1. Introduction
Coconut palm (
Cocos nucifera L.) is one of the most economically important trees in the world. It generates employment and income in many countries, where its fruit is either eaten raw or processed into manufactured products and by-products. However, coconut production is affected by many biotic and abiotic factors. Coconut stem-bleeding (CSB), caused by the fungus
Ceratocystis paradoxa (De Seynes) Höhn (anamorph
Thielaviopsis paradoxa), is one of the most notorious biotic diseases of coconut. CSB is known to occur in coconut-producing areas worldwide. The disease was first reported in Sri Lanka [
1] and caused severe damage to PB-121 hybrids in Indonesia. In the state of Sergipe, Brazil, it was first detected in early 2004 [
2]. It was discovered in China for the first time in 2009, where it was found to occur in nearly all coconut gardens [
3]. The disease has gradually become the primary concern of producers and researchers due to its rapid spread and great lethality to coconut. Affected plants perish within 3–4 months after the first appearance of stem symptoms.
The symptoms of CSB are described as follows: The affected trunk areas exhibit dark discoloration and a reddish-brown or rust-colored liquid bleeding from the stem cracks that may turn blackish when dried, resulting in rotting when moist; there is a reduced frequency of leaf emergence and a reduced young leaf size, with stem-thinning occurring near the canopy as the disease progresses; and the leaves eventually become brownish-yellow and fragile [
2,
3]. Besides the trunk and leaves,
C. paradoxa also infects the coconut fruit, with the infected pericarp becoming black and soft and producing a strong fruity aroma [
4].
The ascomycete
C. paradoxa not only infects palm trees, such as
Cocos nucifera,
Phoenix dactylifera,
Borassus flabellifer,
Butia capitata,
Elaeis guineensis, and
Areca catechu, but also infects other plants, including
Ananas comosus,
Mangifera indica,
Musa paradisiaca,
Solanum muricatum,
Theobroma cacao, and
Zea mays, resulting in large agricultural losses [
3]. The biological characteristics of the pathogen have thus far been described [
5], and the partial and temporal dynamics of stem-bleeding have been recorded for clarifying the disease progression and characterizing the production losses from the disease [
6]. In addition, some CSB disease management studies have been reported, including physical, chemical, and biological control measures [
7,
8,
9,
10]. However, a detailed understanding of the pathogenesis and infection dynamics of this pathogen is lacking. While a previous report on the transformation of
C. paradoxa using
Agrobacterium exists [
11], the pathogenesis of
C. paradoxa was not explored.
Highly efficient transformation would be beneficial for a detailed investigation of the infection characteristics and pathogenesis of
C. paradoxa and its interaction with the host. Green fluorescent protein (GFP) expression has proven to be a useful tool for such analyses in filamentous fungi [
12,
13] and has been successfully expressed in numerous ascomycetes, including
Colletotrichum acutatum,
Verticillium fungicola Acremonium chrysosporium, and
Sordari macrospora [
14,
15]. In this study, the first protoplast preparation and polyethylene glycol (PEG)-mediated transformation system for
C. paradoxa was established, and the expression vector pCT74-sGFP was transformed into the fungus to obtain a recombinant fungus labeled with GFP. Our protocol will inform research that involves monitoring the infection progression of this fungus in the coconut and should also inform the further exploration of the mechanisms of
C. paradoxa pathogenicity.
3. Discussion
As the causal agent of coconut stem-bleeding (CSB), C. paradoxa causes great economic losses to the coconut industry. Understanding the invasion, colonization, and localization mechanisms of C. paradoxa in coconut could facilitate the development of new strategies for disease control in the future. In the present study, we successfully transformed the hygromycin B resistance gene and a ‘reporter’ gfp gene into C. paradoxa, obtaining stable transformants of C. paradoxa that efficiently expressed GFP.
Protoplast quantity is one of the key factors influencing transformation efficiency. The results of this study demonstrated that transformants were rarely obtained when the number of protoplasts was lower than 1 × 10
6/mL
−1. This corroborates the consensus that a high concentration of protoplasts is required for successful transformation [
16,
17]. The concentration of the plasmid pCT74-sGFP also influenced the transformation results, with 500 ng/μL of pCT74-sGFP means approximately 2 μg of DNA being required. Thus, the preparation of a sufficient quantity of protoplasts is necessary for successful transformation, but a high plasmid concentration is equally important.
