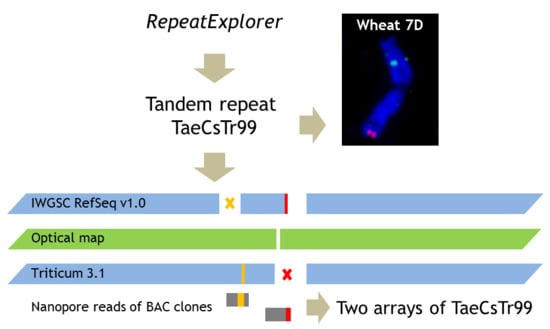
The Dark Matter of Large Cereal Genomes: Long Tandem Repeats
Abstract
1. Introduction
2. Results and Discussion
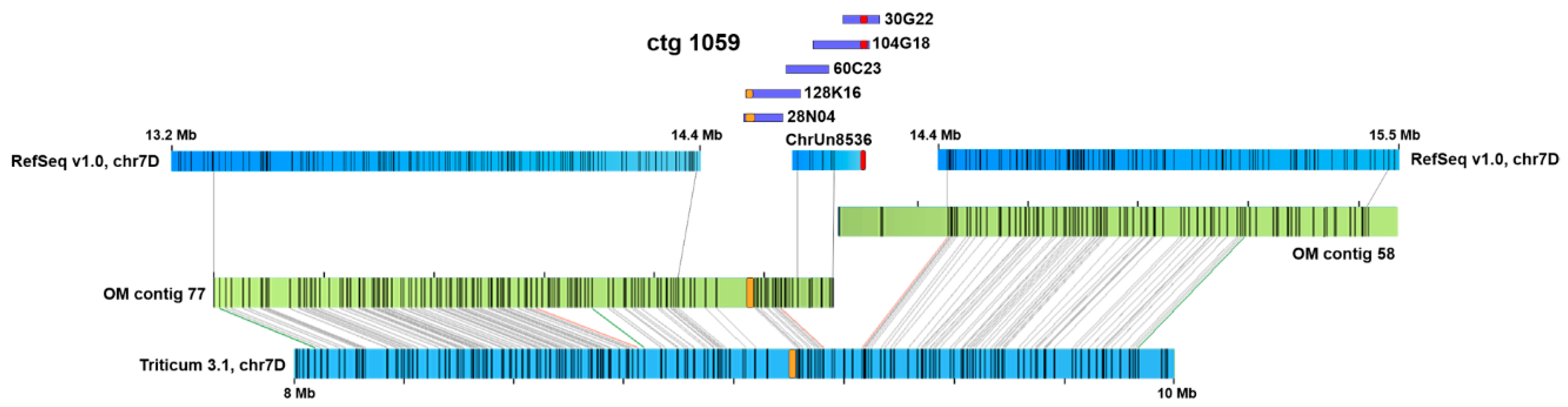
2.1. Chromosome-Specific Tandem Repeats in Wheat
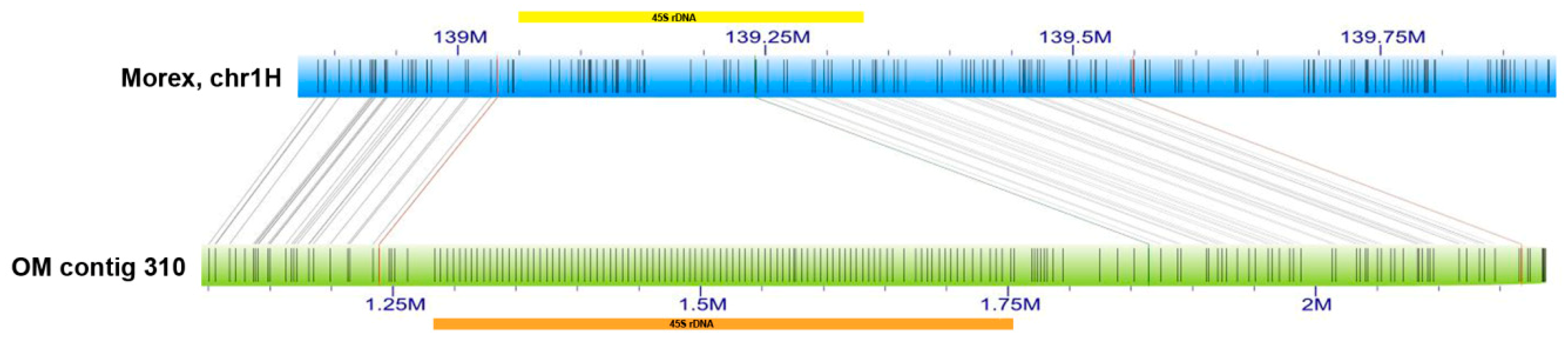
2.2. Minor 45S rDNA Locus in Barley Chromosome 1H
3. Materials and Methods
3.1. De Novo Identification of Wheat Tandem Repeats
3.2. In Situ Hybridisation
3.3. Reconstruction of Barley 1H rDNA Unit
3.4. Application of Optical Maps
3.5. Nanopore Sequencing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 7DS | Short arm of wheat chromosome 7D |
| BAC | Bacterial Artificial Chromosome |
| BNG mapping | Bionano genome mapping |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DAPI | 4,6-diamidino-2-phenylindole |
| FISH | Fluorescence in situ hybridisation |
| IGS | Intergenic spacer |
| ISH | In situ hybridisation |
| IWGSC | International Wheat Genome Sequencing Consortium |
| MDA | Multiple Displacement Amplification |
| OM | Optical map |
| ONT | Oxford Nanopore Technologies |
| PacBio | Pacific Biosciences |
| rDNA | Ribosomal DNA |
| SMRT | Single-molecule real-time |
| SRA | Sequence read archive |
References
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Stein, N.; (IPK, Gatersleben, Germany). Personal communication, 2018.
- Doležel, J.; Čížková, J.; Šimková, H.; Bartoš, J. One major challenge of sequencing large plant genomes is to know how big they really are. Int. J. Mol. Sci. 2018, 19, 3554. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef]
- Martis, M.M.; Zhou, R.; Haseneyer, G.; Schmutzer, T.; Vrána, J.; Kubaláková, M.; König, S.; Kugler, K.G.; Scholz, U.; Hackauf, B.; et al. Reticulate evolution of the rye genome. Plant Cell 2013, 25, 3685–3698. [Google Scholar] [CrossRef]
- Mayer, K.F.X.; Martis, M.; Hedley, P.E.; Šimková, H.; Liu, H.; Morris, J.A.; Steuernagel, B.; Taudien, S.; Roessner, S.; Gundlach, H.; et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 2011, 23, 1249–1263. [Google Scholar] [CrossRef]
