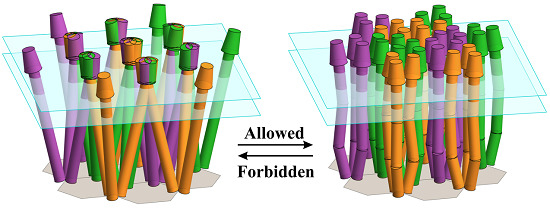
Spatial Restrictions in Chemotaxis Signaling Arrays: A Role for Chemoreceptor Flexible Hinges across Bacterial Diversity
Abstract
1. Introduction
2. Results
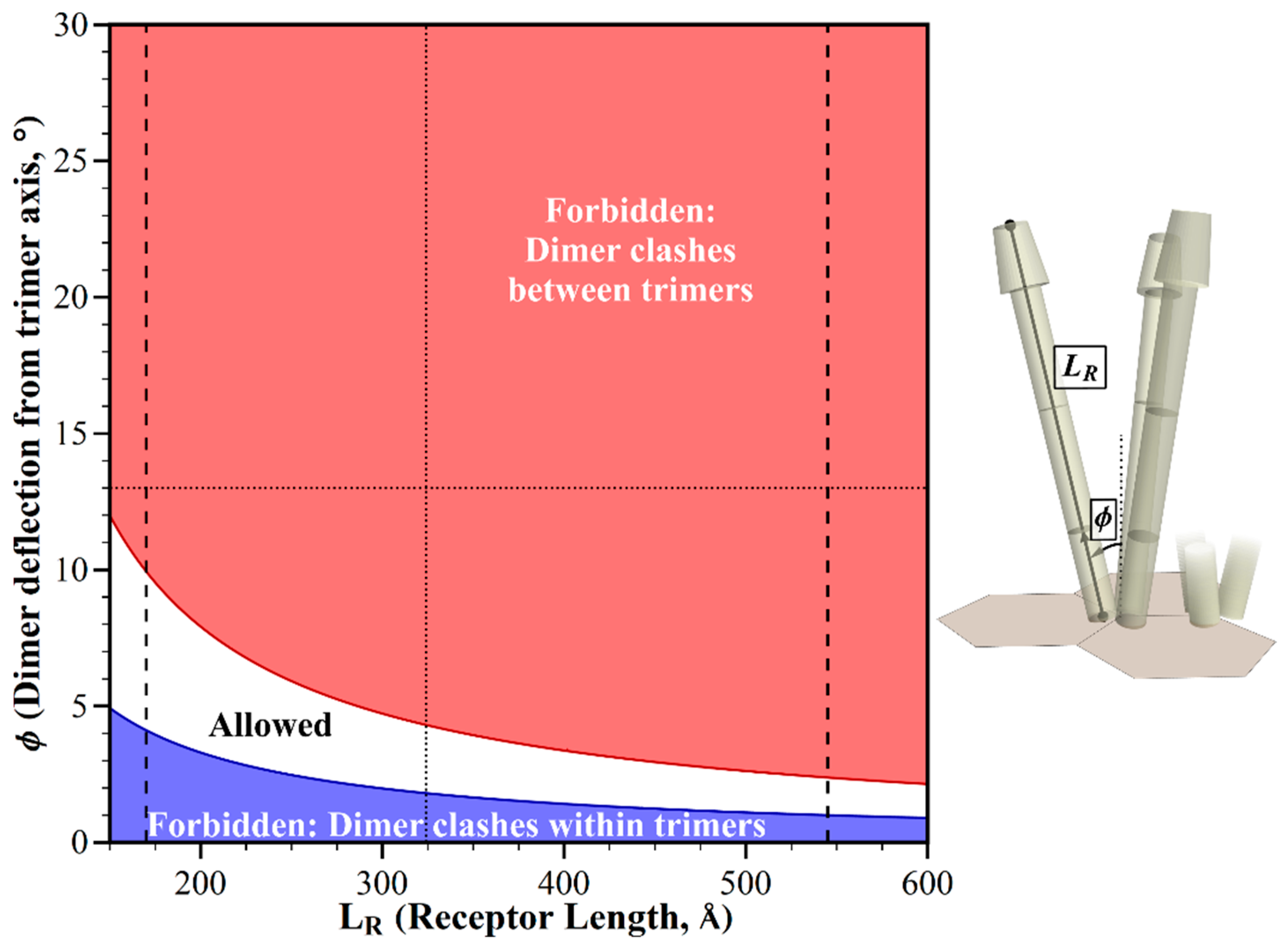
2.1. Geometric Restrictions as A Function of Chemoreceptor Length
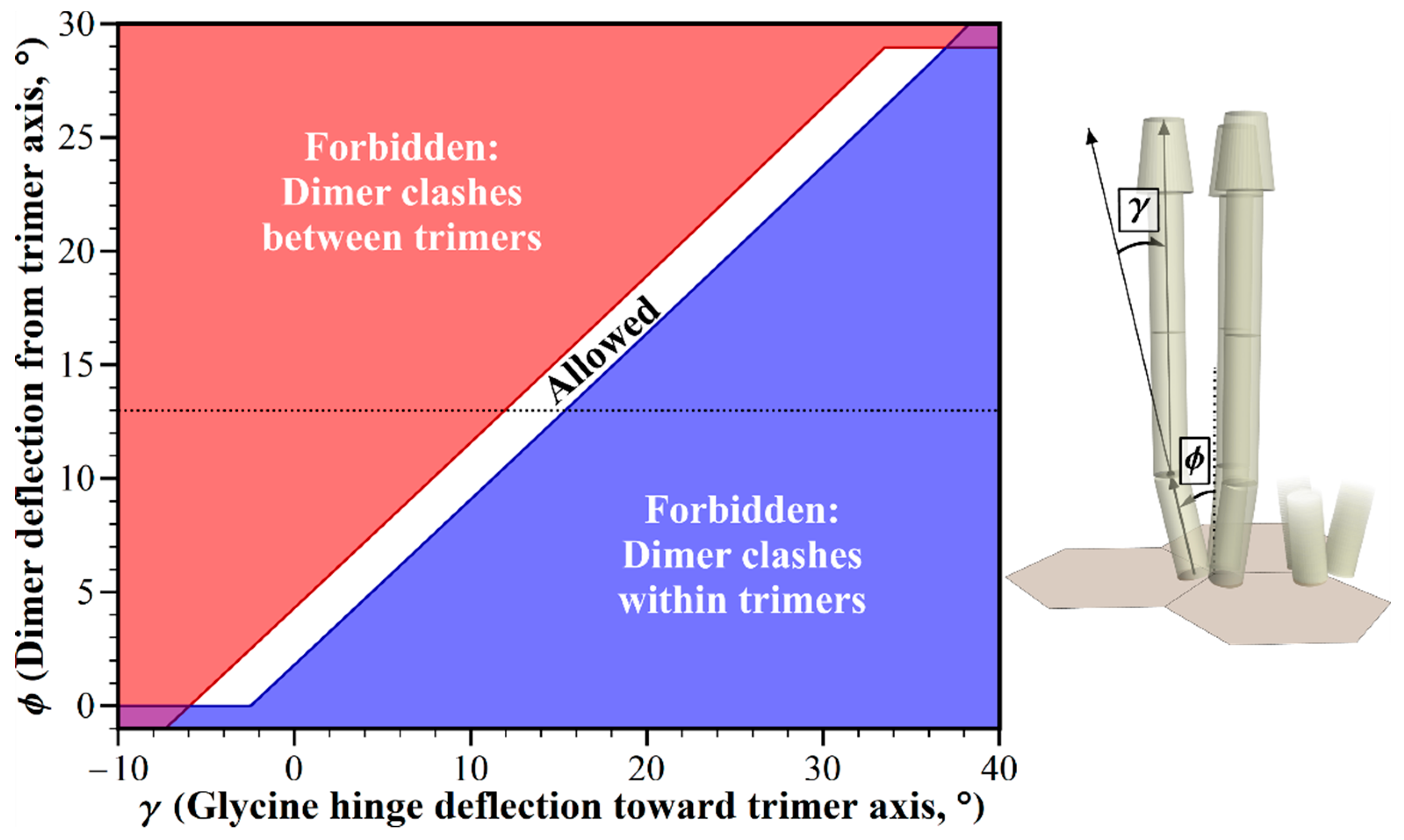
2.2. Geometric Restrictions as A Function of Dimer Deflection from the Trimer Central Axis
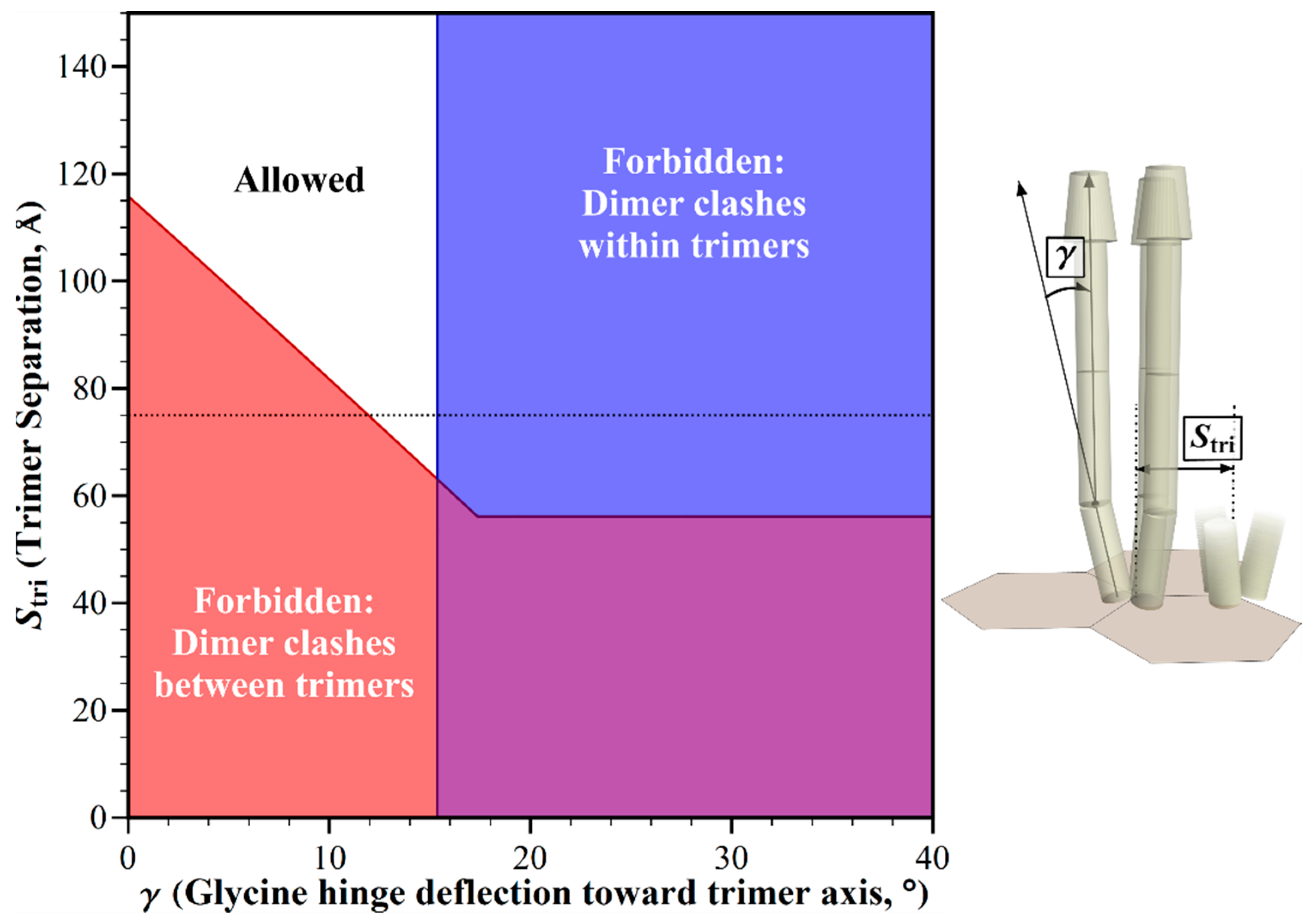
2.3. Structural Clashes as A Function of Trimer Separation
2.4. Avoiding Structural Clashes in the Absence of Receptor Bends
3. Discussion
3.1. Flexible Hinges in Chemoreceptors
3.2. Varying Other Geometric Parameters
3.3. Heterogeneous and Dynamic Receptor Bending
3.4. Other Roles for Flexible Hinges?
4. Materials and Methods
4.1. Geometric Modeling of Core Signaling Complexes and Arrays
4.2. Estimating the Shortest and Longest Chemoreceptors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HAMP | Histidine kinases, Adenylyl cyclases, Methyl-accepting chemoreceptors, and Phosphatases |
References
- Hazelbauer, G.L.; Falke, J.J.; Parkinson, J.S. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 2008, 33, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hazelbauer, G.L. Core unit of chemotaxis signaling complexes. Proc. Natl. Acad. Sci. USA 2011, 108, 9390–9395. [Google Scholar] [CrossRef] [PubMed]
- Briegel, A.; Li, X.; Bilwes, A.M.; Hughes, K.T.; Jensen, G.J.; Crane, B.R. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 3766–3771. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, B.; Morado, D.R.; Jani, S.; Manson, M.D.; Margolin, W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. USA 2012, 109, E1481–E1488. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fleetwood, A.D.; Bayas, C.; Bilwes, A.M.; Ortega, D.R.; Falke, J.J.; Zhulin, I.B.; Crane, B.R. The 3.2 A resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry 2013, 52, 3852–3865. [Google Scholar] [CrossRef] [PubMed]
