Abstract
With the improper application of fungicides, Phytophthora sojae begins to develop resistance to fungicides, and biological control is one of the potential ways to control it. We screened two strains of Bacillus; Bacillus amyloliquefaciens JDF3 and Bacillus subtilis RSS-1, which had an efficient inhibitory effect on P. sojae. They could inhibit mycelial growth, the germination of the cysts, and the swimming of the motile zoospores. To elucidate the response of P. sojae under the stress of B. amyloliquefaciens and B. subtilis, and the molecular mechanism of biological control, comparative transcriptome analysis was applied. Transcriptome analysis revealed that the expression gene of P. sojae showed significant changes, and a total of 1616 differentially expressed genes (DEGs) were detected. They participated in two major types of regulation, namely “specificity” regulation and “common” regulation. They might inhibit the growth of P. sojae mainly by inhibiting the activity of ribosome. A pot experiment indicated that B. amyloliquefaciens and B. subtilis enhanced the resistance of soybean to P. sojae, and their control effects of them were 70.7% and 65.5%, respectively. In addition, B. amyloliquefaciens fermentation broth could induce an active oxygen burst, NO production, callose deposition, and lignification. B. subtilis could also stimulate the systemic to develop the resistance of soybean by lignification, and phytoalexin.
1. Introduction
Soybean Phytophthora root and stem rot, caused by Phytophthora sojae has been reported worldwide, for instance, in Asia, Africa, Europe, North America, South America, and so on [1]. P. sojae is a destructive soil-borne oomycete pathogen, which is not easy to control and it, therefore, results in great soybean (Glycine max) yield reductions. Therefore, the prevention and control of P. sojae has been widely studied by researchers. At present, the prevention and control of P. sojae is mainly achieved by rationally planting soybean-resistant varieties, spraying effective fungicides, and improving field management measures [2]. However, with the improper application of fungicides, P. sojae begins to develop resistance to fungicides, such as metalaxyl, fluorophenylalanine, mefenoxam, zoxamide, phenylamide, and oxathiapiprolin [3,4,5,6,7], and some of the researchers have begun to study their resistance mechanisms. The results showed that the resistance of Phytophthora to metalaxyl and mefenoxam was related to RNA polymerase I or its subunit RPA190 [8], and the β-tubulin and oxysterol binding protein from P. sojae is related to the resistance of Zoxamide and oxathiapiprolin [4,7]. Moreover, the resistance of P. sojae to pharmacy has led to a decrease in the control effect of soybean root rot, which poses a significant potential risk to soybean cultivation. Biological control is one of the effective ways to address this risk [2,9,10]. Wagner et al. [11] found that Pseudomonas can colonize well in soybean roots and inhibit P. sojae. Moreover, Pseudomonas has a good application prospect in controlling soybean Phytophthora blight. According to Godfrey et al. [12] P. aureofaciens can also be used as a potential biological control option for P. sojae. Fermentation filtrates of Pseudomonas sp. BS1 inhibited the zoosporangium formation and zoospore germination of P. sojae, and the inhibition rate of stock solutions reaches 95.9% [13]. Sowanpreecha and Rerngsamran [14] found that the antibacterial protein from P. aeruginosa RS1 can effectively inhibit P. palmivora, and the molecular weight of the antibacterial protein is about 20 kDa to 54 kDa. According to LC/MS, these proteins may be catalase, chitin-binding protein, and protease. Additionally, some proteins from Pseudomonas can stimulate the defense reaction of soybean against P. sojae, and the over-expression harpin-encoding gene hrpZm from P. syringae in soybean enhances tolerance to Phytophthora root and stem rot [15]. Soybean endophytic bacteria, such as Enterobacter, Acinetobacter, Pseudomonas, Ochrobactrum, and Bacillus, associated with the nodules of soybean, can promote the growth of soybeans and inhibit the pathogenicity of P. sojae [16]. Arfaoui et al. [17] found that Paenibacillus sp. and Streptomyces sp. had a good control effect on P. sojae race 4 in vitro. Research shows that the borrelidin from Streptomyces sp. neau-D50 plays an important role in the inhibition of P. sojae, and the antifungal activity of borrelidin was mediated by the inhibition of threonyl-tRNA synthetase (ThrRS) via the formation of the ThrRS-borrelidin complex [18].
Some Bacillus, such as B. subtilis and B. pumilus, also have good control effects against P. sojae [2,12]. B. altitudinis JSCX-1 can inhibit P. sojae by inhibiting the mycelial growth and zoospore germination. Biocontrol efficiency of B. altitudinis JSCX-1 against P. sojae was 49.28%, and it reduces the Pathogenicity of P. sojae via increasing the ROS production and callose deposition on soybean, and up-regulating the expression of genes in the salicylate-responsive pathway, such as GmPR1a [2]. It has been reported that Bacillus can produce some of the secondary metabolites, lipopeptides, and antibacterial proteins, such as aminopeptidase and chitinase, which are the main antibacterial components [9,19]. When B. amyloidosis is induced by methyl salicylate secreted by plants, it produces antibiotic Bacillus peptide and weathered mycin to help plants to resist Fusarium oxysporum [13]. It is important to reveal the inhibitory mechanism of Bacillus on P. sojae, and this will help us to develop Bacillus into bio-fertilizers and bio-pesticide. Transcriptomics has become one of the most widely used high-throughput sequencing technologies due to its low-cost and simple processing compared with proteomics and metabonomics [20]. Transcriptome sequencing technology has been extensively used in the interaction between oomycetes and plant hosts, especially between P. sojae and soybean. This technology has proven to be a useful tool for identifying oomycetes and genes associated with infection [21,22,23,24]. In 2012, the oomycete transcriptomics database was established by Tripathy et al. [21], and it includes transcript sequences from P. sojae and its host soybean (P. sojae mycelia, healthy soybean, and P. sojae-infected soybean) as well as Illumina transcript sequences from five Hyaloperonospora arabidopsidis libraries. In addition, it also has a complete set of Sanger EST sequences from P. sojae, P. infestans, and H. arabidopsidis, grown under various conditions, and these resources have promoted the rapid development of the research on the pathogenic mechanism of Phytophthora. Ye et al. [22] used transcriptomics to exploit the candidate virulence gene, and some effector genes (RxLR, CRN, and NLP) were found to play an important role in the interaction between P. sojae and soybean. Differentially expressed genes are analyzed with transcriptomics when the zoospore of P. sojae is released at a cold temperature, and some genes encoding phosphatidylinositol kinase and Ca2+/calmodulin-dependent protein kinase are involved in the zoospore release [23]. Chen et al. [24] applied RNA-Seq technology to reveal infection-related gene expression changes in P. capsici, and a large number of genes associated with pathogenicity were identified, including 98 predicted effector genes (RXLR and CRN effectors). The transcriptional dynamics of P. infestans during sequential stages of hemibiotrophic infection of tomato was carried out, and transcriptome data indicated that enzyme inhibitors, proteases, and glycosyl hydrolases are associated with pathogenicity, including the induction of putative cytoplasmic and apoplastic effectors [25].
Transcriptome analysis was also occasionally applied to the study of biological control mechanisms [26,27]. Our research uses transcriptomics to elucidate the response of P. sojae under the stress of B. amyloliquefaciens and B. subtilis, and this will contribute to the biological control molecular mechanism of P. sojae and lead to an in-depth understanding of the interactions between rhizosphere bacterial and soil-borne oomycete pathogens.