- Mayer, K.F.X.; Rogers, J.; Doležel, J.; Pozniak, C.; Eversole, K.; Feuillet, C.; Gill, B.; Friebe, B.; Lukaszewski, A.J.; Sourdille, P.; et al. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Chaisson, M.J.; Wilson, R.K.; Eichler, E.E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 2015, 16, 627–640. [Google Scholar] [CrossRef]
- Zimin, A.V.; Puiu, D.; Hall, R.; Kingan, S.; Clavijo, B.J.; Salzberg, S.L. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef]
- Handa, H.; Kanamori, H.; Tanaka, T.; Murata, K.; Kobayashi, F.; Robinson, S.J.; Koh, C.S.; Pozniak, C.J.; Sharpe, A.G.; Paux, E.; et al. Structural features of two major nucleolar organizer regions (NORs), Nor-B1 and Nor-B2, and chromosome-specific rRNA gene expression in wheat. Plant J. 2018, 96, 1148–1159. [Google Scholar] [CrossRef]
- Symonová, R.; Ocalewicz, K.; Kirtiklis, L.; Delmastro, G.B.; Pelikánová, Š.; Garcia, S.; Kovařík, A. Higher-order organisation of extremely amplified, potentially functional and massively methylated 5S rDNA in European pikes (Esox sp.). BMC Genom. 2017, 18, 391. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Gerlach, W.L.; Dennis, E.S.; Swift, H.; Peacock, W.J. Molecular and Chromosomal Organization of DNA Sequences Coding for the Ribosomal RNAs in Cereals. Chromosoma 1980, 78, 293–311. [Google Scholar] [CrossRef]
- Rayburn, A.L.; Gill, B.S. Use of biotin-labeled probes to map specific DNA sequences on wheat chromosomes. Heredity 1985, 76, 78–81. [Google Scholar] [CrossRef]
- Mukai, Y.; Endo, T.R.; Gill, B.S. Physical mapping of the 18S.26S rRNA multigene family in common wheat: Identification of a new locus. Chromosoma 1991, 100, 71–78. [Google Scholar] [CrossRef]
- Leitch, I.J.; Leitch, A.R.; Heslop-Harrison, J.S. Physical mapping of plant DNA sequences by simultaneous in situ hybridization of two differently labelled fluorescent probes. Genome 1991, 34, 329–333. [Google Scholar] [CrossRef]