- Pinas, G.E.; Frank, V.; Vaknin, A.; Parkinson, J.S. The source of high signal cooperativity in bacterial chemosensory arrays. Proc. Natl. Acad. Sci. USA 2016, 113, 3335–3340. [Google Scholar] [CrossRef] [PubMed]
- Akkaladevi, N.; Bunyak, F.; Stalla, D.; White, T.A.; Hazelbauer, G.L. Flexible hinges in bacterial chemoreceptors. J. Bacteriol. 2018, 200, 1–16. [Google Scholar] [CrossRef]
- Coleman, M.D.; Bass, R.B.; Mehan, R.S.; Falke, J.J. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry 2005, 44, 7687–7695. [Google Scholar] [CrossRef]
- Alexander, R.P.; Zhulin, I.B. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 2007, 104, 2885–2890. [Google Scholar] [CrossRef]
- Briegel, A.; Ortega, D.R.; Tocheva, E.I.; Wuichet, K.; Li, Z.; Chen, S.; Müller, A.; Iancu, C.V.; Murphy, G.E.; Dobro, M.J.; et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 2009, 106, 17181–17186. [Google Scholar] [CrossRef] [PubMed]
- Weis, R.M.; Hirai, T.; Chalah, A.; Kessel, M.; Peters, P.J.; Subramaniam, S. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 2003, 185, 3636–3643. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Khursigara, C.M.; Subramaniam, S.; Hazelbauer, G.L. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol. Microbiol. 2011, 79, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Zhulin, I.B.; Krell, T. Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, L.E.; Zhulin, I.B. Four-helix bundle: A ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics 2005, 21, iii45–iii48. [Google Scholar] [CrossRef] [PubMed]
- Dunin-Horkawicz, S.; Lupas, A.N. Comprehensive analysis of HAMP domains: Implications for transmembrane signal transduction. J. Mol. Biol. 2010, 397, 1156–1174. [Google Scholar] [CrossRef] [PubMed]
- Boldog, T.; Hazelbauer, G.L. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 2004, 13, 1466–1475. [Google Scholar] [CrossRef]