2. Results and Discussion
2.1. Inhibition of P. sojae by B. amyloliquefaciens and B. subtilis
P. sojae was cultured for five days with B. subtilis RSS-1 and B. amyloliquefaciens JDF3. Compared with the control, the growth of P. sojae was significantly inhibited, and the inhibition rates were 71.4% and 67.1%, respectively (Figure 1A). Under the microscope, it can be observed that, compared with the normal hyphae, the mycelium of P. sojae hyphae showed excessive branching, swelling, and deformity under the stress of two strains of Bacillus (Figure 1B). A. calcoaceticus had the strong inhibitory activity (71.14%) against the P. sojae, which caused fracture, lysis, and the formation of a protoplast ball at the end of hyphae [16]. Zhang et al. [28] reported that melatonin resulted in a branched hyphae, blunt hyphal tip, and a decrease of lipid droplets in P. infestans. Benzothiazole is an antimicrobial secondary metabolite volatilized by microbes, and may act as potential leading compound for the development of new oomycete fungicides [29]. Abnormal swellings and branches were developed along the hyphae after exposure to benzothiazole, and the polarized distribution of newly synthesized walls was also disrupted. The abnormal changes in hyphae was due to the disorganized cytoskeleton, and mycelium-related genes were involved in regulation. The fermentation broth of B. amyloliquefaciens JDF3 and B. subtilis RSS-1 have a good inhibitory effect on P. sojae (Figure 2), and the EC50 values for P. sojae are 4.06% and 2.79%, respectively. The regression equation of fermentation liquid RSS-1, and JDF3 were Y1 = 1.1059X1 + 4.4103 (R2 = 0.9602) and Y2 = 1.2539X2 + 4.5508 (R2 = 0.9933), respectively. In addition, their fermentation broth can also inhibit the germination of cysts (Figure 2) and the swimming of motile zoospores (Figure 3), indicating that these two strains of Bacillus have a better potential for controlling P. sojae.
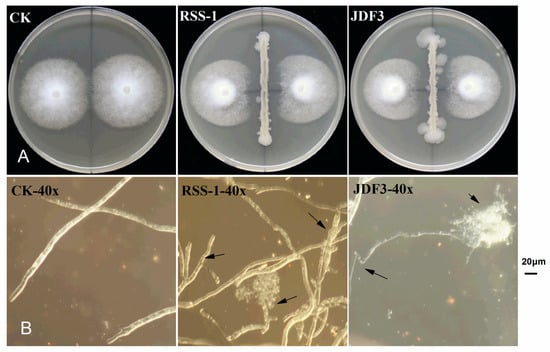
Figure 1.
Dual culture in vitro bacterial-fungal assays and morphological changes of mycelium under biocontrol stress. Mycelium morphology was photographed at a 40-fold microscope, and the arrows indicate abnormal hyphae.
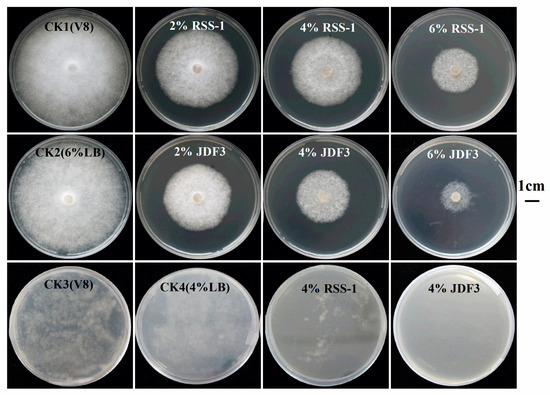
Figure 2.
Bacillus fermentation broth inhibited the growth and germination of cyst of P. sojae. The plates, which contained 2, 4, and 6% Bacillus fermentation broth, were used. The CK1 (10% V8), CK3 (10% V8), CK2 (10% V8 + 6% LB), and CK4(V8 + 4%LB) was used as negative and positive control. The third line was the results of Bacillus fermentation broth inhibiting the germination of cysts.
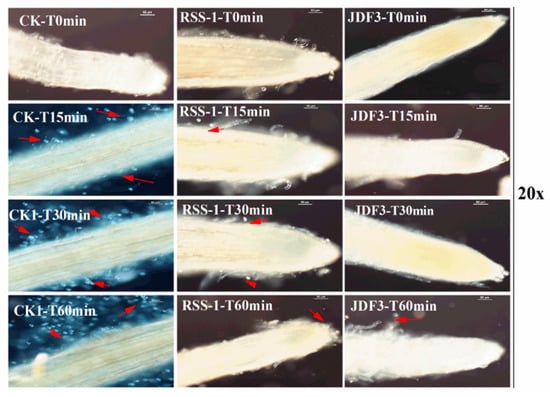
Figure 3.
Bacillus fermentation broths inhibit the swimming of zoospores. The first column was the results of the adsorption of zoospores on the soybean root after they were placed in the suspension of untreated zoospores for 60 min. The second and third columns were the results of the adsorption of zoospores on the soybean root after they were placed in the suspension of zoospores, which were added into Bacillus fermentation broths at 200 μL/20 mL for 10 s. The results were recorded with a 20-fold microscope.
2.2. Transcriptome Sequencing Quality Analysis
In order to study the molecular mechanism of two strains of Bacillus inhibiting P. sojae, a transcriptome analysis in response to biotic stress was carried out. P. sojae (CK), B. subtilis vs. P. sojae (BST), and B. amyloliquefaciens vs. P. sojae (BAT), in triplicate, was created, with a sequencing data volume of 6G, and raw reads (150bp, single-end) were obtained. The transcriptome statistics of the nine processed samples are shown in Table 1, and the raw read is in the range of 44,290,410~72,226,262. After the sequencing, the results were evaluated, the low-quality reads were filtered out, and the number of Clean Reads obtained was in the range of 43,117,708~68,948,564. The average number of Clean Reads per sample was 49,584,800. The sequenced read was compared with the P. sojae v3.0, and the mapped reads were in the range of 10,625,344~342,99,648. The GC content varied from 48.25–57.73%. In addition, the Q30 value was greater than 83%, indicating that RNA-Seq sequencing is of a good quality and can be used for the analysis of bioinformatics.

Table 1.
Summary of the quality of transcriptomic sequencing data of P. sojae.
2.3. Analysis of Principal Components
To determine the replication of the transcriptome samples, we performed principal component analysis (PCA), and the results are shown in Figure 4. The PCA results revealed that the transcriptome samples were clearly divided into three groups: the control group (CK1, CK2, CK3), B. amyloliquefaciens treatment group (BAT1, BAT2, BAT3), and B. subtilis treatment group (BST1, BST2, BST3). Three replicate samples within each group were brought together to form an independent population. According to the numerical values of the sample gene expression in the first principal component (PC1) and the second principal component (PC2), a two-dimensional coordinates map of the principal component is drawn. PC1 (64.5%) and PC2 (13.8%) revealed a change in gene expression of 9 samples of 78.3% and showed good agreement between sample biological replicates [22,24].
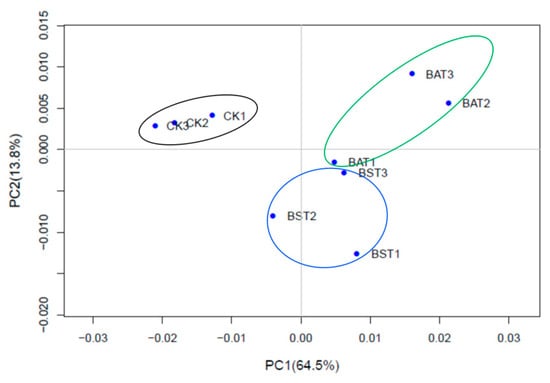
Figure 4.