- Mukai, Y.; Nakahara, Y.; Yamamoto, M. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 1993, 36, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Gill, B.S. New 18S. 26S ribosomal RNA gene loci: Chromosomal landmarks for the evolution of polyploid wheats. Chromosoma 1994, 103, 179–185. [Google Scholar] [CrossRef]
- Brandes, A.; Röder, M.S.; Ganal, M.W. Barley telomeres are associated with two different types of satellite DNA sequences. Chromosome Res. 1995, 3, 315–320. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 2010, 11, 378. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Martis, M.M.; Klemme, S.; Banaei-Moghaddam, A.M.; Blattner, F.R.; Macas, J.; Schmutzer, T.; Scholz, U.; Gundlach, H.; Wicker, T.; Šimková, H.; et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 2012, 109, 13343–13346. [Google Scholar] [CrossRef]
- Lam, E.T.; Hastie, A.; Lin, C.; Ehrlich, D.; Das, A.K.; Austin, M.D.; Deshpande, P.; Cao, H.; Nagarajan, N.; Xiao, M.; et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 2012, 30, 771–777. [Google Scholar] [CrossRef]
- Staňková, H.; Hastie, A.R.; Chan, S.; Vrána, J.; Tulpová, Z.; Kubaláková, M.; Visendi, P.; Hayashi, S.; Luo, M.C.; Batley, J.; et al. BioNano genome mapping of individual chromosomes supports physical mapping and sequence assembly in complex plant genomes. Plant Biotechnol. J. 2016, 14, 1523–1531. [Google Scholar] [CrossRef]
- Luo, MC.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rodriguez, J.C.; Deal, K.R.; Avni, R.; Distelfeld, A.; McGuire, P.E.; Dvorak, J.; Luo, MC. Improved genome sequence of wild emmer wheat Zavitan with the aid of optical maps. G3 (Bethesda) 2019, 9, 619–624. [Google Scholar] [CrossRef]
- Tulpová, Z.; Toegelová, H.; Lapitan, N.L.V.; Peairs, F.B.; Macas, J.; Novák, P.; Lukaszewski, A.J.; Kopecký, D.; Mazáčová, M.; Vrána, J.; et al. Accessing a Russian wheat aphid resistance gene in bread wheat by long-read technologies. Plant Genome 2019, 12, 1–11. [Google Scholar] [CrossRef]
