Blume, B.; Nürnberger, T.; Nass, N.; Scheel, D. Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 2000, 12, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Brown, I.; Adams, S.; Knight, M.; Ainslie, A.; Mansfield, J. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 2000, 23, 441–450. [Google Scholar] [CrossRef]
- Consonni, C.; Bednarek, P.; Humphry, M.; Francocci, F.; Ferrari, S.; Harzen, A.; Ver Loren van Themaat, E.; Panstruga, R. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 Mutant. Plant Physiol. 2010, 152, 1544–1561. [Google Scholar] [CrossRef]
- Cui, F.; Wu, H.; Safronov, O.; Zhang, P.; Kumar, R.; Kollist, H.; Salojärvi, J.; Panstruga, R.; Overmyer, K. Arabidopsis MLO2 is a negative regulator of sensitivity to extracellular ROS. Plant Cell Environ. 2018, 41, 782–796. [Google Scholar] [CrossRef]
- Kim, M.C.; Panstruga, R.; Elliott, C.; Müller, J.; Devoto, A.; Yoon, H.W.; Park, H.C.; Cho, M.J.; Schulze-Lefert, P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 2002, 416, 447–451. [Google Scholar] [CrossRef]
- Bhat, R.A.; Miklis, M.; Schmelzer, E.; Schulze-Lefert, P.; Panstruga, R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 2005, 102, 3135–3140. [Google Scholar] [CrossRef]
- Jung, H.W.; Hwang, B.K. Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol. Plant Microbe Interact. 2000, 13, 136–142. [Google Scholar] [CrossRef]
- Choi, H.W.; Lee, D.H.; Hwang, B.K. The pepper calmodulin gene CaCaM1 is involved in reactive oxygen species and nitric oxide generation required for cell death and the defense response. Mol. Plant Microbe Interact. 2009, 22, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kim, D.S.; Chung, E.H.; Hwang, B.K. Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase 1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation dependent manner. Plant Physiol. 2014, 165, 76–91. [Google Scholar] [CrossRef]
- Pike, S.M.; Zhang, X.C.; Gassmann, W. Electrophysiological characterization of the Arabidopsis avrRpt2-specific hypersensitive response in the absence of other bacterial signals. Plant Physiol. 2005, 138, 1009–1017. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, W.; Smigel, A.; Tsai, Y.C.; Braam, J.; Berkowitz, G.A. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the activation of calmodulin or a calmodulin-like protein. Plant Physiol. 2008, 148, 818–828. [Google Scholar] [CrossRef]
- Meng, X.; Yu, Y.; Zhao, J.; Cui, N.; Song, T.; Yang, Y.; Chen, H. The two translationally controlled tumor protein genes, CsTCTP1 and CsTCTP2, are negative modulators in the Cucumis sativus defense response to Sphaerotheca fuliginea. Front. Plant Sci. 2018, 9, 544. [Google Scholar] [CrossRef]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W.; et al. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly Luciferase Complementation Imaging Assay for Protein-Protein Interactions in Plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- He, P.; Chintamanani, S.; Chen, Z.; Zhu, L.; Kunkel, B.N.; Alfano, J.R.; Tang, X.; Zhou, J.M. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 2004, 37, 589–602. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Panstruga, R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 2005, 33, 389–392. [Google Scholar] [CrossRef]
- Eichmann, R.; Hückelhoven, R. Accommodation of powdery mildew fungi in intact plant cells. J. Plant Physiol. 2008, 165, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Piffanelli, P.; Zhou, F.S.; Casais, C.; Orme, J.; Jarosch, B.; Schaffrath, U.; Collins, N.C.; Panstruga, R.; Schulze-Lefert, P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002, 129, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Hückelhoven, R.; Dechert, C.; Kogel, K.H. Non-host resistance of barley is associated with a hydrogen peroxide burst at sites of attempted penetration by wheat powdery mildew fungus. Mol. Plant Pathol. 2001, 2, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Mellersh, D.G.; Foulds, I.V.; Higgins, V.J.; Heath, M.C. H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. 2002, 29, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Nürnberger, T.; Brunner, F. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 2002, 5, 318–324. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J. 2012, 72, 843–855. [Google Scholar] [CrossRef]
- Kim, N.H.; Choi, H.W.; Hwang, B.K. Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol. Plant Microbe Interact. 2010, 23, 1069–1082. [Google Scholar] [CrossRef]
- Kim, D.S.; Choi, H.W.; Hwang, B.K. Pepper mildew resistance locus O interacts with pepper calmodulin and suppresses Xanthomonas AvrBsT-triggered cell death and defense responses. Planta 2014, 240, 827–839. [Google Scholar] [CrossRef]
- Hu, X.; Jiang, M.; Zhang, A.; Lu, J. Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 2005, 223, 57–68. [Google Scholar] [CrossRef]
- Hu, X.; Jiang, M.; Zhang, J.; Zhang, A.; Lin, F.; Tan, M. Calcium-calmodulin is required for abscisic acid-induced antioxidant defense and functions both upstream and downstream of H2O2 production in leaves of maize (Zea mays) plants. New Phytol. 2007, 173, 27–38. [Google Scholar] [CrossRef]
- Sang, J.; Zhang, A.; Lin, F.; Tan, M.; Jiang, M. Cross-talk between calcium-calmodulin and nitric oxide in abscisic acid signaling in leaves of maize plants. Cell Res. 2008, 18, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2018, 69, 3465–3476. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Kim, Y.J.; Lee, S.C.; Hong, J.K.; Hwang, B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, H.M.; Murali, M.; Anup, C.P.M.; Melvin, P.; Sharada, M.S. Salicylic acid seed priming instigates defense mechanism by inducing PR-Proteins in Solanum melongena L. upon infection with Verticillium dahliae Kleb. Plant Physiol. Biochem. 2017, 117, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.H.; Elmohamedy, R.S.R. Induction of Defense-Related Physiological and Antioxidant Enzyme Response against Powdery Mildew Disease in Okra (Abelmoschus esculentus L.) Plant by Using Chitosan and Potassium Salts. Mycobiology 2017, 45, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, S.; Sero, A.; Pratelli, R.; Pilot, G.; Chen, J.; Sardi, M.I.; Parsa, S.A.; Kim, D.Y.; Acharya, B.R.; Stein, E.V.; et al. A Membrane Protein/Signaling Protein Interaction Network for Arabidopsis Version AMPv2. Front. Physioogyl. 2010, 1, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKYW46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Chen, F.; Cheng, Z.; Wang, Y.; Yang, P.; Zhang, Y.; Cha, Z. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: Effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 2013, 64, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Miraz, P.; Kennedy, R.; Banniza, S.; Wei, Y. Dual trypan-aniline blue fluorescence staining methods for studying fungus-plant interactions. Biotech. Histochem. 2010, 85, 99–105. [Google Scholar]
- Klaus, H.; Andreas, H. Aniline Blue and Calcofluor White Staining of Callose and Cellulose in the, Streptophyte Green Algae Zygnema and Klebsormidium. Bio Protoc. 2016, 6. [Google Scholar] [CrossRef]
- Herburger, K.; Holzinger, A. Localization and Quantification of Callose in the Streptophyte Green Algae Zygnema and Klebsormidium: Correlation with Desiccation Tolerance. Plant Cell Physiol. 2015, 56, 2259–2270. [Google Scholar] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).