Cell-Free, Embryo-Specific sncRNA as a Molecular Biological Bridge between Patient Fertility and IVF Efficiency
Abstract
1. Introduction
2. Results and Discussion
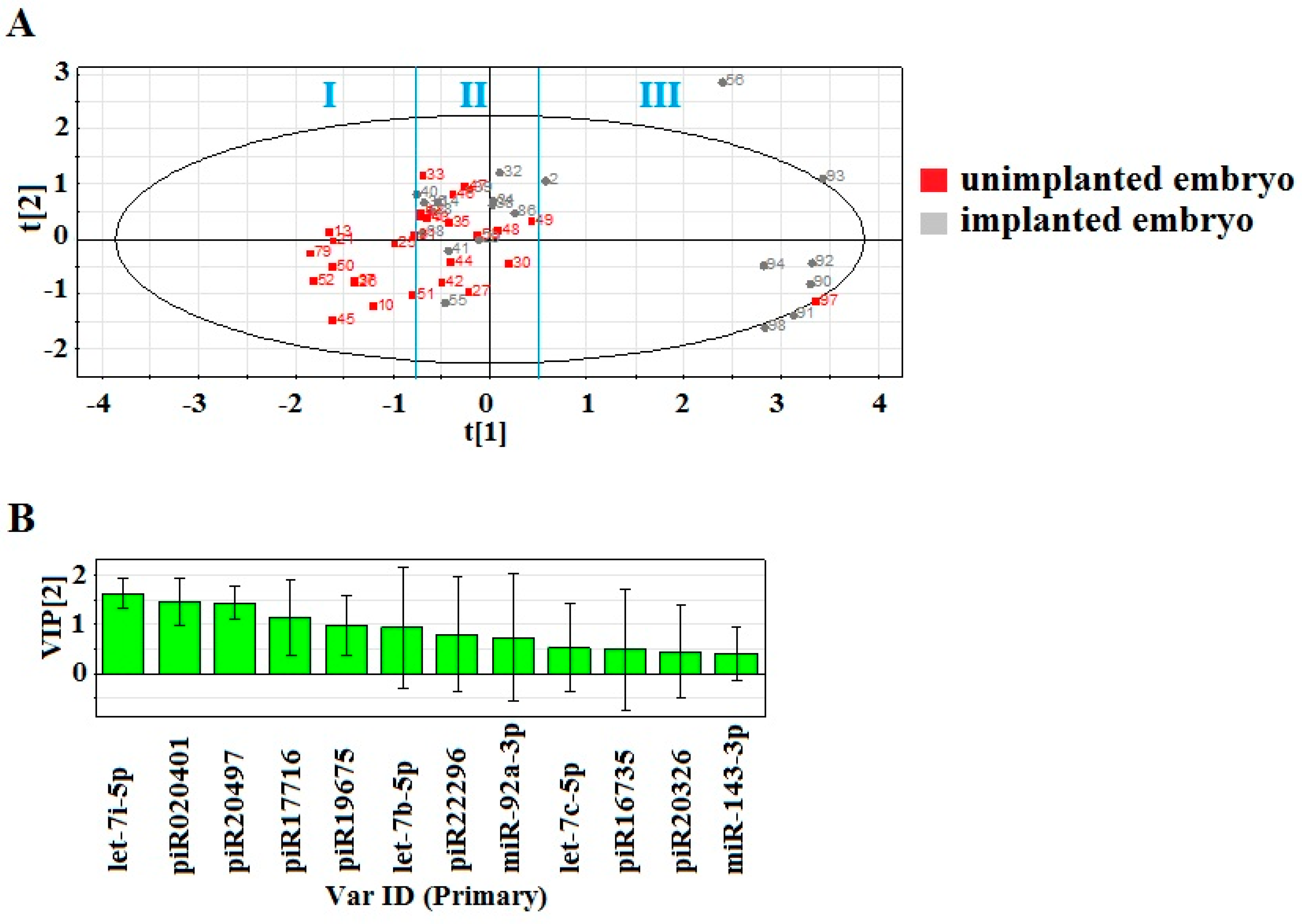
2.1. Identification of Embryo-Specific sncRNAs
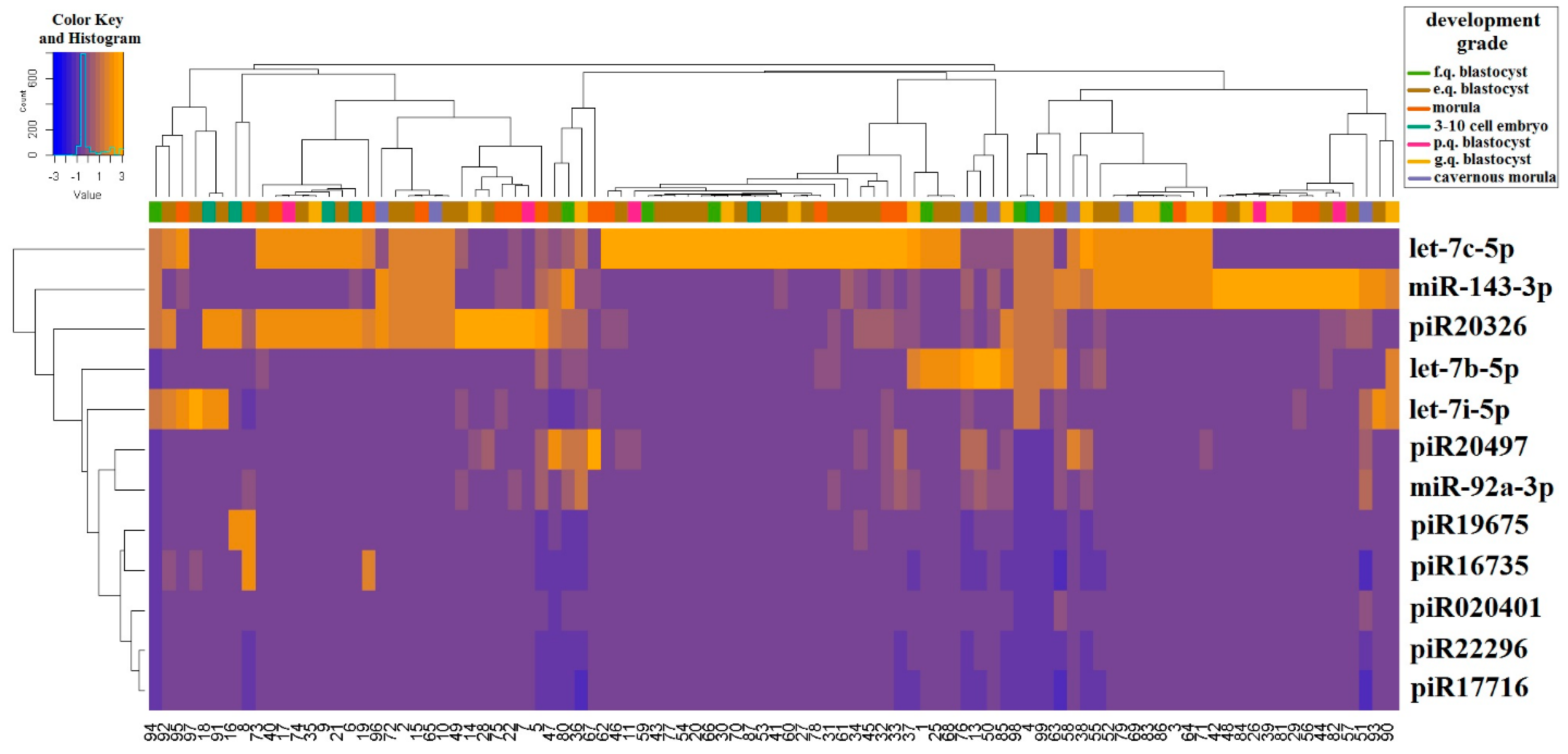
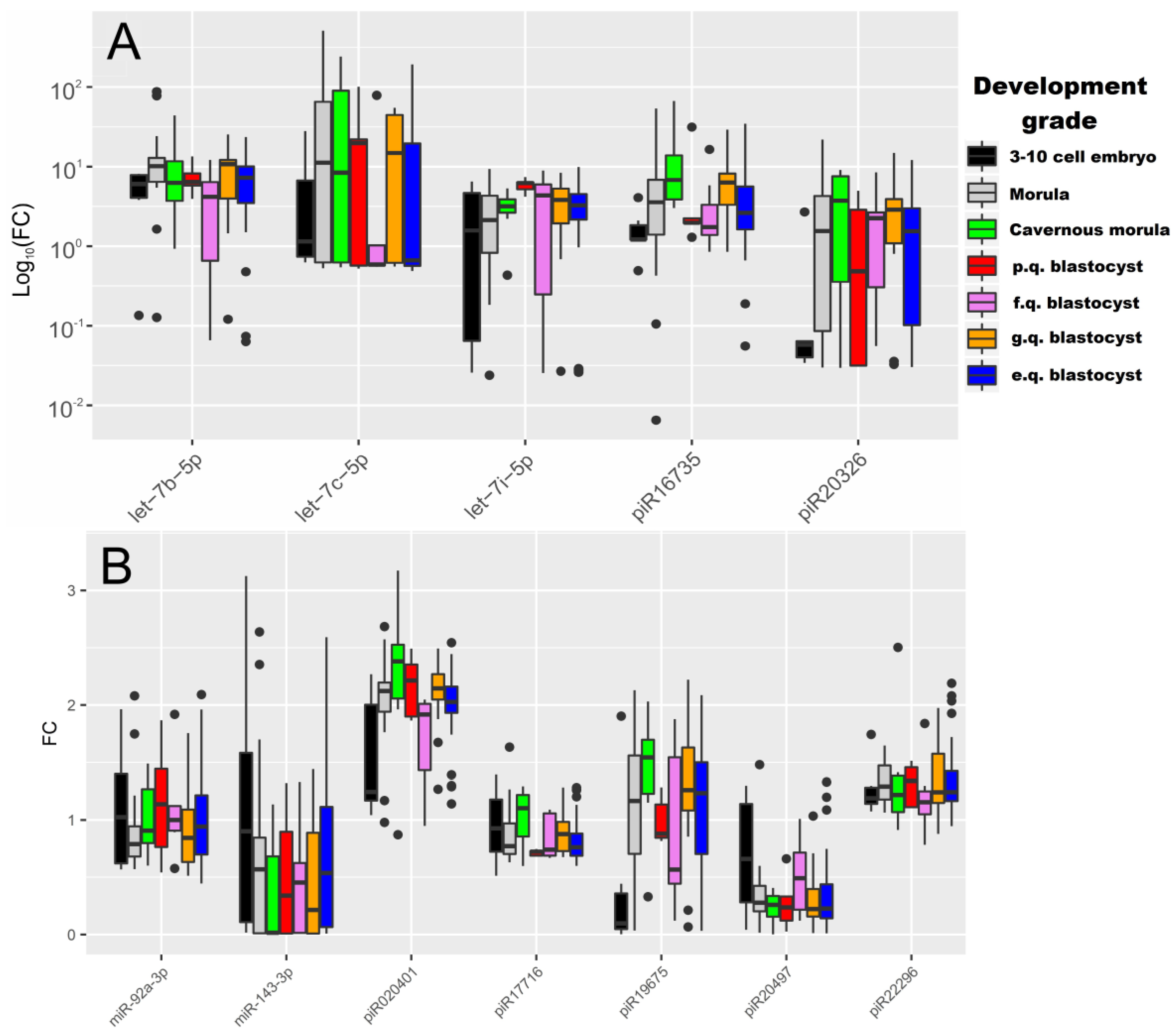
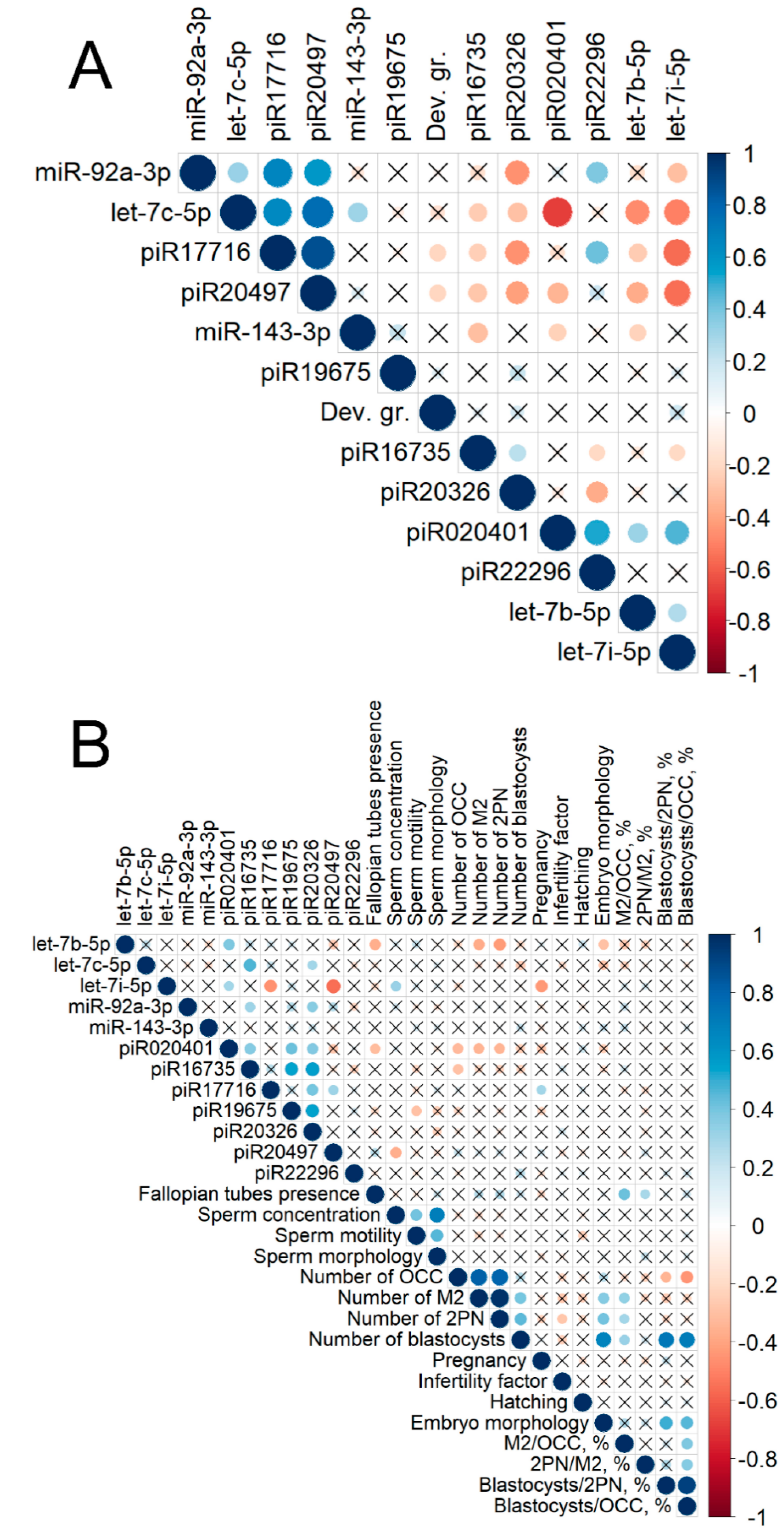
2.2. Real Time PCR Analysis of sncRNA Revealed by NGS