Principal component analysis of the P. sojae samples.* The black circle was the control 1 (CK1), control 2 (CK2), control 3 (CK3). The blue circle was P. sojae samples treated with B. subtilis RSS-1, and BST1, BST2, and BST3 are three biological repeats. The green circle was P. sojae samples treated with B. amyloliquefaciens, and BAT1, BAT2, and BAT3 are three biological repeats.
2.4. Effect of B. amyloliquefaciens and B. subtilis on the Gene Expression of P. sojae and KEGG Enrichment Analysis
Under the inhibition of two strains of Bacillus, the expression of P. sojae gene showed significant changes. Under the stress of B. subtilis RSS-1, 728 differentially expressed genes (DEGs) were up-regulated, while 584 DEGs were down-regulated. The inhibition of the B. amyloliquefaciens JDF3 strain also caused a significant differential expression of the gene, with 332 and 340 genes up-regulated and down-regulated, respectively. Comparing the two strains of Bacillus that were treated, it was found that the expression of the P. sojae genes was different, and the total number of DEGs was 422, with 154 up-regulated and 268 down-regulated (Figure 5). To learn more about the DEGs between BST and BAT, we showed the differential expression of transcripts in a Venn diagram (Figure 6). In the stressed P. sojae, there were 368 transcripts in the intersection, which were generally affected by the biocontrol stress of B. subtilis and B. amyloliquefaciens, and there were 944 and 304 that were transcript-specific, respectively. Thus, we divided the differentially expressed transcripts into two major types of regulation, namely, “specificity” regulation and “common regulation”.
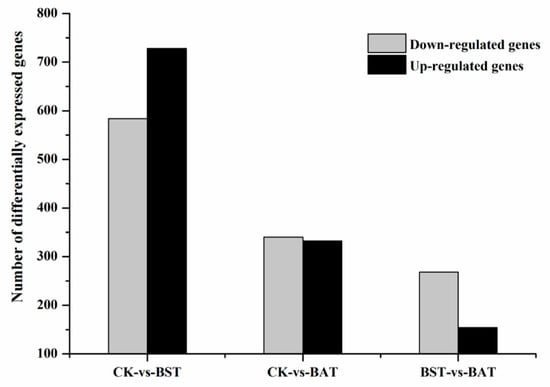
Figure 5.
Number of differentially expressed genes of P. sojae challenged with B. amyloliquefaciens and B. subtilis at 25 °C for five days.
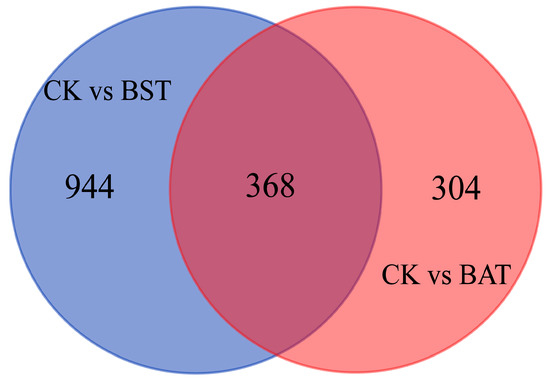
Figure 6.
Venn diagram illustrating relationship between the differentially expressed genes of P. sojae challenged with B. amyloliquefaciens and B. subtilis at 25 °C for five days. Intersection is a “common regulation”, and the others are “specific regulation”.
The commonly regulated genes were enriched in ribosome (ko03010), starch and sucrose metabolism (ko00500), tyrosine metabolism (ko00350), tryptophan metabolism (ko00380), pyruvate metabolism (ko00620), amino sugar and nucleotide sugar metabolism (ko00520), etc. (Figures S1 and S2). The genes enriched in the “ribosome (ko03010)” pathway were all down-regulated. In the BST group, 26 genes were down-regulated, accounting for 26% of the pathway. They involved small ribosomal subunits (S8e, S10e, S20e, S26e, S27e, S27Ae, and S28e, etc.) and large subunits (L9e, L26e, L13Ae, L34Ae, L10Ae, and L36Ae and LP1, LP2e, etc.). In the BAT group, 33 genes were down-regulated, accounting for 33% of the pathway, involving small ribosomal subunits (S4e, S10e, S19e, S20e, S24e, S26e, S27e, S27Ae, and S28e, etc.) and large subunits (L9e, L13Ae, L14e, L34Ae, L35Ae, and L37Ae, etc.). The down-regulated expression of the ribosomal size subunit gene may be caused by the inhibition of the secondary metabolites of Bacillus, and the abnormal synthesis of ribosomal proteins may cause the cell proliferation process to be blocked [30,31,32,33]. Moreover, the protein transport pathway (ko03060) was also affected, and PHYSODRAFT_511553 may be involved in the process of protein folding, sorting and degradation.
Sphingolipid acts as a component of the cell membrane, and in the sphingolipid metabolism (ko00600) pathway, the down-regulation of genes affects many functions of the cell membrane, such as membrane transport [34]. In the starch and sucrose metabolism pathway (ko00500) in the above two treatments, the down-regulation DEGs was 10 (in the BST group, Table S2) and 11 (BAT group, Table S3), which account for approximately 6% of the total number of genes in the pathway. This pathway is closely related to the utilization of carbohydrates, energy production and the growth of P. sojae, resulting in the inhibition of the growth of P. sojae [35,36]. In addition, in the BST group in the “specific regulation” pathway, five down-regulated expression genes were enriched in the fatty acid elongation (ko00062) pathway, while five down-regulated genes were enriched in the fatty acid metabolism (ko01212) pathway, affecting the lipid metabolism pathway. These genes, which were involved in ko04130 (Figure S1, soluble N-ethylmaleimide-sensitive factor attachment protein receptor, SNARE), might play an important role in asexual development, sexual reproduction, and pathogenesis like the gene of PsYKT6 in this family [37].
In the BAT group, the other glycan degradation (ko00511) and regulation of autophagy (ko04140) pathways may affect the cell wall production and autophagy regulation of P. sojae [38,39]. The down-regulated expression of the regulation of the autophagy pathway may affect the growth and development, sporulation and pathogenicity of P. sojae [39], and similar phenomena are observed [40]. Autophagy is ubiquitous in eukaryotic cells and plays an important role in material turnover, which encapsulates damaged organelles or long-lived proteins to form autophagic vesicles [41,42]. After being fused with lysosomes or vacuoles, the substance is degraded and recycled [42]. Two genes (Table S2), which are involved in the pathways of MAPK (ko04011), possibly participated in hyphal growth, zoosporogenesis, cell wall integrity, and pathogenicity like the gene of PsSAK1 and PsMPK7 in this family [43,44].
2.5. GO Enrichment Analysis
In order to identify the response of P. sojae to these two strains of Bacillus, we performed a GO functional enrichment analysis on DEGs. The GO enrichment analysis describes the classification of gene functions and the relationships between these genes. The GO functions of DEGs were divided into three groups: The biological process, the cellular component, and the molecular function [26,27]. In the BST samples, 217 up-regulated and 242 down-regulated differentially expressed genes were annotated, respectively. Therefore, a total of 459 DEGs were annotated (Table S4). Among them, 234 DEGs (130 up, 104 down) belong to the biological process (BP) category, 189 DEGs (65 up, 124 down) belong to the cellular component (CC) category, and 36 DEGs (22 up, 14 down) belong to the molecule functional (MF) category (Figure 7A and Table S4).