Due to fungal cell wall differences, the enzyme type and digestion conditions can affect protoplast production. In this study, the cell wall of C. paradoxa was digested with different protoplast release buffers at the same temperature (31 °C), but the duration of the treatment and the quantity of the protoplasts obtained differed between the buffers. A comparison of the six treatment results indicated that adequate protoplasts could only be obtained after digestion with protoplast release buffer 2 for 90 min, while the other treatments (protoplast release buffer 1, 3, 4, 5, and 6) were either overly time-consuming or produced lower yields.
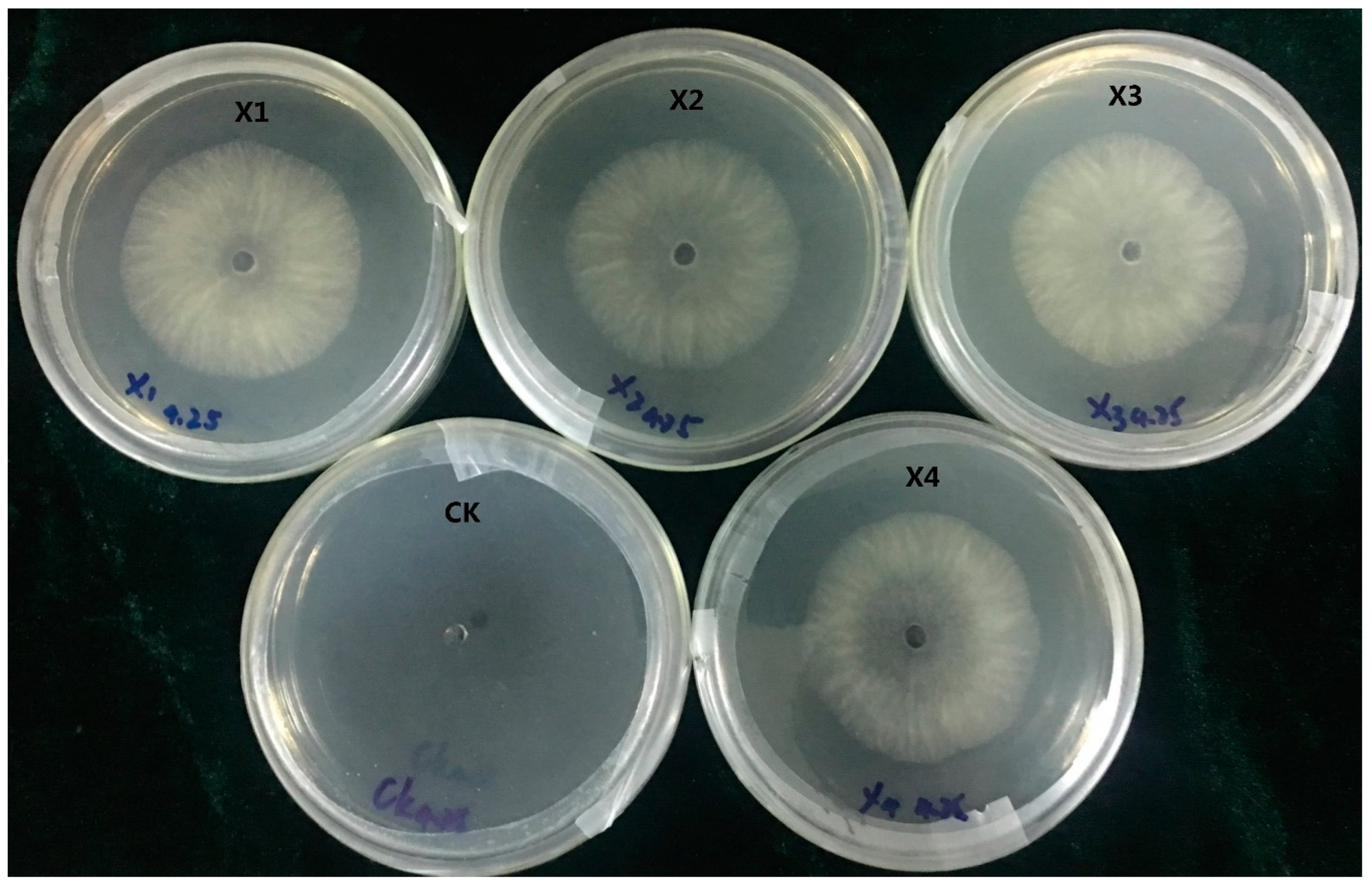
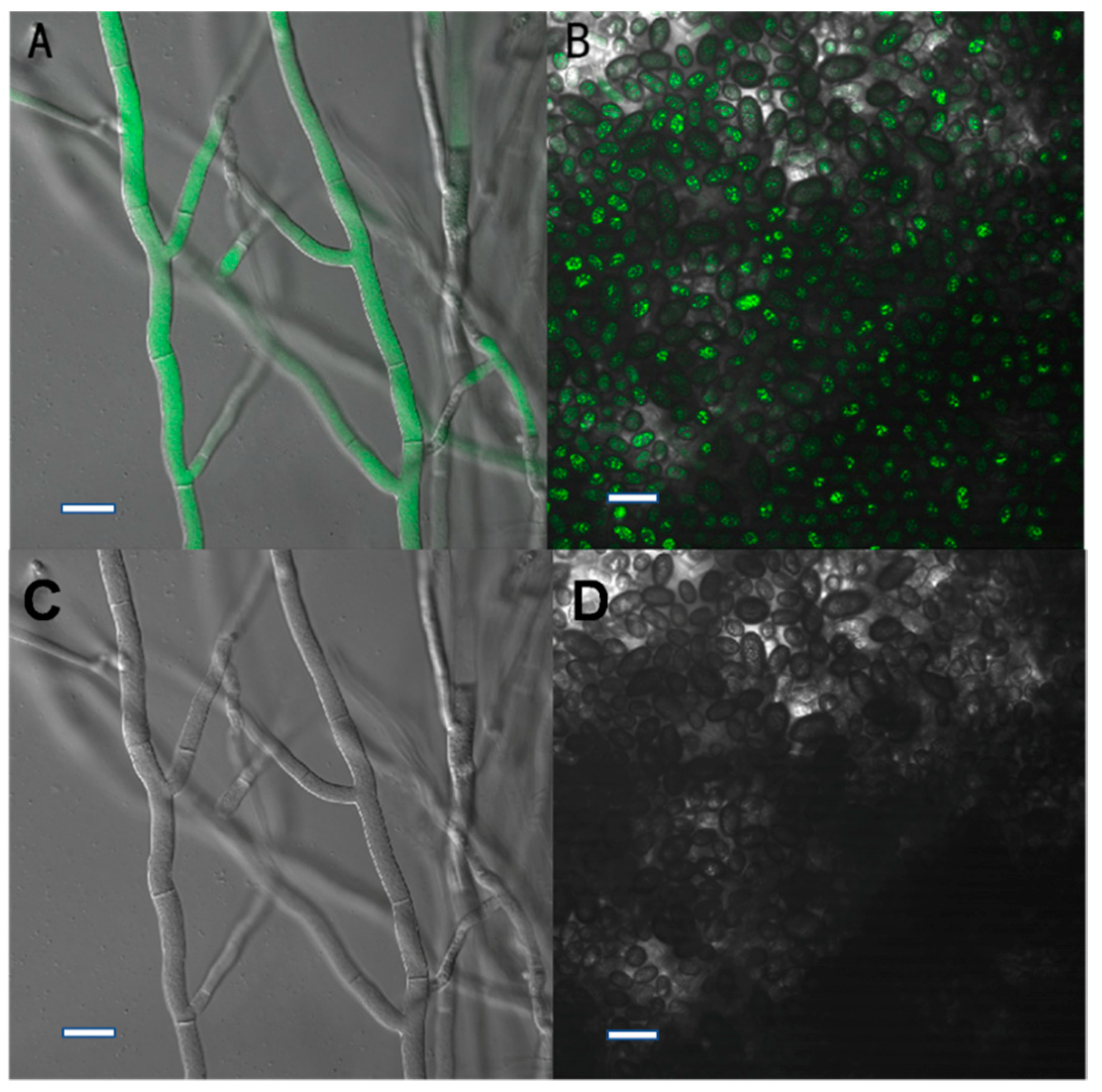
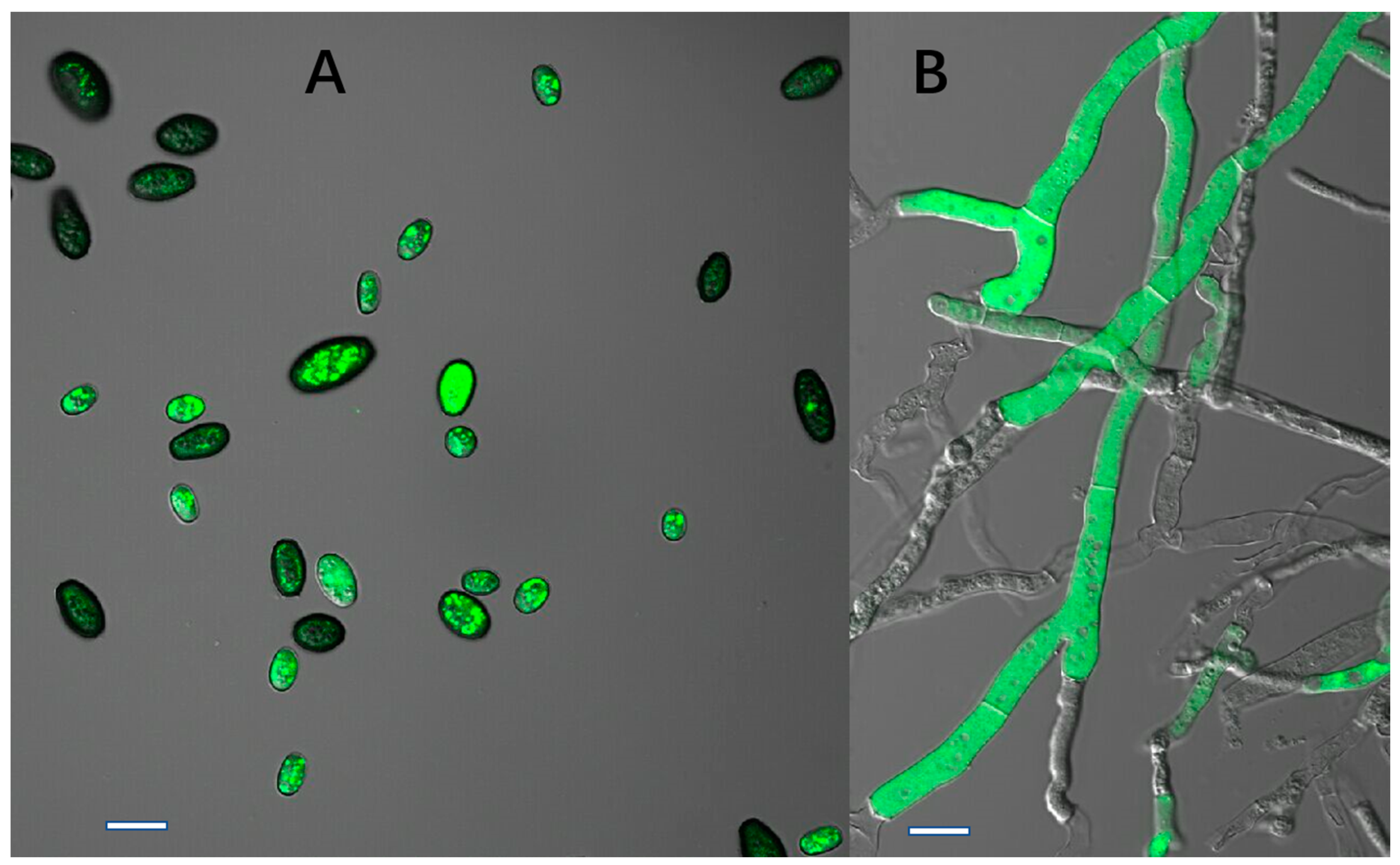
After subculturing seven times on PDA plates lacking HmB, the four transformants (X1, X2, X3, and X4) could still grow on HmB PDA plates and exhibited high green fluorescence inside the hyphae and conidia, which is indicative of genetic stability. The results indicated that the gfp gene had been successfully transformed into C. paradoxa. Furthermore, the pathogenicity test demonstrated that the transformants caused typical disease symptoms that were the same as the wild-type strain, which suggested that the pathogenicity of the transformant strain had not changed. This verified that they can be used for studying C. paradoxa infection in coconut palms.