2.3. Identification of piRNA Targets
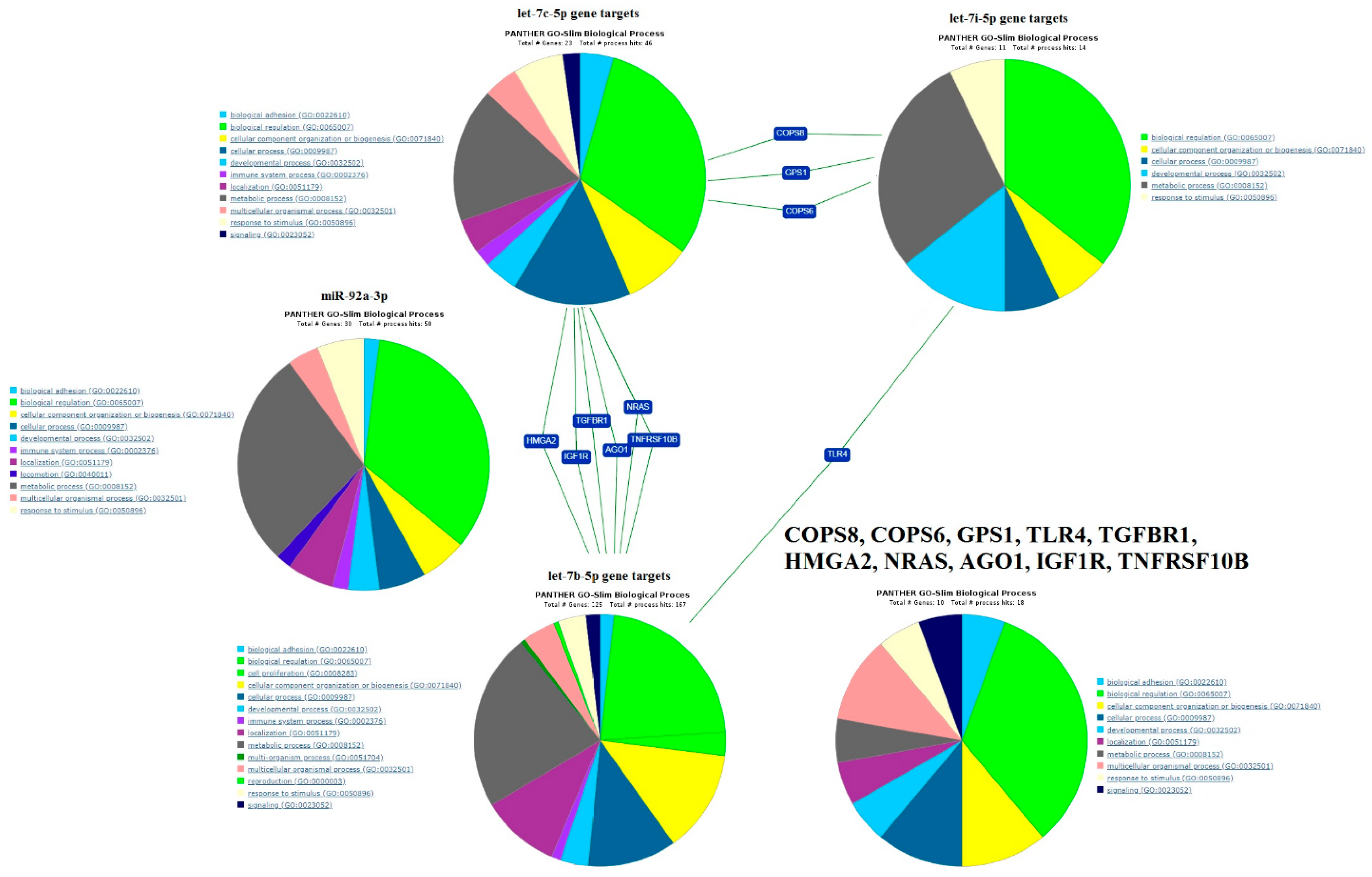
2.4. Functional Annotation of miRNA Target Genes
3. Materials and Methods
3.1. Patients
- Group I: 22 patients who had ovarian stimulation and SET in a stimulated cycle with a negative pregnancy blood test.
- Group II: 18 patients who had ovarian stimulation and SET in a stimulated cycle with a positive pregnancy blood test.
- Group III: 3 patients from group I who had implantation failure in the previous stimulated cycle and frozen-thawed (FT) embryo transfer with a positive result. All embryos were thawed at the blastocyst stage. The endometrium was prepared with the exogenous administration of oral micronized estradiol forms and progesterone. FT embryo transfer was performed on day 19–20 of the menstrual cycle. The endometrium thickness at the day of embryo transfer was 9–12 mm.