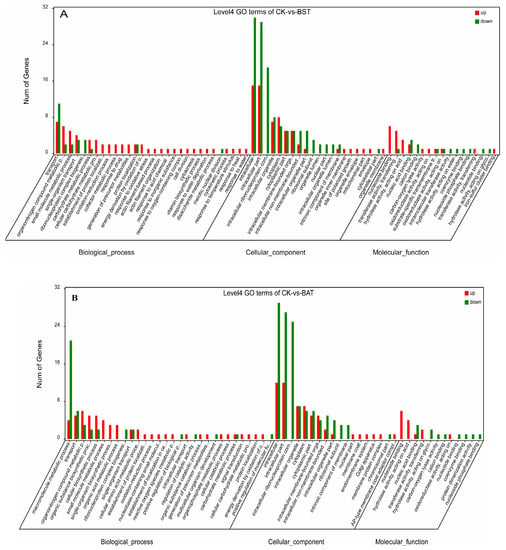
Figure 7.
Gene Ontology (GO) enrichment analysis for differentially expressed gene (DEG).
In the BP, most GO terms were related to transportation, the organonitrogen compound metabolic process, ribonucleoprotein complex biogenesis, and the response to various stresses (e.g., the response to oxidative stress, oxygen-containing compound, temperature stimulus, inorganic substances, heat, water deprivation, etc.). For example, a higher frequency of biological process subcategories is a single-organism cellular process, the regulation of biological process, a cellular response to the stimulus, a macromolecule metabolic process, a heterocycle metabolic process, a small molecule metabolic process, and an emergency response to various external stresses.
For the “cell component” GO category, the higher-frequency subcellular subclasses were the intracellular, intracellular part, intracellular ribonucleoprotein complex, intracellular organelle, cytoplasm, organelle lumen, ribosomal subunit, etc., most of which showed a downward trend, accounting for 65.6%. The results of the “molecular function” GO enrichment showed that the higher frequency molecular functions are nucleoside binding, transferase activity, the transfer of phosphorus-containing groups, and hydrolase activity, and so on. GO enrichment analysis showed that in the BAT sample, there were 148 DEGs annotated in biological processes, of which approximately 56.8% of DEGs were up-regulated and 43.2% of DEGs were down-regulated. In cellular components, 177 DEGs (57 up, 120 down) were annotated and 25 DEGs (14 up, 11 down) were annotated in the molecular function (Figure 7B, Table S5).
In biological processes, DEGs were highly enriched in macromolecule metabolic processes, transports, organonitrogen compound metabolic processes, and biosynthetic processes. Similar to the BST group, the BAT sample has a higher frequency of cellular component subclasses, which are mainly involved in the intracellular, intracellular part, intracellular ribonucleoprotein complex and its membrane function. In the molecular functional classification, DEGs are enriched in nucleoside binding, transferase activity, the transfer of phosphorus-containing groups, and hydrolase activity. Transcriptome analysis was also occasionally applied to the study of biological control mechanisms [26,27]. Rhizoctonia solani was treated with the Serratia proteamaculans and S. plymuthica, resulting in a difference of 462 and 242 genes, respectively, and these gene enrichment assays and functional classification of DEGs were annotated by the GO term [26]. The up-regulated GO terms were the threonine-type endopeptidase activity, oxidation-reduction process, biosynthetic process, pyruvate metabolism, and proteolysis involved in the cellular protein catabolic process, while the under-regulated GO terms included the DNA-dependent transcription, purine ribonucleotide metabolic process, nucleoside-triphosphatase activity, ribosome biogenesis, and ribosome and DNA binding. Saccharothrix yanglingensis was used for the biological control of Valsa mali. The transcriptome revealed that most of the glycoside hydrolase genes were down-regulated, including three key causative genes and the pectinase gene. The GO term revealed that the DEGs were down-regulated and involved in the dcatalytic activity, transmembrane transport, and hydrolase activity [27]. A transcriptome analysis was carried out to mine the downstream genes, whose expression levels were affected by melatonin [28]. After treating them with melatonin, the carbohydrate metabolism, lipid metabolism and amino acid metabolism of P. infestans was altered in the transcriptome data. The down-regulation of the rate-limiting enzyme 6-phosphofructokinase and fatty acid synthase would reduce or suppress the carbohydrate and lipid metabolism. Mei et al. [29] applied the combination of transcriptomic and proteomic approaches to explore the novel antifungal mechanisms of Benzothiazole against P. capsici. A total of 1,071 DEGs were identified, and they were related to the biological regulation, growth, organelle part, and cell part, and the single-organism process was down-regulated.
GO analysis demonstrated that four stress response genes, two detoxification-related genes, two-cell membrane genes, and three cytoskeletal organization genes were significantly down-regulated. Two genes involved in the apoptosis of cell were significantly up-regulated by benzothiazole, and the results of transmission electron micrographs confirm the apoptosis of P. capsici. Phenazine-1-carboxylic acid (PCA)-producing Pseudomonas spp. have been found to be effective against numerous plants pathogenic organisms, including bacteria, fungi, and oomycetes, such as the causal agent of the bacterial blight of rice, and the oomycetes P. infestans [45,46]. Ye et al. [46] used transcriptomics to exploit the alteration in P. infestans in response to phenazine-1-carboxylic acid production by P. fluorescens, and the transcriptome analysis provided a better understanding of the molecular mechanisms, which might in turn contribute to developing and optimizing a biocontrol strategy against the late blight of potato. Transcriptomic analyses implied that the percentages of all P. infestans’ genes significantly changed due to P. fluorescens, and exogenically applied PCA increased as time progressed, from 50 to 61% and from to 32 to 46%. When applying an absolute cut-off value of ≧3-fold change, 207 genes were found to be remarkably differentially expressed. Gene ontology analysis revealed that the key functional genes of the DEGs are involved in major functions, like phosphorylation mechanisms, transmembrane transport, and oxidoreduction activities.
2.6. Validation of RNA-Seq Sequencing
To verify the reliability of the RNA-Seq data, we randomly selected 14 genes in the BAT group and 13 genes in the BST group for quantitative RT-PCR (qRT-PCR) differential expression verification. Fold changes from qRT-PCR were compared with RNA-Seq expression analysis data, and the results are shown in Figure 8. The qRT-PCR results are in agreement with the RNA-Seq high-throughput sequencing data, indicating a similar expression pattern of up- and down-regulated genes, using RNA-Seq sequencing, and qRT-PCR tests.
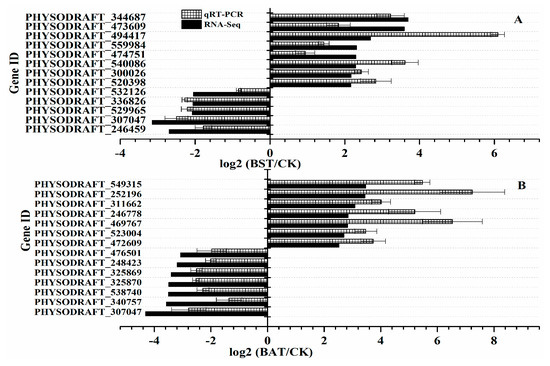
Figure 8.
Comparison of gene expression patterns obtained using RNA-Seq and qRT-PCR validation. The Y-axis shows genes in three tissues validated in this study; X-axis shows the log2 ratio of expression in BST and BAT versus CK.