GFP as a reporter has been widely used in fungi [
18,
19,
20]. The advantage of real-time observation renders it a great tool for the analysis of living cells and organisms. However, its application is limited in plant pathogen analyses due to the lower number of transformed species or inefficient expression and low fluorescence [
21]. The successful expression of GFP is usually based on a stronger promoter or a high copy number viral vector [
22]. The ToxA promoter has proven valuable for the strong expression of GFP in ascomycetes with diverse lifestyles [
23]. Our results indicated that this promoter can be used to obtain high GFP expression in
C. paradoxa.
This study is the first to report on GFP expression in C. paradoxa. The transformants can be used for the study of fungal colonization, localization, and invasion in plants. Furthermore, the protoplast preparation and transformation method described here can be used as a tool for efficiently constructing a random insertional transformant library, gene functional analysis, and the exploration of the pathogenic mechanisms of C. paradoxa.
4. Materials and Methods
4.1. Strain and Plant Material
The C. paradoxa X-3314 strain (Collection No. CCTCC AF2014002) was isolated from an infected coconut stem and stored at the Coconut Research Institute of Chinese Academy of Tropical Agricultural Sciences (Wenchang, Hainan province, China). The Escherichia coli strain DH5a was used in this study. The plant material was coconut fruits of “Malayan Red Dwarf”.
4.2. Fungal Transformation Vector
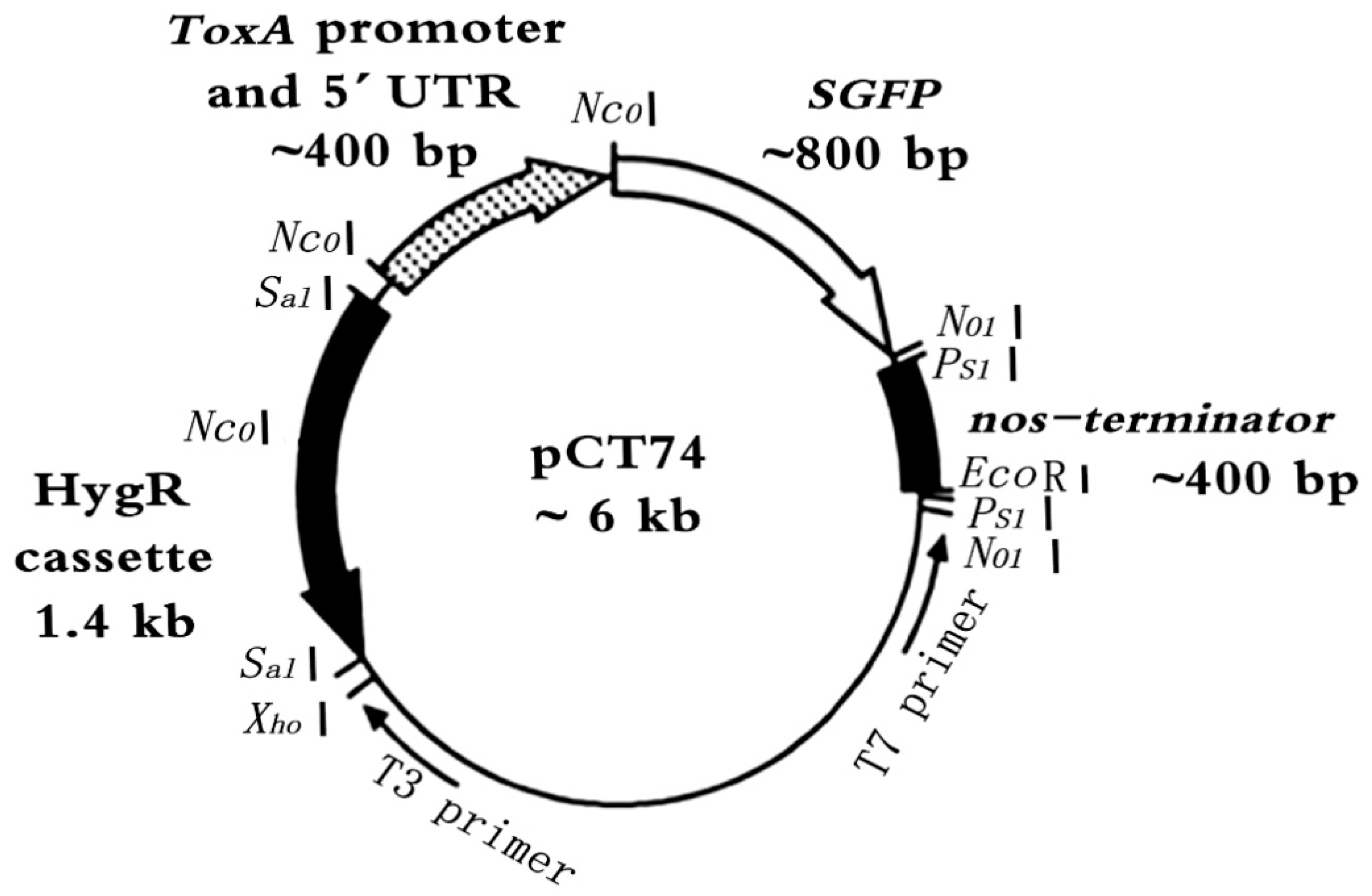
A pCT74-sGFP plasmid (
Figure 7) was constructed and stored at the Functional Genomics Center, Fujian Agriculture and Forestry University. The transformation construct relies on antibiotic resistance conferred by a modified form of the
E. coli hygromycin B phosphotransferase (
hph) gene for transformation selection [
24]. PCT74 further employs the necrosis-inducing host-selective toxin gene ToxA promoter from