- Tulpová, Z.; Luo, M.C.; Toegelová, H.; Visendi, P.; Hayashi, S.; Vojta, P.; Paux, E.; Kilian, A.; Abrouk, M.; Bartoš, J.; et al. Integrated physical map of bread wheat chromosome arm 7DS to facilitate gene cloning and comparative studies. N. Biotechnol. 2019, 48, 12–19. [Google Scholar] [CrossRef]
- Beier, S.; Himmelbach, A.; Colmsee, C.; Zhang, X.Q.; Barrero, R.A.; Zhang, Q.; Li, L.; Bayer, M.; Bolser, D.; Taudien, S.; et al. Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Sci. Data 2017, 4, 170044. [Google Scholar] [CrossRef]
- Leitch, I.J.; Heslop-Harrison, J.S. Physical mapping of the 18S-5.8S-26S rRNA genes in barley by in situ hybridization. Genome 1992, 35, 1013–1018. [Google Scholar] [CrossRef]
- Szakács, É.; Kruppa, K.; Molnár-Láng, M. Analysis of chromosomal polymorphism in barley (Hordeum vulgare L. ssp. vulgare) and between H. vulgare and H. chilense using three-color fluorescence in situ hybridization (FISH). J. Appl. Genet. 2013, 54, 427–433. [Google Scholar] [CrossRef]
- Shoaib, M.; Baconnais, S.; Mechold, U.; Le Cam, E.; Lipinski, M.; Ogryzko, V. Multiple displacement amplification for complex mixtures of DNA fragments. BMC Genom. 2008, 9, 415. [Google Scholar] [CrossRef]
- Zhang, D.; Chan, S.; Sugerman, K.; Lee, J.; Lam, E.T.; Bocklandt, S.; Cao, H.; Hastie, A.R. CRISPR-bind: A simple, custom CRISPR/dCas9-mediated labeling of genomic DNA for mapping in nanochannel arrays. bioRxiv 2018. preprint. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acid Res. 1979, 7, 1869–1886. [Google Scholar] [CrossRef]
- Berkman, P.J.; Skarshewski, A.; Lorenc, M.T.; Lai, K.; Duran, C.; Ling, E.Y.; Stiller, J.; Smits, L.; Imelfort, M.; Manoli, S.; et al. Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS. Plant Biotechnol. J. 2011, 9, 768–775. [Google Scholar] [CrossRef]
- Karafiátová, M.; Bartoš, J.; Doležel, J. Localization of low-copy DNA sequences on mitotic chromosomes by FISH. In Plant cytogenetics. Methods and Protocols; Kianian, S.F., Kianian, P.M.A., Eds.; Humana Press: New York, NY, USA, 2016; Volume 1429, pp. 49–64. [Google Scholar]