2.7. Effect of B. amyloliquefaciens and B. subtilis on the Control of the Phytophthora Root Rot of Soybean
As shown in Figure 9, in the CK group, soybean seedlings have obvious lesions, and some of the soybean stalks and leaves were dry. However, the mildly susceptible potted soybean seedlings, under the hypocotyls showed a yellow-wild phenotype, and the base of the stem and the main root had mild brown lesions. However, for soybean seedlings treated with B. subtilis RSS-1 and B. amyloliquefaciens JDF3, only a small part of the hypocotyls showed yellowish or brownish black spots, and the rhizome had mild lesions. Their control effects of them were 65.5% and 70.7%, respectively. To further understand the effect of Bacillus on the disease resistance mechanism of soybean, we used the qPCR method to evaluate the expression of genes involved in soybean disease resistance (PR1, PR2, PR3, PR10, and LOX7) [47,48]. The sequence of quantitative primers is shown in Table S1. Compared with the control group, soybean treated with B. subtilis RSS-1 had significantly (p < 0.01) increased the expression levels of genes PR1, PR10, and LOX7, while the expression levels of genes PR2 and PR3 were slightly increased. After being treated with B. amyloliquefaciens JDF3, five disease resistance genes of the soybeans showed a significant up-regulation expression, except for PR2 (3 and 6 h) and LOX7 (12 h). Therefore, the two strains of Bacillus had an efficient control capability in relation to soybean phytophthora root, which was not only related to the inhibition of the growth of P. sojae, but also closely connected with the stimulation of soybean resistance [47,48], such as promoting the synthesis genes of phytoalexin and chitinase up-regulation [49].
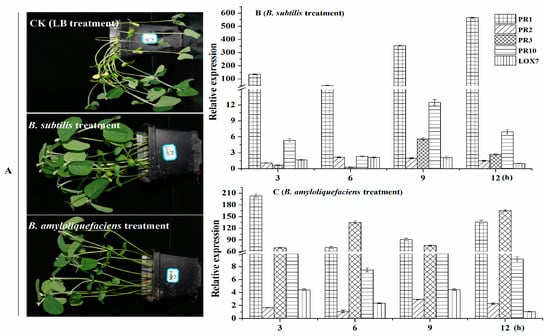
Figure 9.
B. amyloliquefaciens and B. subtilis enhanced the resistance of soybean to P. sojae. (A) A soybean was given a 1 mL liquid culture of Bacillus (OD value = 0.8−1.0) five times. (B and C) The degrees of resistance genes expression in roots of 4-day old soybean seedlings were within 3−12 h after first root irrigation.
2.8. B. amyloliquefaciens Fermentation Broth (BAFB) Activates Soybean Plant Defense Responses
As shown in Figure 10A, the negative control did not cause significant changes in the leaves, while the phenotype caused by B. subtilis RSS-1 fermentation broth was similar to that of the negative control. BAFB caused a hypersensitive reaction (HR) in tobacco leaves, which caused the HR phenotype to be consistent with the SA positive control (+). The above results indicate that BAFB might induce plant defense responses. In addition, previous studies have confirmed that protein elicitor (PeBA1) and lipopeptides secreted by B. amyloliquefaciens can induce plant defense responses [50,51]. Moreover, BAFB induced an active oxygen burst, and the DAB staining shown in Figure 10B. A negative control sample was injected on the left side of the tobacco leaves, which showed milky white after decolorization, while the injection site and veins showed a light-brown color under the microscope. Tobacco leaves treated with 4, 2, and undiluted B. subtilis RSS-1 fermentation broths (BSFB) were essentially no different, compared to the negative control treated with the LB liquid medium. The tobacco leaves treated with BAFB diluted two times and undiluted BAFB showed a brown color, which was significantly different from the white color. Microscopically, the tobacco leaves treated with BAFB were dark brown and the color was significantly darker than that of the control, indicating that the BAFB induced a large amount of active oxygen bursting.
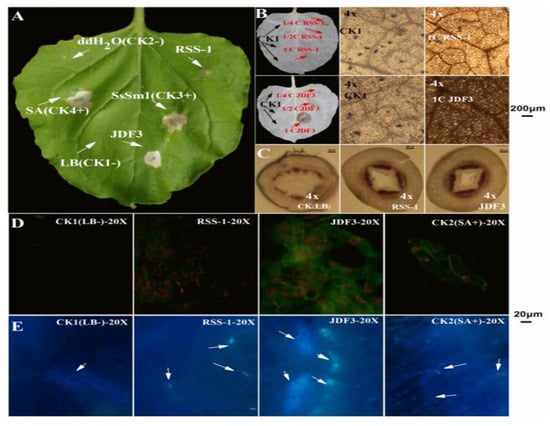
Figure 10.
B. amyloliquefaciens fermentation broth (BAFB) activates soybean plant defense responses. RSS-1 and JDF3 were on behalf of fermentation broth of B. amyloliquefaciens JDF3 and B. subtilis RSS-1, which were degerming with millipore filter (0.22 μm). An LB liquid medium and dd H2O were separately injected as negative controls (−), and salicylic acid (SA) and SsSm1, an elicitor protein was injected as a positive control (+). B and C were taken under a four-fold microscope, while D and E were recorded with a 20-fold microscope. The green fluorescence was generated by the combination of the NO and detection probe, and the orange fluorescence was generated by chloroplast stimulated by a fluorescence microscope (Nikon, Tokyo, Japan, NIS-elements D, Nikon, Tokyo, Japan) light source (D).
After staining with CK soybean stalks, lignin binds to phloroglucin and appears dark red. Compared with the CK, the soybeans treated with two strains of Bacillus had a significantly deeper crimson and more numerous red complexes in the stem (Figure 10C). It can be seen from Figure 10C that Bacillus can promote the lignification of soybean, thereby further improving the control effect of soybean disease. NO is an important signaling molecule of soybean, and it plays an important role in response to elicitor induction to promote plant stress resistance [52]. An NO fluorescent probe kit was used to detect whether BSFB and BAFB could induce NO signal molecules in soybean single cells. No fluorescence signal was detected in the soybean single cells treated with the negative control, whereas the fluorescent signal in the soybean single cell sample treated with the positive control SA was strong (Figure 10D). The single-cell fluorescence signal of soybean treated with BSFB was slightly stronger than that of the negative control, which was significantly weaker than the positive control SA, indicating that it significantly failed to induce the production of soybean NO signaling molecules. However, the single-cell fluorescence signal of soybean treated with BAFB was significantly stronger than that of the negative control, and its effect was similar to that of the positive control SA. This phenomenon indicates that it can significantly stimulate the accumulation of NO in the soybean, and BAFB may affect the resistance of soybean. Additionally, BAFB induces callose deposition in soybean, and a yellowing nursery was observed (Figure 10E). The soybean sprouts treated with BAFB showed brighter blue particles under the microscope. Callose also plays an important role in the disease resistance process of plants such as soybean, and its deposition in plants can help plants respond to pathogen infection and abiotic stress (physical, chemical, environmental factors, etc.) [2,9,52].