Pyrenophoratritici repentis to drive the expression of the synthetic GFP genes (sGFP) [
22,
25].
4.3. Media and Reagents
The media used in this test included complete liquid medium (CM) (6 g yeast extract, 10 g sucrose, 6 g casein acid hydrolysate, 15 g agar, 1000 mL ddH2O), potato dextrose agar (PDA) solid medium (200 g potato, 20 g sucrose, 15 g agar, 1000 mL ddH2O), starch yeast liquid medium (SYM) (2 g yeast extract, 10 g starch, 3 g sucrose, 1000 mL ddH2O), Luria-Bertani (LB) solid medium (5 g yeast extract, 10 g peptone, 10 g NaCl, 15 g agar, 100 mg/mL ampicillin Na (Amp, Takara), 1000 mL ddH2O), TB3 liquid medium (3 g yeast extract, 3 g casamino acid, 20% sucrose, 1000 mL ddH2O), TB3 solid medium (3 g yeast extract, 3 g casamino acid, 20% sucrose, 15 g agar, 1000 mL ddH2O).
STC buffer (44 g sorbitol, 10 mL l.0 mol/L Tris-HCL (pH 8.0), 10 mL 1 mol/L CaCl2, 200 μL streptomycin, add ddH2O up to 200 mL, and sterile water filtered through a 0.22-μm filter (Jet Biofil FPV203000)). PTC buffer (4 g PEG3350, 10 mL STC, 10 μL streptomycin nitrate, heat-dissolved at 60°C for 20 s and sterile filtered), osmotic pressure stabilizer (1.0 mol/L mannitol, 1.0 mol/L sorbitol), protoplast release buffer 1 (1% lysing enzyme (Sigma-Aldrich, St. Louis, MO, US, CB8709259) with 1.0 mol/L mannitol); protoplast release buffer 2 (1% Driselase (Sigma-Aldrich, cat. No. CB0296810) with 1.0 mol/L mannitol in STC); protoplast release buffer 3 (mixture of 1% Driselase and 1% lysing enzyme with 1.0 mol/L mannitol in STC); protoplast release buffer 4 (1% lysing enzyme with 1.0 mol/L sorbitol in STC); protoplast release buffer 5 (1% Driselase with 1.0 mol/L sorbitol in STC); protoplast release buffer 6 (mixture of 1% Driselase and 1% lysing enzyme with 1.0 mol/L sorbitol in STC) were used to prepare protoplasts from mycelia. The hygromycin B (HmB) was purchased from Takara Co. The PCR primers used in this test were synthesized by Sangon Biotech (Shanghai), Co., Ltd., Shanghai, China.