3.2. RNA Isolation from Embryo Culture Medium and Blastocoele Fluid
3.3. sncRNA Deep Sequencing
3.4. Reverse Transcription and Quantitative Real-Time PCR
3.5. Data Processing
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gardner, D.K.; Balaban, B. Assessment of human embryo development using morphological criteria in an era of time-lapse, algorithms and ‘OMICS’: Is looking good still important? Mol. Hum. Reprod. 2016, 22, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Gardner, D.K.; Schoolcraft, W.B. In-vitro culture of human blastocysts. In Towards Reproductive Certainty: Infertility and Genetics Beyond 1999: The Plenary Proceedings of the 11th World Congress on In Vitro Fertilization and Human Reproductive Genetics; Parthenon Press: New York, NY, USA, 1999; pp. 378–388. [Google Scholar]
- Gardner, D.K.; Lane, M.; Stevens, J.; Schoolcraft, W.B. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil. Steril. 2001, 76, 1175–1180. [Google Scholar] [CrossRef]
- Katz-Jaffe, M.G.; Linck, D.W.; Schoolcraft, W.B.; Gardner, D.K. A proteomic analysis of mammalian preimplantation embryonic development. Reproduction 2005, 130, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Poli, M.; Ori, A.; Child, T.; Jaroudi, S.; Spath, K.; Beck, M.; Wells, D. Characterization and quantification of proteins secreted by single human embryos prior to implantation. EMBO Mol. Med. 2015, 7, 1465–1479. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.J.; Yang, C.-X.; Ross, J.W. Small RNA regulation of reproductive function. Mol. Reprod. Dev. 2014, 81, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Chua, J.H.; Armugam, A.; Jeyaseelan, K. MicroRNAs: Biogenesis, function and applications. Curr. Opin. Mol. Ther. 2009, 11, 189–199. [Google Scholar]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Hirakata, S.; Siomi, M.C. piRNA biogenesis in the germline: From transcription of piRNA genomic sources to piRNA maturation. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 82–92. [Google Scholar] [CrossRef]
- Karakaya, C.; Guzeloglu-Kayisli, O.; Uyar, A.; Kallen, A.N.; Babayev, E.; Bozkurt, N.; Unsal, E.; Karabacak, O.; Seli, E. Poor ovarian response in women undergoing in vitro fertilization is associated with altered microRNA expression in cumulus cells. Fertil. Steril. 2015, 103, 1469–1476.e3. [Google Scholar] [CrossRef]
- Machtinger, R.; Rodosthenous, R.S.; Adir, M.; Mansour, A.; Racowsky, C.; Baccarelli, A.A.; Hauser, R. Extracellular microRNAs in follicular fluid and their potential association with oocyte fertilization and embryo quality: An exploratory study. J. Assist. Reprod. Genet. 2017, 34, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Yao, Z.; Wang, H.; Feng, R.; Wang, H.; Zhao, X.; Xing, Q.; Jin, L.; He, L.; Wu, L.; et al. Identification of MicroRNAs in Human Follicular Fluid: Characterization of MicroRNAs That Govern Steroidogenesis in Vitro and Are Associated With Polycystic Ovary Syndrome in Vivo. J. Clin. Endocrinol. Metab. 2013, 98, 3068–3079. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.; Vento, M.E.; Ragusa, M.; Barbagallo, D.; La Ferlita, A.; Di Emidio, G.; Borzi, P.; Artini, P.G.; Scollo, P.; Tatone, C.; et al. MicroRNAs Are Stored in Human MII Oocyte and Their Expression Profile Changes in Reproductive Aging. Biol. Reprod. 2016, 95, 131. [Google Scholar] [CrossRef] [PubMed]
- Abd El Naby, W.S.; Hagos, T.H.; Hossain, M.M.; Salilew-Wondim, D.; Gad, A.Y.; Rings, F.; Cinar, M.U.; Tholen, E.; Looft, C.; Schellander, K.; et al. Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote 2013, 21, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; Chuva de Sousa Lopes, S.M.; van der Westerlaken, L.A.J.; Zischler, H.; Butter, F.; Roelen, B.A.J.; et al. Piwi Proteins and piRNAs in Mammalian Oocytes and Early Embryos. Cell Rep. 2015, 10, 2069–2082. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.-F.; Hou, C.-C.; Yang, W.-X. Small non-coding RNAs and their associated proteins in spermatogenesis. Gene 2016, 578, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016, 143, 635–647. [Google Scholar] [CrossRef]
- Svoboda, P.; Flemr, M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010, 11, 590–597. [Google Scholar] [CrossRef]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Yang, Q.; Gu, W.-W.; Gu, Y.; Yan, N.-N.; Mao, Y.-Y.; Zhen, X.-X.; Wang, J.-M.; Yang, J.; Shi, H.-J.; Zhang, X.; et al. Association of the peripheral blood levels of circulating microRNAs with both recurrent miscarriage and the outcomes of embryo transfer in an in vitro fertilization process. J. Transl. Med. 2018, 16, 186. [Google Scholar] [CrossRef] [PubMed]
- Capalbo, A.; Ubaldi, F.M.; Cimadomo, D.; Noli, L.; Khalaf, Y.; Farcomeni, A.; Ilic, D.; Rienzi, L. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil. Steril. 2016, 105, 225–235.e3. [Google Scholar] [CrossRef] [PubMed]
- Noli, L.; Capalbo, A.; Dajani, Y.; Cimadomo, D.; Bvumbe, J.; Rienzi, L.; Ubaldi, F.M.; Ogilvie, C.; Khalaf, Y.; Ilic, D. Human Embryos Created by Embryo Splitting Secrete Significantly Lower Levels of miRNA-30c. Stem Cells Dev. 2016, 25, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, E.M.; Shelton, D.N.; Sparks, A.E.T.; Devor, E.; Christenson, L.; Van Voorhis, B.J. MicroRNA expression in the human blastocyst. Fertil. Steril. 2013, 99, 855–861.e3. [Google Scholar] [CrossRef] [PubMed]