3. Materials and Methods
3.1. Strains and Culture Conditions
Two bacteria with potential biocontrol effects, B. amyloliquefaciens JDF3 and B. subtilis RSS-1, were isolated, identified, and preserved by the mycology and plant fungal disease laboratory of college of plant protection, Anhui agricultural university. P. sojae P6497 was presented by Professor Wang Yuanchao of the Nanjing Agricultural University. The strain was cultured in a 10% (v/v) V8 solid medium for five days and used for the confrontation culture of B. amyloliquefaciens JDF3 and B. subtilis RSS-1. P. sojae discs were obtained with a 9 mm cork borer, then the two bacteria dishes were placed in the circles on the left and right sides of the dish diagram. The B. amyloliquefaciens JDF3 and B. subtilis RSS-1 single colonies, cultured in an LB solid medium at 37 °C for 24 h, were taken with an inoculation needle, and inoculation was performed by streaking 6 cm along the axis of the symmetry of the two circles. The culture dish was placed in a 25 °C incubator and cultured for five days. Each treatment was inoculated with 30 dishes, 10 dishes were grouped together, and finally, three biological replicates were obtained. The dishes of P. sojae, inoculated by the above method, without the inoculation of Bacillus, were used as the control. According to the published methods [2,17], inhibition rates (I) of mycelium growth was calculated as I = [(R1 − R2)/R1] × 100, where R1 is the control mycelium growth between two discs edge. R2 is the mycelium growth toward the Bacillus. The method for determining the EC50 value and the bacteriostatic rate of the fermentation broth refers to Cui et al. [53]. The fermentation broth of B. amyloliquefaciens JDF3 and B. subtilis RSS-1 was centrifuged at 10,000 rpm for 10 min, and the supernatant was filtered with 0.22 μm filter membrane. The ability of inhibiting germination of P. sojae cysts were evaluated according to the previous literature [44,54], and the plates, which contained 4% fermentation broth of Bacillus, were used. CK3 (V8) and CK4(V8 + 4%LB) were used as negative and positive controls.
3.2. RNA Extraction, cDNA Library Preparation. and Transcriptome Sequencing
Referring to the method reported by Gkarmiri et al. [26], under sterile conditions, a scalpel was used to scrape the P. sojae hygrofilament, with a width of 1.5 ± 0.2 cm, along the outermost axis of the dish, along the axis of symmetry. Into one group, 10 dishes were mixed. Liquid nitrogen was used to freeze them, and they were prepared for RNA extraction. RNA extraction was performed using the takara RNA extraction kit, following the method provided in the kit. The concentration and purity of the RNA samples were quantified using a spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific, Waltham, MA, USA), and the RNA degradations of nine samples were assessed in 1% agarose gels. The RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA), following the manufacturer’s recommendations. The effective concentration of the library was accurately quantified using QPCR to ensure the library quality, and then cDNA library sequencing was conducted with an Illumina high-throughput sequencing platform (HiSeqTM2500) by Guangzhou Saizhe Biotechnology Co., Ltd. (Guangzhou, China).
3.3. RNA-Seq Data Analysis
High-quality clean data can be obtained by the quality control of the raw data output from the sequencing platform, deleting the linker sequence and sequencing primer sequences, and filtering low-quality read lengths. The clean data were compared to the P. sojae 3rd edition reference genome by the software, TopHat2, and the mapped data were assembled using the software, Cufflinks. The assembly results of multiple samples were combined with Cuffmerge. The expression and transcripts of genes were quantified using the Cuffdiff module, and the expression level of the gene was calculated by locating the reads each 1 million maps to the reads per megascript of the transcript per Million fragments mapped (FPKM). The differential expression analysis and calculation of the genes were performed using the EBSeq software, and the false discovery rate (FDR) of each transcript was counted. The fold change (FC) of the gene expression was calculated from different samples, using FDR and log2FC to screen differential genes. The treatment group was considered to have a significant differential expression relative to the control, and the screening conditions were FDR < 0.05 and |log2FC| > 1.
3.4. Gene Annotation
The Gene Ontology (GO) annotation is based on the significant enrichment of GO functions to analyze DEGs and related gene modules for bioinformatics analysis. Referring to the annotation information in the non-redundant (Nr) database, the Blast2 GO software (version 3.0, https://www.blast2go.com/, BioBam Bioinformatics S.L., Valencia, Spain) was used to perform GO Annotation on the core genes in the differentially expressed genes and the co-expressed gene modules, and the WEGO software was used to annotate and statistical the GO function classification of all genes. It covers three aspects of biology: the cellular component, the molecular function, and the biological process. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (available online: https://www.kegg.jp/) can systematically classify and annotate the metabolic pathways of genes and classify and study genes and their expression information at a general level. The KO-BAS software (version 2.0, KOBAS, Surrey, UK) was used to detect the enrichment of DEGs in the KEGG pathway, and the biological function of the specific gene of P. sojae was considered and evaluated at a macro level.
3.5. Quantitative Real-Time PCR (qRT-PCR)
The remaining P. sojae and soybean samples were taken for RNA extraction and purification treatment experiments and then reverse transcribed into cDNA for use. Twenty-seven DEGs were randomly selected and verified by relative quantitative expression using fluorescence quantitative PCR. To verify their resistance changes after stimulation by B. subtilis RSS-1 and B. amyloliquefaciens JDF3, the genes (PR1, PR2, PR3, PR10, and LOX7) of soybean [53,54] were used for qRT-PCR. The ratio of the degree of gene expression at different time periods to the degree of gene expression of the untreated sample was used to evaluate the soybean resistance; that is, the relative expression level of the resistance gene. The genes, Physo1_1 108986 (PsActinA for P. sojae) [51,52] and EFla (Actin for soybean) [53,54], were used as actin for qRT-PCR. Moreover, all the qRT-PCR primers were designed with the software Premier 6.0, and synthesized by Nanjing Qingke Biological Co., Ltd. (Nanjing, China). The primer sequences used are shown in Table S5, and the qRT-PCR verification test for the differential gene expression were carried out according to the method described in the takara kit. The qRT-PCR system (25 μL) is as follows: 2 μL of template cDNA, 12.5 μL of SYBR green mixture, 8.5 μL of dd H2O, and 1 μL of each of the upstream and downstream primers. The reaction procedure was as follows: 95 °C, 30 s; 39 cycles (95 °C for 5 s, 60 °C for 30 s). The above reaction procedure was carried out using ABIPRISM 7300 (Applied Biosystems, Foster, CA, USA), and the obtained product was obtained after the PCR amplification reaction was completed. To obtain the dissolution curve, the temperature was increased from 65 °C to 95 °C.
3.6. Effect of B. amyloliquefaciens and B. subtilis on the Control of Phytophthora Root Rot of Soybean
Soybean greenhouse cultivation referred to the method reported by Lu et al. [2], with slight modifications. Four days after sowing, the soybean seedlings were given a one milliliter liquid culture of B. subtilis RSS-1 or B. amyloliquefaciens JDF3 (OD value = 0.8–1.0) to a soybean every two days, and roots were implemented a total of five times. The effects of B. amyloliquefaciens and B. subtilis on the control of the Phytophthora root rot of soybean were evaluated with the method described by Lu et al. [2] and Pawlowski et al. [49]. The control effects (CE) was calculated as CE = [(disease incidence control-disease incidence treatment)/disease incidence control] × 100%, and disease incidence was based on whether the disease/pathogen was not observed (rating = 0) or observed (rating =1) and converted to percent incidence. Severity ratings were recorded from 0 to 5 using a pre-transformed scale (0 = no visible detection, 1 = 1–10%, 2 = 11–35%, 3 = 35–65%, 4 = 66–90%, and 5 = 91–100% of the stem or sampled area covered).
3.7. BAFB Activates Soybean Defense Responses
The fermentation broth was centrifuged at 10,000 rpm for 10 min, and the supernatant was filtered with a 0.22 μm filter membrane. To verify whether B. subtilis RSS-1 and B. amyloliquefaciens JDF3 could induce plant resistance, we injected Bacillus fermentation broth into Nicotiana benthamiana leaves. An LB liquid medium and dd H2O were separately injected as negative controls (−), and salicylic acid (SA) [55] and SsSm1 [56], an elicitor protein, was injected as a positive control (+). The evaluation of the active oxygen burst was carried out with DAB staining [50]. The lignin was detected with a phloroglucinol kit based on instruction from Qingdao JSK Biotechnology Co., Ltd. (Qingdao, Shangdong, China). An NO fluorescent probe kit (Biyuntian Biotechnology Co., Ltd.,Shanghai, China) was used to detect the NO molecules in soybean single cells. Moreover, the callose staining referred to the method reported by Lu et al. [2].