4.4. Sensitivity of C. paradoxa to HmB
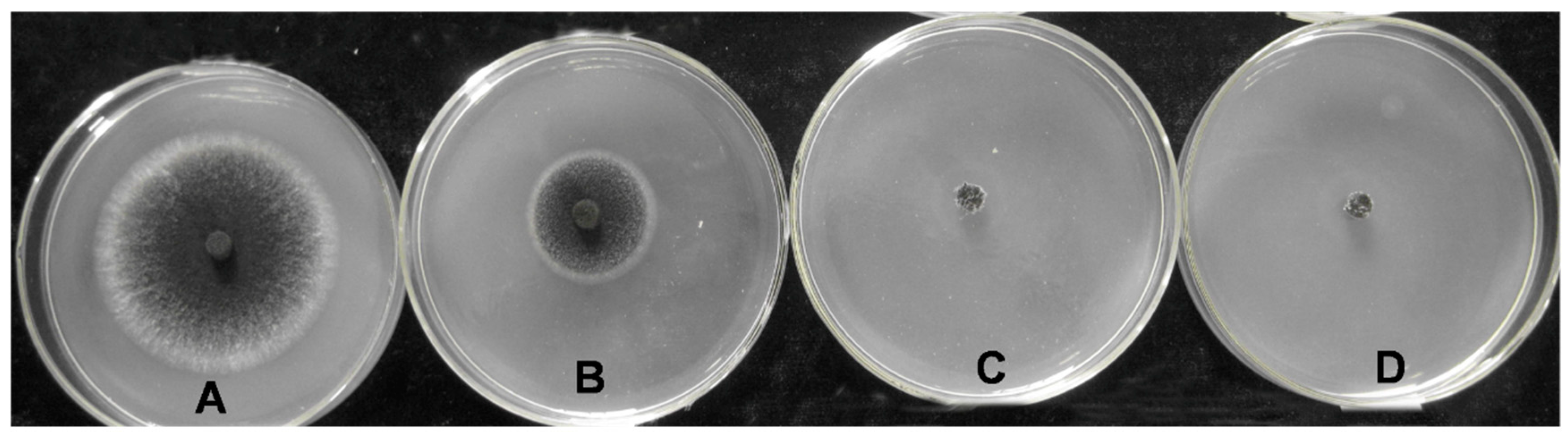
Hygromycin B (HmB) ranging from 10–40 μg/mL was used for testing the sensitivity of C. paradoxa to the antibiotic. Agar blocks (5 mm × 5 mm) containing mycelia were cut from the edge of the C. paradoxa colonies grown on PDA plates and transferred into the center of new PDA plates supplemented with different concentrations of hygromycin B and incubated at 25 °C, PDA plates without HmB served as a control. The effect of HmB on C. paradoxa growth was analyzed by measuring the diameter of growing mycelial on the plates after 5 days. The test was carried out in triplicate.
4.5. Plasmid Extraction of pCT74-sGFP
The E. coli DH5α construct containing pCT74 was spread onto LB plates containing ampicillin Na (Amp, 100 μg/mL). A single colony was inoculated into LB liquid medium containing 2 μL 100 mg/mL Amp and incubated with shaking at 37 °C overnight. Plasmid extraction was achieved using a plasmid Mini Kit (Takara, Japan). Plasmids were detected by 1.5% agarose gel electrophoresis and the plasmid DNA concentration was measured using a Micro-Ultraviolet Spectrophotometer (NanoDrop 2000, Thermo Scientific, Waltham, MA, USA).