- No, J.; Zhao, M.; Lee, S.; Ock, S.A.; Nam, Y.; Hur, T.-Y. Enhanced in vitro maturation of canine oocytes by oviduct epithelial cell co-culture. Theriogenology 2018, 105, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Liu, Y.; Tong, X.H.; Luo, L.; Ma, J.; Chen, Z. The optimum number of oocytes in IVF treatment: An analysis of 2455 cycles in China. Hum. Reprod. 2013, 28, 2728–2734. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, X.; Wang, Q.; Zhang, S.; Jiang, L.; Zhang, C.; Ge, Z. Optimum oocyte retrieved and transfer strategy in young women with normal ovarian reserve undergoing a long treatment protocol: A retrospective cohort study. J. Assist. Reprod. Genet. 2015, 32, 1459–1467. [Google Scholar] [CrossRef]
- La Ferlita, A.; Battaglia, R.; Andronico, F.; Caruso, S.; Cianci, A.; Purrello, M.; Di Pietro, C. Non-Coding RNAs in Endometrial Physiopathology. Int. J. Mol. Sci. 2018, 19, 2120. [Google Scholar] [CrossRef]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosom. Res. 2018, 26, 25–43. [Google Scholar] [CrossRef]
- Russell, S.; Patel, M.; Gilchrist, G.; Stalker, L.; Gillis, D.; Rosenkranz, D.; LaMarre, J. Bovine piRNA-like RNAs are associated with both transposable elements and mRNAs. Reproduction 2017, 153, 305–318. [Google Scholar] [CrossRef]
- Sobala, A.; Hutvagner, G. Transfer RNA-derived fragments: Origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2011, 2, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Bühler, M.; Spies, N.; Bartel, D.P.; Moazed, D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 2008, 15, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef] [PubMed]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A Broadly Conserved Pathway Generates 3′UTR-Directed Primary piRNAs. Curr. Biol. 2009, 19, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Iwasaki, Y.W.; Lin, Z.Y.-C.; Imamura, M.; Seki, N.M.; Sasaki, E.; Saito, K.; Okano, H.; Siomi, M.C.; Siomi, H. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 2014, 20, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Kang, J.-Y.; Gou, L.-T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.-W.; Chen, R.; Fu, X.-D.; et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef]
- Roth, L.M.; Michal, M.; Michal, M.; Cheng, L. Protein expression of the transcription factors DMRT1, TCLF5, and OCT4 in selected germ cell neoplasms of the testis. Hum. Pathol. 2018, 82, 68–75. [Google Scholar] [CrossRef]
- Duquette, P.M.; Lamarche-Vane, N. Rho GTPases in embryonic development. Small GTPases 2014, 5, e972857. [Google Scholar] [CrossRef]
- Ringrose, L.; Paro, R. Epigenetic Regulation of Cellular Memory by the Polycomb and Trithorax Group Proteins. Annu. Rev. Genet. 2004, 38, 413–443. [Google Scholar] [CrossRef] [PubMed]
- Lund, A.H.; van Lohuizen, M. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 2004, 16, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Van der Vlag, J.; Otte, A.P. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 1999, 23, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.; Bender, W.; Kingston, R.E. Stabilization of Chromatin Structure by PRC1, a Polycomb Complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef]
- Deshpande, A.M.; Akunowicz, J.D.; Reveles, X.T.; Patel, B.B.; Saria, E.A.; Gorlick, R.G.; Naylor, S.L.; Leach, R.J.; Hansen, M.F. PHC3, a component of the hPRC-H complex, associates with 2A7E during G0 and is lost in osteosarcoma tumors. Oncogene 2007, 26, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Ohkumo, T.; Masutani, C.; Eki, T.; Hanaoka, F. Deficiency of the Caenorhabditis elegans DNA polymerase eta homologue increases sensitivity to UV radiation during germ-line development. Cell Struct. Funct. 2006, 31, 29–37. [Google Scholar] [CrossRef][Green Version]
- Bürglin, T.R. Homeodomain Subtypes and Functional Diversity; Springer: Dordrecht, The Netherlands, 2011; pp. 95–122. [Google Scholar] [CrossRef]
- Mo, J.-S.; Park, H.W.; Guan, K.-L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef]
- Hers, I.; Vincent, E.E.; Tavaré, J.M. Akt signalling in health and disease. Cell. Signal. 2011, 23, 1515–1527. [Google Scholar] [CrossRef]
- Varelas, X.; Sakuma, R.; Samavarchi-Tehrani, P.; Peerani, R.; Rao, B.M.; Dembowy, J.; Yaffe, M.B.; Zandstra, P.W.; Wrana, J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008, 10, 837–848. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Baek, K.-H. TGF-β signaling pathway mediated by deubiquitinating enzymes. Cell. Mol. Life Sci. 2019, 76, 653–665. [Google Scholar] [CrossRef]
- Song, J.L.; Nigam, P.; Tektas, S.S.; Selva, E. microRNA regulation of Wnt signaling pathways in development and disease. Cell. Signal. 2015, 27, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, N.; Celik-Ozenci, C. FOXO1, FOXO3, AND FOXO4 are differently expressed during mouse oocyte maturation and preimplantation embryo development. Gene Expr. Patterns 2015, 18, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Perkel, K.J.; Madan, P. Spent culture medium analysis from individually cultured bovine embryos demonstrates metabolomic differences. Zygote 2017, 25, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Brison, D.R.; Houghton, F.D.; Falconer, D.; Roberts, S.A.; Hawkhead, J.; Humpherson, P.G.; Lieberman, B.A.; Leese, H.J. Identification of viable embryos in IVF by non-invasive measurement of amino acid turnover. Hum. Reprod. 2004, 19, 2319–2324. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.G.; Esposito, D.; Bagni, R.K.; McCormick, F. Dragging Ras Back in the Ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Gerttula, S.; Jin, Y.S.; Anderson, K. V Zygotic expression and activity of the Drosophila Toll gene, a gene required maternally for embryonic dorsal-ventral pattern formation. Genetics 1988, 119. [Google Scholar]