4. Conclusions
B. amyloliquefaciens JDF3 and B. subtilis RSS-1 could inhibit mycelial growth, the germination of the cysts, and the swimming of the motile zoospores. Comparative transcriptomics was applied to elucidate the molecular mechanism of the biological control. A total of 1616 DEGs were detected. Most DEGs participated in inhibiting the activity of ribosome. A pot experiment indicated that B. amyloliquefaciens and B. subtilis enhanced the resistance of soybean to P. sojae, and their control effects were 70.7% and 65.5%, respectively. In addition, B. amyloliquefaciens fermentation broth could induce an active oxygen burst, NO production, callose deposition, and lignification.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/12/2908/s1.
Author Contributions
Conceptualization: D.L. and Z.G.; methodology: J.H., K.L., and D.L.; software: W.W., and J.H.; writing—original draft preparation: K.L., and D.L.; writing—reviewing and editing: D.L., and X.L.; supervision: Z.G.
Funding
This work was supported by National Natural Science Foundation of China (grant no.31671977), and Excellent Young Talents Fund Program of Higher Education Institutions of Anhui Province (grant no. gxyqZD2018120; grant no. gxyqZD2016516).
Acknowledgments
Thanks to National Natural Science Foundation of China and Provincial Education Board of Anhui.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, D.; Li, P.; Hu, J.; Li, K.; Zhao, Z.; Wang, W.; Zhang, J.Y.; Ding, X.; Gao, Z. Genetic Diversity among Isolates of Phytophthora sojae in Anhui Province of China based on ISSR-PCR Markers. J. Am. Soc. Hortic. Sci. 2018, 143, 304–309. [Google Scholar] [CrossRef]
- Lu, X.X.; Zhou, D.M.; Chen, X.; Zhang, J.F.; Huang, H.W.; Wei, L.H. Isolation and characterization of Bacillus altitudinis JSCX-1 as a new potential biocontrol agent against Phytophthora sojae in soybean. Plant Soil 2017, 416, 53–66. [Google Scholar] [CrossRef]
- Bhat, R.G.; McBlain, B.A.; Schmitthenner, A.F. The inheritance of resistance to metalaxyl and to fluorophenylalanine in matings of homothallic phytophthora sojae. Mycol. Res. 1993, 97, 865–870. [Google Scholar] [CrossRef]
- Cai, M.; Miao, J.; Song, X.; Lin, D.; Bi, Y.; Chen, L.; Liu, X.L.; Tyler, B.M. C239S Mutation in the β-Tubulin of Phytophthora sojae confers resistance to zoxamide. Front. Microbiol. 2016, 7, 762. [Google Scholar] [CrossRef] [PubMed]
- Dorrance, A.E. Management of Phytophthora sojae of soybean: A review and future perspectives. Can. J. Plant Pathol. 2018, 40, 210–219. [Google Scholar] [CrossRef]
- Han, Q.; Zhao, H.; Huang, L.; Buchenauer, H.; Zuo, Y.; Kang, Z. Light and electron microscopy studies on the infection of a wild-type and metalaxyl-resistant isolate of phytophthora sojae in soybean hypocotyls. J. Phytopathol. 2011, 159, 368–376. [Google Scholar] [CrossRef]
- Miao, J.; Chi, Y.; Lin, D.; Tyler, B.M.; Liu, X.L. Mutations in ORP1 conferring oxathiapiprolin resistance confirmed by genome editing using CRISPR/Cas9 in Phytophthora capsici and Phytophthora sojae. Phytopathology 2018, 108, 1412–1419. [Google Scholar] [CrossRef] [PubMed]
- Randall, E.; Young, V.; Sierotzki, H.; Scalliet, G.; Birch, P.R.; Cooke, D.E.; Csukai, M.; Whisson, S.C. Sequence diversity in the large subunit of RNA polymerase I contributes to M efenoxam insensitivity in Phytophthora infestans. Mol. Plant Pathol. 2014, 15, 664–676. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42–a review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef]
- Maksimov, I.V.; Abizgil’Dina, R.R.; Pusenkova, L.I. Plant growth promoting rhizobacteria as alternative to chemical crop protectors from pathogens. Appl. Biochem. Microbiol. 2011, 47, 333–345. [Google Scholar] [CrossRef]
- Wagner, A.; Norris, S.; Chatterjee, P.; Morris, P.F.; Wildschutte, H. Aquatic Pseudomonads Inhibit Oomycete Plant Pathogens of Glycine max. Front. Microbiol. 2018, 9, 1007. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, S.A.; Silby, M.W.; Falloon, P.G.; Mahanty, H.K. Biological control of Phytophthora megasperma var. sojae, causal agent of Phytophthora rot of asparagus, by Pseudomonas aureofaciens PA147-2: A preliminary field trial. N. Z. J. Crop Hortic. Sci. 2000, 28, 97–103. [Google Scholar] [CrossRef][Green Version]
- Hou, J.; Bi, S.; Yan, L.; Zuo, Y.; Wang, Y.; Liu, T.; Zhu, J. Biological potential of Pseudomonas sp. BS1 in the control of Phytophthora root rot of soybean. Afr. J. Microbiol. Res. 2012, 6, 3589–3593. [Google Scholar]
- Sowanpreecha, R.; Rerngsamran, P. Biocontrol of Orchid-pathogenic Mold, Phytophthora palmivora, by Antifungal Proteins from Pseudomonas aeruginosa RS1. Mycobiology 2018, 46, 129–135. [Google Scholar] [CrossRef]
- Du, Q.; Yang, X.; Zhang, J.; Zhong, X.; Kim, K.S.; Yang, J.; Xing, G.J.; Li, X.Y.; Jiang, Z.Y.; Li, Q.Y.; et al. Over-expression of the Pseudomonas syringae harpin-encoding gene hrpZm confers enhanced tolerance to Phytophthora root and stem rot in transgenic soybean. Transgenic Res. 2018, 27, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xu, Y.; Lai, X. Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth-promoting properties. Braz. J. Microbiol. 2018, 49, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Arfaoui, A.; Adam, L.R.; Bezzahou, A.; Daayf, F. Isolation and identification of cultivated bacteria associated with soybeans and their biocontrol activity against Phytophthora sojae. BioControl 2018, 63, 607–617. [Google Scholar] [CrossRef]
- Gao, Y.M.; Wang, X.J.; Zhang, J.; Li, M.; Liu, C.X.; An, J.; Jiang, L.; Xiang, W.S. Borrelidin, a potent antifungal agent: Insight into the antifungal mechanism against Phytophthora sojae. J. Agric. Food Chem. 2012, 60, 9874–9881. [Google Scholar] [CrossRef]
- Syed-Ab-Rahman, S.F.; Carvalhais, L.C.; Chua, E.; Xiao, Y.; Wass, T.J.; Schenk, P.M. Identification of soil bacterial isolates suppressing different Phytophthora spp. and promoting plant growth. Front. Plant Sci. 2018, 9, 1502. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies-the next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Tripathy, S.; Deo, T.; Tyler, B.M. Oomycete Transcriptomics Database: A resource for oomycete transcriptomes. BMC Genom. 2012, 13, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Wang, X.; Tao, K.; Lu, Y.; Dai, T.; Dong, S.; Gijzen, M.; Wang, Y. Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol. Plant Microbe Interact. 2011, 24, 1530–1539. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, X.; Rui, H.; Liu, T.; Hou, J. Cold temperature regulation of zoospore release in Phytophthora sojae: The genes that differentially expressed by cold temperature. Russ. J. Genet. 2018, 54, 618–628. [Google Scholar] [CrossRef]