4.6. Protoplast Preparation
The entire procedure detailed below was performed under aseptic conditions. A 5-mm diameter agar disc from the growth margins of the X-3314 colony was placed in the center of a fresh PDA plate. After incubation at 25 °C for 5 days, the colony had grown over and covered the plate. Ten milliliters of sterile water was added to the plate and the conidia of C. paradoxa were collected and adjusted to a concentration of 1 × 107 spores mL−1 with sterilized water using a hemocytometer prior to inoculation. Approximately 6 mL of l × 106 spores mL−1 conidia was inoculated into the SYM medium and distributed into three flasks, with 400 mL of medium per flask. The flasks were incubated at 25 °C for 24 h under shaking at 60 rpm/min. The mycelia were collected using two layers of micro-cloth (Calbiochem, Cambridge, MA, USA) and washed with sterilized water twice, followed by washing three times with 15 mL osmotic pressure stabilizer, and then dried on filter paper. One gram of mycelia was collected and thoroughly suspended in protoplast release buffer 2 at 31 °C with shaking at 90 rpm/min for 90 min. Three layers of micro-cloth were placed in a 50-mL sterilized centrifuge tube used for filtering the lysis solution, after which the filtrates were centrifuged at 4000 rpm for 10 min at 4 °C. After discarding the supernatant, the pellet was washed by gently re-suspending it in 20 mL STC and the same STC wash procedure was repeated twice. The concentration of the protoplasts was then adjusted to 6.0 × 107–3.0 × 108/mL with STC. Finally, 20 μL DMSO was added to the protoplast suspension, which was then distributed into 2 mL centrifuge tubes with 150 μL of suspension per tube. The tubes were then stored at −80 °C until use.
4.7. Protoplast Transformation
Ten microliters of pCT74-sGFP plasmid DNA was added to 150 μL of prepared protoplast suspension and gently mixed, and then incubated at 25 °C for 30 min. Following this, 600 μL PTC was added and the solution was incubated at room temperature (25 °C ± 1 °C) for 30 min, after which 6 mL TB3 was added to the reaction tube (50 mL) and incubated at 25 °C with shaking (90 rpm) overnight. The treatment tube with liquid TB3 was mixed with 100 mL solid TB3 medium (that had been melted at 50 °C) containing 80 μL of a 40 μg/mL HmB stock solution and poured into 15 cm diameter plates, which were then incubated at 25 °C for 3–4 d in preparation for transformant growth detection.
4.8. Screening for HmB Resistance and Stability Test
The potential transformant colonies growing on HmB plates were used to inoculate selective PDA medium containing 80 μL of a 40 μg/mL HmB stock solution, with one plate for each potential transformant. These plates were then incubated at 25 °C for 4~5 days. To confirm the stability of the transformants, they were inoculated on PDA plates lacking HmB and incubated at 25 °C for 3 d. The mycelia from the edge of the colonies were inoculated on fresh PDA plates for seven successive subcultures, and then transferred to selective PDA plates containing 80 μL of a 40 μg/mL HmB stock solution in order to observe the growth under selective conditions. The fluorescence of the transformants was monitored using a confocal microscope (Nikon C-HGFIE) with VC40 oil objective, and fluorescence was detected at 488 nm.
4.9. PCR Detection
The primers were as follows: ToxAsGFPF, 5’TGGAATCCATGGAGGAGTTC3’; ToxAsGFPR, 5’CTTGTACAGCTCGTCCATGC3’. To determine whether the
gfp gene had integrated into the genome, a pair of primers (ToxAsGFPF and ToxAsGFPR’) were designed based on the ToxA promoter sequence and the sGFP sequence. Polymerase Chain Reaction (PCR) was used to confirm the integration of the GFP gene insert, with a predicted length of the amplified sequence containing the GFP of about 1.1 kb. The genomic DNA of the transformants and wild-type strain was extracted using a modified cetyltrimethylammonium bromide (CTAB) method [
26]. The pCT74-sGFP plasmid was regarded as a positive control. The PCR reaction system contained: 2.5 μL 10×PCR buffer, 2.5 μL 2.5 mmol/L dNTPs, 1.0 μL primerF, 1.0 μL primerR, and 0.5 μLExtag. The PCR reaction procedure was as follows: 95°C for 3 min, followed by 35 cycles of 94 °C for 40 s, 64 °C for 40 s, 72 °C for 80 s, and a final extension at 72 °C for 10 min.
4.10. Pathogenicity Detection of the Transformants
The wild strain X-3314 and positive transformants were cultured on PDA medium at 25 °C for 5 d. Coconut fruits (“Malayan Red Dwarf”) were wounded and inoculated with a 1.5 × 105 spores·mL−1 suspension of X-3314 or transformants. Wound incisions were made with a sterilized cork borer. The experiment was repeated in triplicate.