- Vijay, K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int. Immunopharmacol. 2018, 59, 391–412. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Jun, J.H. Identification of differentially expressed microRNAs in outgrowth embryos compared with blastocysts and non-outgrowth embryos in mice. Reprod. Fertil. Dev. 2019, 31, 645. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 5 June 2019).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Dayal, V. An Introduction to R for Quantitative Economics; SpringerBriefs in Economics; Springer India: New Delhi, India, 2015; ISBN 978-81-322-2339-9. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
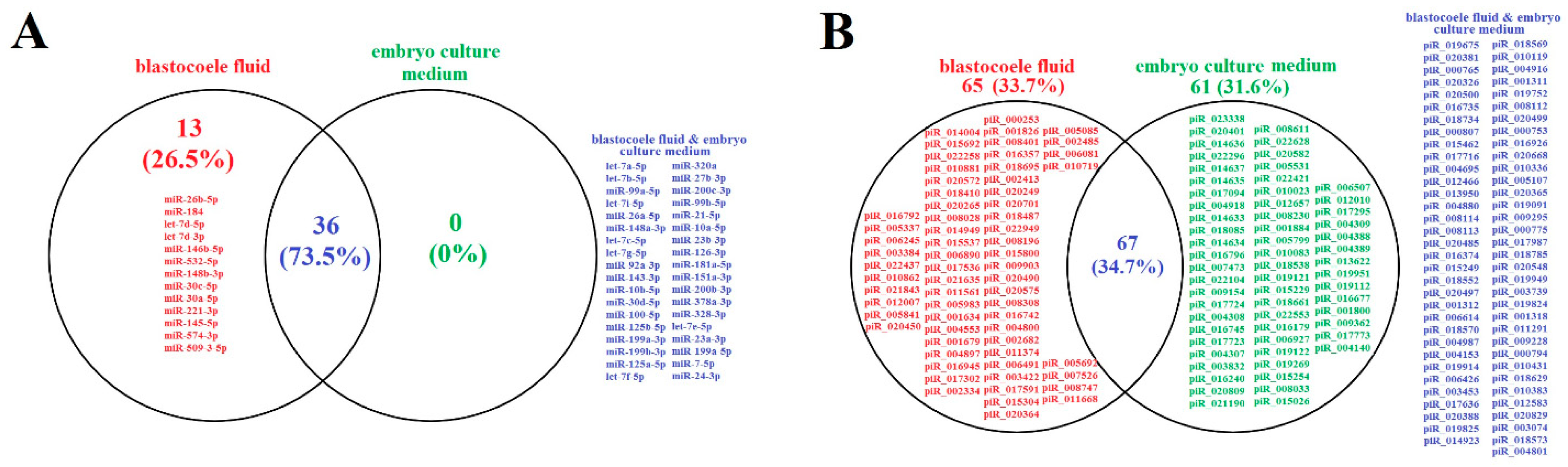
| sncRNA | NGS Read Count (Blastocoele Liquid/Embryo Culture Medium) | 5′–3′ Sequence of Sense Primer, Tm (Melting Temperature) | PCR (Embryo Culture Medium) |
|---|---|---|---|
| hsa-let-7b-5p | 527/282 | Hs_let-7b_1 miScript Primer Assay, Cat.No. MS00003122, Tm = 55 °C | + |
| hsa-let-7a-5p | 537/258 | Hs_let-7a_2 miScript Primer Assay, Cat.No. MS00031220, Tm = 55 °C | Ct > 35 |
| hsa-miR-99a-5p | 351/145 | Hs_miR-99a_2 miScript Primer Assay, Cat.No. MS00032158, Tm = 55 °C | Ct > 35 |
| hsa-miR-148a-3p | 194/136 | Hs_miR-148a_1 miScript Primer Assay, Cat.No. MS00003556, Tm = 55 °C | Ct > 35 |
| hsa-let-7i-5p | 239/114 | Hs_let-7i_1 miScript Primer Assay, Cat.No. MS00003157, Tm = 55 °C | + |
| hsa-miR-26a-5p | 209/111 | Hs_miR-26a_2 miScript Primer Assay, Cat.No. MS00029239, Tm = 55 °C | Ct > 35 |
| hsa-let-7c-5p | 192/110 | Hs_let-7c_1 miScript Primer Assay, Cat.No. MS00003129, Tm = 55 °C | + |
| hsa-let-7g-5p | 145/89 | Hs_let-7g_2 miScript Primer Assay, Cat.No. MS00008337, Tm = 55 °C | Ct > 35 |
| hsa-miR-92a-3p | 144/91 | Hs_miR-92_1 miScript Primer Assay, Cat.No. MS00006594, Tm = 55 °C | + |
| hsa-miR-143-3p | 135/81 | Hs_miR-143_1 miScript Primer Assay, Cat.No. MS00003514, Tm = 55 °C | + |
| hsa-miR-125b-5p | 69/57 | Hs_miR-125b_1 miScript Primer Assay, Cat.No. MS00006629, Tm = 55 °C | Ct > 35 |
| hsa-miR-100-5p | 77/50 | Hs_miR-100_2 miScript Primer Assay, Cat.No. MS00031234, Tm = 55 °C | Ct > 35 |
| hsa-miR-125a-5p | 63/36 | Hs_miR-125a_1 miScript Primer Assay, Cat.No. MS00003423, Tm = 55 °C | Ct > 35 |
| hsa-miR-320a | 57/30 | Hs_miR-320a_1 miScript Primer Assay, Cat.No. MS00014707, Tm = 55 °C | Ct > 35 |
| hsa-miR-27b-3p | 56/28 | Hs_miR-27b_2 miScript Primer Assay, Cat.No. MS00031668, Tm = 55 °C | Ct > 35 |
| hsa-miR-200c-3p | 47/28 | Hs_miR-200c_1 miScript Primer Assay, Cat.No. MS00003752, Tm = 55 °C | Ct > 35 |
| Hsa-piR020401, DQ598029 | 0/23,808 | GGCTGGTCTCGAACTCCTGACCTCAGGT, Tm = 45 °C | + |
| Hsa-piR023338, DQ601914 | 0/27,727 | TAGTCCCAGCTACTTGGGAGGCTGAGGCA, Tm = 45 °C | + |
| Hsa-piR019675, DQ596992 | 2708/2645 | GCAATAACAGGTCTGTGATGCCCTTAGA, Tm = 53 °C | + |
| Hsa-piR016735, DQ593039 | 1676/1240 | CCTGGGAATACCGGGTGCTGTAGGCTTA, Tm = 50 °C | + |
| Hsa-piR017716, DQ594453 | 620/680 | TTCCCTGGTGGTCTAGTGGTTAGGATTCGGC, Tm = 45 °C | + |
| Hsa-piR020326, DQ597916 | 1810/2540 | GGCATTGGTGGTTCAGTGGTAGAATTCTCGC, Tm = 60 °C | + |
| Hsa-piR020497, DQ598177 | 291/350 | TGTAGCTCAGTGGTAGAGCGCGTGCT, Tm = 45 °C | + |
| Hsa-piR022296, DQ600515 | 0/10,609 | TACTCAGGAGGCTGAGGCAGGAGAATGGC, Tm = 45 °C | + |
| sncRNA | Group 1–Group 2 | p-Value | Group 1, Me(Q1, Q3) * | Group 2, Me(Q1, Q3) * |
|---|---|---|---|---|
| let-7b-5p | “3–10 cell embryon”–“morula” | 0.04 | 6.16 (4.08, 7.86) | 10.1 (6.42, 12.86) |
| “morula”–“fair quality blastocyst” | 0.02 | 10.1 (6.42, 12.86) | 4.18 (1.94, 6.71) | |
| “morula”–“excellent quality blastocyst” | 0.05 | 10.1 (6.42, 12.86) | 7.24 (3.48, 10.07) | |
| let-7i-5p | “morula”–“poor quality blastocyst” | 0.02 | 2.2 (0.87, 4.33) | 6.2 (5.19, 6.34) |
| “cavernous morula”–“poor quality blastocyst” | 0.01 | 3.17 (2.67, 3.89) | 6.2 (5.19, 6.34) | |
| “poor quality blastocyst”–“excellent quality blastocyst” | 0.01 | 6.2 (5.19, 6.34) | 3.26 (2.18, 4.52) | |
| piR020401 | “morula”–“fair quality blastocyst” | 0.02 | 2.12 (1.94, 2.2) | 1.92 (1.43, 2.01) |
| “fair quality blastocyst”–“good quality blastocyst” | <0.001 | 1.92 (1.43, 2.01) | 2.14 (2.05, 2.27) | |
| piR16735 | “3–10 cell embryon”–“cavernous morula” | <0.001 | 1.21 (1.21, 1.88) | 6.78 (3.97, 14.92) |
| “3–10 cell embryon”–“good quality blastocyst” | 0.01 | 1.21 (1.21, 1.88) | 6.3 (3.51, 8.2) | |
| “cavernous morula”–“fair quality blastocyst” | 0.04 | 6.78 (3.97, 14.92) | 1.74 (1.41, 3.85) | |
| “cavernous morula”–“excellent quality blastocyst” | 0.02 | 6.78 (3.97, 14.92) | 2.61 (1.62, 5.62) | |
| piR17716 | “poor quality blastocyst”–“good quality blastocyst” | 0.04 | 0.72 (0.69, 0.73) | 0.88 (0.73, 0.98) |
| piR19675 | “3–10 cell embryo”–“good quality blastocyst” | 0.02 | 0.1 (0.05, 0.36) | 1.26 (1.08, 1.63) |
| “3–10 cell embryo”–“excellent quality blastocyst” | 0.03 | 0.1 (0.05, 0.36) | 1.23 (0.7, 1.5) | |
| piR20326 | “3–10 cell embryo”–“good quality blastocyst” | 0.02 | 0.06 (0.04, 0.06) | 2.88 (1.09, 3.92) |
| Genbank ID | PiRBase ID for piRNAs | Location in hg38 * | RefSeq Gene | Repeat Information | Potential Gene Targets |
|---|---|---|---|---|---|
| hsa_piR_017716|gb|DQ594453|Homo | piR-hsa-24672, aliases piR-60565, PIR55564 | chr6_GL000255v2_alt:247775-247806: + | Name: tRNA-Glu-GAG, Family: tRNA, Class: tRNA | ||
| hsa_piR_020401|gb|DQ598029|Homo | piR-hsa-28244, aliases piR-36095 PIR59140 | chr6_GL000253v2_alt:2984874-2984902: − | LY6G5C NM_025262 | Name: AluSx, Family: Alu, Class: SINE | PAK3,CXorf38,FRRS1L,C9orf85,EEF1D,CACNA2D1,RBM28,POLH,TRIM52,FER,NAF1,NSUN7,LIAS,PHC3,RABL2B,KIAA1755,ZNF13,PHF20,ZHX3,TCFL5,CARF |