- Muñoz-Amatriaín, M.; Lonardi, S.; Luo, MC.; Madishetty, K.; Svensson, J.T.; Moscou, M.J.; Wanamaker, S.; Jiang, T.; Kleinhofs, A.; Muehlbauer, G.J.; et al. Sequencing of 15 622 gene-bearing BACs clarifies the gene-dense regions of the barley genome. Plant J. 2015, 84, 216–227. [Google Scholar] [CrossRef]
- Šimková, H.; Šafář, J.; Kubaláková, M.; Suchánková, P.; Číhalíková, J.; Robert-Quatre, H.; Azhaguvel, P.; Weng, Y.; Peng, J.; Lapitan, N.L.V.; et al. BAC Libraries from wheat chromosome 7D: Efficient tool for positional cloning of aphid resistance genes. J. Biomed. Biotechnol. 2011, 2011, 302543. [Google Scholar] [CrossRef]

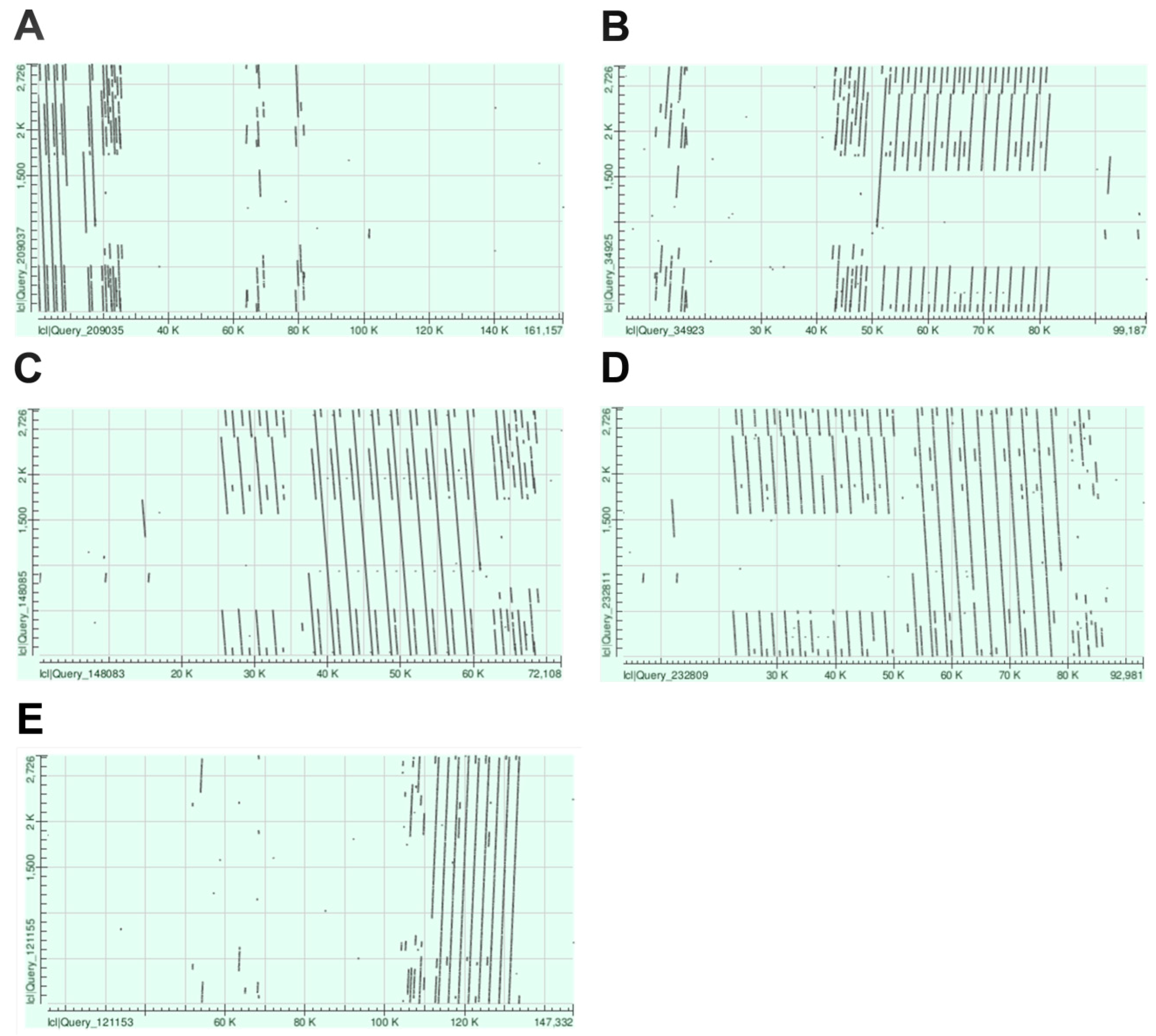
| Tandem Repeat | Monomer Size | Distribution |
|---|---|---|
| TaeCsTr163 | 1390 bp | 7D subtelomere |
| TaeCsTr230 | 1242 bp | 7D subtelomere |
| TaeCsTr99 | 2726 bp | 7D subtelomere |
| TaeCsTr111 | 1167 bp | All chromosomes Dispersed |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapustová, V.; Tulpová, Z.; Toegelová, H.; Novák, P.; Macas, J.; Karafiátová, M.; Hřibová, E.; Doležel, J.; Šimková, H. The Dark Matter of Large Cereal Genomes: Long Tandem Repeats. Int. J. Mol. Sci. 2019, 20, 2483. https://doi.org/10.3390/ijms20102483
Kapustová V, Tulpová Z, Toegelová H, Novák P, Macas J, Karafiátová M, Hřibová E, Doležel J, Šimková H. The Dark Matter of Large Cereal Genomes: Long Tandem Repeats. International Journal of Molecular Sciences. 2019; 20(10):2483. https://doi.org/10.3390/ijms20102483
Chicago/Turabian StyleKapustová, Veronika, Zuzana Tulpová, Helena Toegelová, Petr Novák, Jiří Macas, Miroslava Karafiátová, Eva Hřibová, Jaroslav Doležel, and Hana Šimková. 2019. "The Dark Matter of Large Cereal Genomes: Long Tandem Repeats" International Journal of Molecular Sciences 20, no. 10: 2483. https://doi.org/10.3390/ijms20102483
APA StyleKapustová, V., Tulpová, Z., Toegelová, H., Novák, P., Macas, J., Karafiátová, M., Hřibová, E., Doležel, J., & Šimková, H. (2019). The Dark Matter of Large Cereal Genomes: Long Tandem Repeats. International Journal of Molecular Sciences, 20(10), 2483. https://doi.org/10.3390/ijms20102483