- Kim, K.K.; Yokota, H.; Kim, S.-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 1999, 400, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, C.K.; Himes, B.A.; Alvarez, F.J.; Ma, J.; Zhao, G.; Perilla, J.R.; Schulten, K.; Zhang, P. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. eLife 2015, 4, e08419. [Google Scholar] [CrossRef]
- Yang, W.; Alvarado, A.; Glatter, T.; Ringgaard, S.; Briegel, A. Baseplate variability of Vibrio cholerae chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 2018, 115, 13365–13370. [Google Scholar] [CrossRef]
- Bardy, S.L.; Maddock, J.R. Polar explorations: Recent insights into the polarity of bacterial proteins. Curr. Opin. Microbiol. 2007, 10, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, D.; McEvoy, A.L.; Shroff, H.; Crooks, G.E.; Wingreen, N.S.; Betzig, E.; Liphardt, J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009, 7, e1000137. [Google Scholar] [CrossRef] [PubMed]
- Pedetta, A.; Massazza, D.A.; Herrera Seitz, M.K.; Studdert, C.A. Mutational replacements at the “glycine hinge” of the Escherichia coli chemoreceptor Tsr support a signaling role for the C-helix residue. Biochemistry 2017, 56, 3850–3862. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.A.; Armitage, J.P.; Sansom, M.S.P. Mechanism of bacterial signal transduction revealed by molecular dynamics of Tsr dimers and trimers of dimers in lipid vesicles. PLoS Comput. Biol 2012, 8, e1002685. [Google Scholar] [CrossRef] [PubMed]
- Swain, K.E.; Falke, J.J. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: A disulfide mapping study. Biochemistry 2007, 46, 13684–13695. [Google Scholar] [CrossRef] [PubMed]
- Kitanovic, S.; Ames, P.; Parkinson, J.S. Mutational analysis of the control cable that mediates transmembrane signaling in the Escherichia coli serine chemoreceptor. J. Bacteriol. 2011, 193, 5062–5072. [Google Scholar] [CrossRef] [PubMed]