| hsa_piR_016735|gb|DQ593039|Homo | piR-hsa-23317, aliases piR-33151 PIR54150 | chr6:4428052-4428083: + | Name: 5S, Family: rRNA, Class: rRNA | ||
| hsa_piR_019675|gb|DQ596992|Homo | piR-hsa-27282, aliases piR-35058 PIR58103 | chr14:89875053-89875081: − | EFCAB11 NM_001284269;NM_145231; NM_001284267 | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | MDN1,MLKL,METTL22, ARHGEF10L |
| chr21:8211110-8211138: + | RNA18SN2 NR_146146; RNA18SN4 NR_146119; RNA18SN3 NR_146152; RNA18SN5 NR_003286; RNA45SN2 NR_146144; LOC100507412 NR_038958; RNA18SN1 NR_145820 | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | |||
| chr21:8255319-8255347: + | RNA28SN5 NR_003287 | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | |||
| chr21:8394145-8394173: + | LOC100507412 NR_038958; RNA28SN5 NR_003287; RNA18SN2 NR_146146; RNA18SN5 NR_003286; RNA45SN3 NR_146151; RNA18SN3 NR_146152; RNA18SN4 NR_146119; RNA18SN1 NR_145820; | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | |||
| chr21:8438355-8438383: + | RNA45SN1 NR_145819; RNA18SN2 NR_146146; RNA45SN5 NR_046235; RNA18SN5 NR_003286; RNA18SN3 NR_146152; RNA18SN4 NR_146119; RNA18SN1 NR_145820; | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | |||
| chr22_KI270733v1_random:127410-127438: + | RNA45SN1 NR_145819; RNA45SN5 NR_046235; RNA45SN2 NR_146144; RNA45SN4 NR_146117; RNA18SN5 NR_003286; RNA18SN3 NR_146152; RNA18SN4 NR_146119; RNA18SN2 NR_146146; RNA18SN1 NR_145820; | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | |||
| chr22_KI270733v1_random:172491-172519: + | RNA18SN3 NR_146152; RNA18SN5 NR_003286; RNA18SN4 NR_146119; RNA18SN2 NR_146146; RNA18SN1 NR_145820; | Name: SSU-rRNA_Hsa, Family: rRNA, Class: rRNA | |||
| hsa_piR_020497|gb|DQ598177|Homo | piR-hsa-28392, aliases piR-36243 PIR59288 | chr6:28863725-28863757: −, chr6_GL000250v2_alt:129309-129341: −, chr6_GL000251v2_alt:354265-354297: −, chr6_GL000252v2_alt:129327-129359: −, chr6_GL000253v2_alt:129283-129315: −, chr6_GL000254v2_alt:129319-129351: −, chr6_GL000255v2_alt:129307-129339: −, chr6_GL000256v2_alt:172970-173002: − | Name: tRNA-Ala-GCY, Family: tRNA, Class: tRNA | ||
| hsa_piR_020326|gb|DQ597916|Homo | piR-hsa-28131, aliases piR-35982 PIR59027 | chr1:16545979-16546010: −, chr1:16861919-16861950: + | Name: tRNA-Gly-GGG, Family: tRNA, Class: tRNA | ||
| chr16:70789505-70789536: + | VAC14 NM_001351157, NM_018052 | Name: tRNA-Gly-GGY, Family: tRNA, Class: tRNA | |||
| chr3:15507990-15508021: + | COLQ NM_080539, NM_005677 | Name: tRNA-Gly-GGG, Family: tRNA, Class: tRNA | |||
| chr6:27902948-27902979: − | Name: tRNA-Gly-GGY, Family: tRNA, Class: tRNA | ||||
| hsa_piR_022296|gb|DQ600515|Homo | piR-hsa-30715, aliases piR-38581 PIR61626 | chr18:13769187-13769216: + | Name: AluY, Family: Alu, Class: SINE | NR6A1,RELL1,IL17RB,POLA2,PLPP4,PRPF3,C3orf70,CDH18,ZNF726,UGCG,IL17RD,MCF2L2,TBC1D24,AMN1,KIN,PLPP4,ZNF124,ACTR8,LPP,CPT1A,ZNF320 | |
| hsa_piR_023338|gb|DQ601914|Homo | piR-hsa-30937, aliases piR-39980 PIR63025 | chr17:42974237-42974266: + | PTGES3L NM_001142654, NM_001142653, NM_001261430; PTGES3L-AARSD1 NM_025267, NM_001136042; | Name: AluSq, Family: Alu, Class: SINE |
| DianaTools microT-CDS Algorithm for the Prediction of miRNA Targets in Both the three prime untranslated regions (3’UTRs) and Coding Sequence (CDS) | |||
| Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway | p-Value | Number of Genes | miRNAs |
| ECM–receptor interaction | < 1 × 10−325 | 15 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Mucin type O-Glycan biosynthesis | 4.21 × 10−11 | 5 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | 8.44 × 10−8 | 4 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Amoebiasis | 2.48 × 10−7 | 11 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Signaling pathways regulating stem cell pluripotency | 1.19 × 10−5 | 16 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Protein digestion and absorption | 0.006789 | 11 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| PI3K-Akt signaling pathway | 0.009286 | 27 | hsa-let-7b-5p, hsa-let-7c-5p |
| Wnt signaling pathway | 0.017555 | 11 | hsa-let-7c-5p |
| DianaTools_TargetScan—Predicts Biological Targets of miRNAs Considering Matches to 3’ UTRs | |||
| Mucin type O-Glycan biosynthesis (hsa00512) | 4.43 × 10−12 | 1 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Valine, leucine, and isoleucine biosynthesis (hsa00290) | 1.79 × 10−8 | 1 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Signaling pathways regulating stem cell pluripotency (hsa04550) | 7.77 × 10−6 | 6 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Biosynthesis of amino acids (hsa01230) | 0.000132 | 2 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| 2-Oxocarboxylic acid metabolism (hsa01210) | 0.000166 | 1 | hsa-let-7b-5p, hsa-let-7c-5p, hsa-let-7i-5p |
| Oocyte meiosis (hsa04114) | 0.008317 | 3 | hsa-let-7b-5p, hsa-let-7c-5p |
| Valine, leucine, and isoleucine degradation (hsa00280) | 0.026219 | 1 | hsa-let-7c-5p |
| Arginine and proline metabolism (hsa00330) | 0.026219 | 2 | hsa-let-7c-5p |
| DianaTools_Tarbase—Database of Experimentally Supported miRNA Targets | |||
| Lysine degradation (hsa00310) | 6.06959 × 10−13 | 20 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Cell cycle (hsa04110) | 1.13021 × 10−11 | 52 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Viral carcinogenesis (hsa05203) | 5.62537 × 10−10 | 60 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Hepatitis B (hsa05161) | 5.35744 × 10−08 | 54 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Oocyte meiosis (hsa04114) | 1.75682 × 10−07 | 41 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Chronic myeloid leukemia (hsa05220) | 2.5576 × 10−7 | 27 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Hippo signaling pathway (hsa04390) | 4.05654 × 10−7 | 50 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Proteoglycans in cancer (hsa05205) | 1.70389 × 10−6 | 64 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Thyroid hormone signaling pathway (hsa04919) | 2.7327 × 10−6 | 39 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Adherens junctions (hsa04520) | 3.42243 × 10−6 | 30 | hsa-let-7i-5p, hsa-miR-92a-3p |