- Chen, X.R.; Xing, Y.P.; Li, Y.P.; Tong, Y.H.; Xu, J.Y. RNA-Seq reveals infection-related gene expression changes in Phytophthora capsici. PLoS ONE 2013, 8, e74588. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, A.P.; Vega-Arreguín, J.C.; Fei, Z.; Ponnala, L.; Lee, S.J.; Matas, A.J.; Patev, S.; Fry, W.E.; Rose, J.K. Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Mol. Plant Pathol. 2016, 17, 29–41. [Google Scholar] [CrossRef]
- Gkarmiri, K.; Finlay, R.D.; Alström, S.; Thomas, E.; Cubeta, M.A.; Högberg, N. Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genom. 2015, 16, 630. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fan, D.; Li, Y.; Chen, Y.; Huang, L.; Yan, X. Transcriptome analysis of valsa mali reveals its response mechanism to the biocontrol actinomycete saccharothrix yanglingensis hhs.015. BMC Microbiol. 2018, 18, 2–13. [Google Scholar] [CrossRef]
- Zhang, S.; Zheng, X.; Reiter, R.J.; Feng, S.; Wang, Y.; Liu, S.; Jin, L.; Li, Z.G.; Ren, M. Melatonin attenuates potato late blight by disrupting cell growth, stress tolerance, fungicide susceptibility and homeostasis of gene expression in Phytophthora infestans. Front. Plant Sci. 2017, 8, 1993. [Google Scholar] [CrossRef]
- Mei, X.Y.; Liu, Y.X.; Huang, H.C.; Du, F.; Huang, L.L.; Wu, J.Q.; Li, Y.W.; Zhu, S.S.; Yang, M. Benzothiazole inhibits the growth of Phytophthora capsici through inducing apoptosis and suppressing stress responses and metabolic detoxification. Pestic. Biochem. Phys. 2019, 154, 7–16. [Google Scholar] [CrossRef]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Yusupova, G.; Yusupov, M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014, 83, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Manning, B.D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell. Biol. 2013, 15, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Donati, G.; Montanaro, L.; Derenzini, M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012, 72, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, S.; Merrill, A.H., Jr. Sphingolipid metabolism and cell growth regulation. FASEB J. 1996, 10, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ye, T.; Gao, Z. Cloning and functional analysis of succinate dehydrogenase gene PsSDHA in Phytophthora sojae. Microb. Pathog. 2017, 108, 40–48. [Google Scholar] [CrossRef]
- Pan, Y.; Ye, T.; Gao, Z. The succinate dehydrogenase PsSDHB is involved in hyphal morphology, chemical stress response and pathogenicity of Phytophthora sojae. Physiol. Mol. Plant Pathol. 2018, 102, 8–16. [Google Scholar] [CrossRef]
- Zhao, W.; Dong, S.; Ye, W.; Hua, C.C.; Meijer, H.J.G.; Dou, D.L.; Govers, F.; Wang, Y.C. Genome-wide identification of Phytophthora sojae SNARE genes and functional characterization of the conserved SNARE PsYKT6. Fungal Genet. Biol. 2011, 48, 241–251. [Google Scholar] [CrossRef]
- Ayers, A.R.; Valent, B.; Ebel, J.; Albersheim, P. Host-Pathogen Interactions: XI. Composition and structure of wall-released elicitor fractions. Plant Physiol. 1976, 57, 766–774. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Wang, W.; Geng, X.; Shi, Y.; Na, R.; Dou, D.L.; Li, H.L. Network and role analysis of autophagy in phytophthora sojae. Sci. Rep. 2017, 7, 1879. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, F.X.; Zhong, N.Q.; Wang, H.Y.; Xia, G.X. The role of autophagy during development of the oomycete pathogen Phytophthora infestans. J. Genet. Genom. 2014, 41, 225–228. [Google Scholar] [CrossRef]
- Pollack, J.K.; Harris, S.D.; Marten, M.R. Autophagy in filamentous fungi. Fungal Genet. Biol. 2009, 46, 1–8. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wang, Y.; Tao, K.; Dong, S.; Huang, Q.; Dai, T.; Wang, Y. PsSAK1, a stress-activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol. Plant Microbe Interact. 2010, 23, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Cao, M.N.; Ye, W.W.; Li, H.Y.; Kong, L.; Zheng, X.; Wang, Y.C. PsMPK7, a stress-associated mitogen-activated protein kinase (MAPK) in phytophthora sojae, is required for stress tolerance, reactive oxygenated species detoxification, cyst germination, sexual reproduction and infection of soybean. Mol. Plant Pathol. 2015, 16, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.K.; Arseneault, T.; Novinscak, A.; Filion, M. Phenazine-1-carboxylic acid production by Pseudomonas fluorescens LBUM636 alters Phytophthora infestans growth and late blight development. Phytopathology 2016, 107, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Roquigny, R.; Novinscak, A.; Arseneault, T.; Joly, D.L.; Filion, M. Transcriptome alteration in Phytophthora infestans in response to phenazine-1-carboxylic acid production by Pseudomonas fluorescens strain LBUM223. BMC Genom. 2018, 19, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Upchurch, R.G.; Ramirez, M.E. Defense-related gene expression in soybean leaves and seeds inoculated with Cercospora kikuchii and Diaporthe phaseolorum var. meridionalis. Physiol. Mol. Plant Pathol. 2010, 75, 64–70. [Google Scholar] [CrossRef]
- Upchurch, R.G.; Ramirez, M.E. Effects of temperature during soybean seed development on defense-related gene expression and fungal pathogen accumulation. Biotechnol. Lett. 2011, 33, 2397–2404. [Google Scholar] [CrossRef]
- Pawlowski, M.L.; Bowen, C.R.; Hill, C.B.; Hartman, G.L. Responses of soybean genotypes to pathogen infection after the application of elicitors. Crop Prot. 2016, 87, 78–84. [Google Scholar] [CrossRef]
- Wang, N.; Liu, M.; Guo, L.; Yang, X.; Qiu, D. A novel protein elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 induces systemic resistance in tobacco. Int. J. Biol. Sci. 2016, 12, 757. [Google Scholar] [CrossRef]
- Yamamoto, S.; Shiraishi, S.; Suzuki, S. Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides. Lett. Appl. Microbiol. 2015, 60, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.B.; Chai, H.Y.; Jiang, L.X. Isolation and partial characterization of an antifungal protein produced by Bacillus licheniformis BS-3. Molecules 2012, 17, 7336–7347. [Google Scholar] [CrossRef] [PubMed]
- Suo, B.; Chen, Q.M.; Wu, W.X.; Wu, D.; Tian, M.; Jie, Y.; Zhang, B.; Wen, J.Z. Chemotactic responses of Phytophthora sojae zoospores to amino acids and sugars in root exudates. J. Gen. Plant Pathol. 2016, 82, 142–148. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell. Physiol. 2017, 59, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.M.; Wei, J.J.; Yao, C.C.; Reng, H.X.; Gao, Z.M. SsSm1, a Cerato-platanin family protein, is involved in the hyphal development and pathogenic process of Sclerotinia sclerotiorum. Plant Sci. 2018, 270, 37–46. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).