- Herrera Seitz, M.K.; Frank, V.; Massazza, D.A.; Vaknin, A.; Studdert, C.A. Bacterial chemoreceptors of different length classes signal independently. Mol. Microbiol. 2014, 93, 814–822. [Google Scholar] [CrossRef] [PubMed]
- Briegel, A.; Ladinsky, M.S.; Oikonomou, C.; Jones, C.W.; Harris, M.J.; Fowler, D.J.; Chang, Y.W.; Thompson, L.K.; Armitage, J.P.; Jensen, G.J. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife 2014, 3, e02151. [Google Scholar] [CrossRef]
- Mauriello, E.M.F.; Jones, C.; Moine, A.; Armitage, J.P. Cellular targeting and segregation of bacterial chemosensory systems. FEMS Microbiol. Rev. 2018, 42, 462–476. [Google Scholar] [CrossRef]
- Vaknin, A.; Berg, H.C. Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 592–596. [Google Scholar] [CrossRef]
- Vaknin, A.; Berg, H.C. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 2007, 366, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Vaknin, A.; Berg, H.C. Direct evidence for coupling between bacterial chemoreceptors. J. Mol. Biol. 2008, 382, 573–577. [Google Scholar] [CrossRef][Green Version]
- Briegel, A.; Ortega, D.R.; Huang, A.N.; Oikonomou, C.M.; Gunsalus, R.P.; Jensen, G.J. Structural conservation of chemotaxis machinery across Archaea and Bacteria. Environ. Microbiol. Rep. 2015, 7, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Wolfram Research, Inc. Mathematica; Version 11.1 ed.; Wolfram Research, Inc.: Champaign, IL, USA, 2017. [Google Scholar]
- Milburn, M.V.; Prive, G.G.; Milligan, D.L.; Scott, W.G.; Yeh, J.; Jancarik, J.; Koshland, D.E., Jr.; Kim, S.H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 1991, 254, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- Tajima, H.; Imada, K.; Sakuma, M.; Hattori, F.; Nara, T.; Kamo, N.; Homma, M.; Kawagishi, I. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J. Biol. Chem. 2011, 286, 42200–42210. [Google Scholar] [CrossRef] [PubMed]
- Lacal, J.; García-Fontana, C.; Muñoz-Martínez, F.; Ramos, J.-L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884. [Google Scholar] [CrossRef] [PubMed]