| FoxO signaling pathway (hsa04068) | 2.15597 × 10−5 | 50 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| ECM–receptor interaction (hsa04512) | 2.47672 × 10−5 | 12 | hsa-let-7i-5p |
| Bacterial invasion of epithelial cells (hsa05100) | 4.61176 × 10−5 | 28 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Glioma (hsa05214) | 7.08143 × 10−5 | 20 | hsa-let-7b-5p, hsa-let-7i-5p |
| Pathways in cancer (hsa05200) | 0.000154251 | 99 | hsa-let-7i-5p, hsa-miR-92a-3p |
| Colorectal cancer (hsa05210) | 0.000634129 | 25 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Thyroid cancer (hsa05216) | 0.000693712 | 12 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Prostate cancer (hsa05215) | 0.001016403 | 35 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Epstein–Barr virus infection (hsa05169) | 0.002168481 | 51 | hsa-let-7b-5p, hsa-let-7i-5p |
| MAPK signaling pathway (hsa04010) | 0.003378721 | 59 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Endocytosis (hsa04144) | 0.003828609 | 62 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| Huntington’s disease (hsa05016) | 0.003922408 | 50 | hsa-let-7b-5p, hsa-miR-92a-3p |
| Small cell lung cancer (hsa05222) | 0.004508148 | 24 | hsa-let-7b-5p, hsa-let-7i-5p |
| TGF-beta signaling pathway (hsa04350) | 0.006182085 | 29 | hsa-let-7b-5p, hsa-let-7i-5p, hsa-miR-92a-3p |
| p53 signaling pathway (hsa04115) | 0.007856274 | 20 | hsa-let-7b-5p, hsa-let-7i-5p |
| Melanoma (hsa05218) | 0.008148048 | 18 | hsa-let-7b-5p, hsa-let-7i-5p |
| Shigellosis (hsa05131) | 0.009612655 | 13 | hsa-miR-92a-3p |
| Bladder cancer (hsa05219) | 0.009747186 | 14 | hsa-let-7b-5p, hsa-let-7i-5p |
| Signaling pathways regulating pluripotency of stem cells (hsa04550) | 0.01296196 | 26 | hsa-miR-92a-3p |
| Estrogen signaling pathway (hsa04915) | 0.01345837 | 17 | hsa-let-7i-5p |
| Transcriptional misregulation in cancer (hsa05202) | 0.01729337 | 45 | hsa-let-7i-5p, hsa-miR-92a-3p |
| Ubiquitin mediated proteolysis (hsa04120) | 0.01776272 | 34 | hsa-let-7b-5p |
| Protein processing in endoplasmic reticulum (hsa04141) | 0.02539808 | 38 | hsa-let-7b-5p, hsa-let-7i-5p |
| RNA transport (hsa03013) | 0.02748059 | 41 | hsa-let-7b-5p |
| Valine, leucine and isoleucine biosynthesis (hsa00290) | 0.03746544 | 2 | hsa-miR-92a-3p |
| Clinical and Demographic Characteristics | Group I (Implantation Failure)—25 Patients | Group II (Successful Implantation)—14 Patients | Group III (Successful Implantation in A Frozen Embryo Transfer Protocol)—4 Patients | p |
|---|---|---|---|---|
| Body mass index (BMI) * | 22.1 (2.0) | 22.2 (1.4) | 22.0 (1.8) | >0.05 |
| Menstrual cycle length (days) * | 29.9 (1.6) | 28.4 (0.9) | 29.0 (2.7) | >0.05 |
| Average age (years) * | 32.3 (3.5) | 32.0 (3.1) | 30.0 (2.4) | >0,05 |
| Follicle-stimulating hormone level on day 2–3 of menstrual cycle (mIU/mL) * | 7.8 (1.4) | 7.2 (1.4) | 5.8 (0.5) | >0.05 |
| Anti-mullerian hormone level (ng/mL) * | 2.4 (1.0) | 2.4 (0.7) | 2.8 (0.5) | >0.05 |
| Antral follicle count on day 2–3 of the menstrual cycle * | 8.2 (1.6) | 7.8 (1.5) | 8.3 (1.0) | >0.05 |
| Male factor infertility ** | 18 patients (64%) had male factor infertility | 14 patients (78%) had male factor infertility | 2 patients (50%) had male factor infertility | >0.05 |
| Primary infertility ** | 13 patients (46%) had primary infertility | 8 patients (44%) had primary infertility | 2 patients (50%) had primary infertility | >0.05 |
| Assisted hatching ** | In 20 (71%) patients, assisted hatching was performed | In 10 (56%) patients, assisted hatching was performed | In 2 (50%) patients, assisted hatching was performed | >0.05 |
| Number of oocytes retrieved * | 7.8 (3.7) | 7.4 (4.5) | 8.0 (3.7) | >0.05 |
| Number of blastocysts * | 1.4 (1.3) | 1.4 (1.2) | 2.8 (2.0) | >0.05 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timofeeva, A.V.; Chagovets, V.V.; Drapkina, Y.S.; Makarova, N.P.; Kalinina, E.A.; Sukhikh, G.T. Cell-Free, Embryo-Specific sncRNA as a Molecular Biological Bridge between Patient Fertility and IVF Efficiency. Int. J. Mol. Sci. 2019, 20, 2912. https://doi.org/10.3390/ijms20122912
Timofeeva AV, Chagovets VV, Drapkina YS, Makarova NP, Kalinina EA, Sukhikh GT. Cell-Free, Embryo-Specific sncRNA as a Molecular Biological Bridge between Patient Fertility and IVF Efficiency. International Journal of Molecular Sciences. 2019; 20(12):2912. https://doi.org/10.3390/ijms20122912
Chicago/Turabian StyleTimofeeva, Angelika V., Vitaliy V. Chagovets, Yulia S. Drapkina, Nataliya P. Makarova, Elena A. Kalinina, and Gennady T. Sukhikh. 2019. "Cell-Free, Embryo-Specific sncRNA as a Molecular Biological Bridge between Patient Fertility and IVF Efficiency" International Journal of Molecular Sciences 20, no. 12: 2912. https://doi.org/10.3390/ijms20122912
APA StyleTimofeeva, A. V., Chagovets, V. V., Drapkina, Y. S., Makarova, N. P., Kalinina, E. A., & Sukhikh, G. T. (2019). Cell-Free, Embryo-Specific sncRNA as a Molecular Biological Bridge between Patient Fertility and IVF Efficiency. International Journal of Molecular Sciences, 20(12), 2912. https://doi.org/10.3390/ijms20122912