- Wiener, M.C.; White, S.H. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of X-ray and neutron diffraction data. III. Complete structure. Biophys. J. 1992, 61, 434–447. [Google Scholar] [CrossRef]
- Hulko, M.; Berndt, F.; Gruber, M.; Linder, J.U.; Truffault, V.; Schultz, A.; Martin, J.; Schultz, J.E.; Lupas, A.N.; Coles, M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 2006, 126, 929–940. [Google Scholar] [CrossRef]
- Airola, M.V.; Watts, K.J.; Bilwes, A.M.; Crane, B.R. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 2010, 18, 436–448. [Google Scholar] [CrossRef]
- Le Moual, H.; Koshland, D.E., Jr. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 1996, 261, 568–585. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stalla, D.; Akkaladevi, N.; White, T.A.; Hazelbauer, G.L. Spatial Restrictions in Chemotaxis Signaling Arrays: A Role for Chemoreceptor Flexible Hinges across Bacterial Diversity. Int. J. Mol. Sci. 2019, 20, 2989. https://doi.org/10.3390/ijms20122989
Stalla D, Akkaladevi N, White TA, Hazelbauer GL. Spatial Restrictions in Chemotaxis Signaling Arrays: A Role for Chemoreceptor Flexible Hinges across Bacterial Diversity. International Journal of Molecular Sciences. 2019; 20(12):2989. https://doi.org/10.3390/ijms20122989
Chicago/Turabian StyleStalla, David, Narahari Akkaladevi, Tommi A. White, and Gerald L. Hazelbauer. 2019. "Spatial Restrictions in Chemotaxis Signaling Arrays: A Role for Chemoreceptor Flexible Hinges across Bacterial Diversity" International Journal of Molecular Sciences 20, no. 12: 2989. https://doi.org/10.3390/ijms20122989
APA StyleStalla, D., Akkaladevi, N., White, T. A., & Hazelbauer, G. L. (2019). Spatial Restrictions in Chemotaxis Signaling Arrays: A Role for Chemoreceptor Flexible Hinges across Bacterial Diversity. International Journal of Molecular Sciences, 20(12), 2989. https://doi.org/10.3390/ijms20122989