NK3R Mediates the EGF-Induced SLα Secretion and mRNA Expression in Grass Carp Pituitary
Abstract
1. Introduction
2. Results
2.1. Molecular Cloning and Tissue Distribution of Grass Carp EGF and EGFR
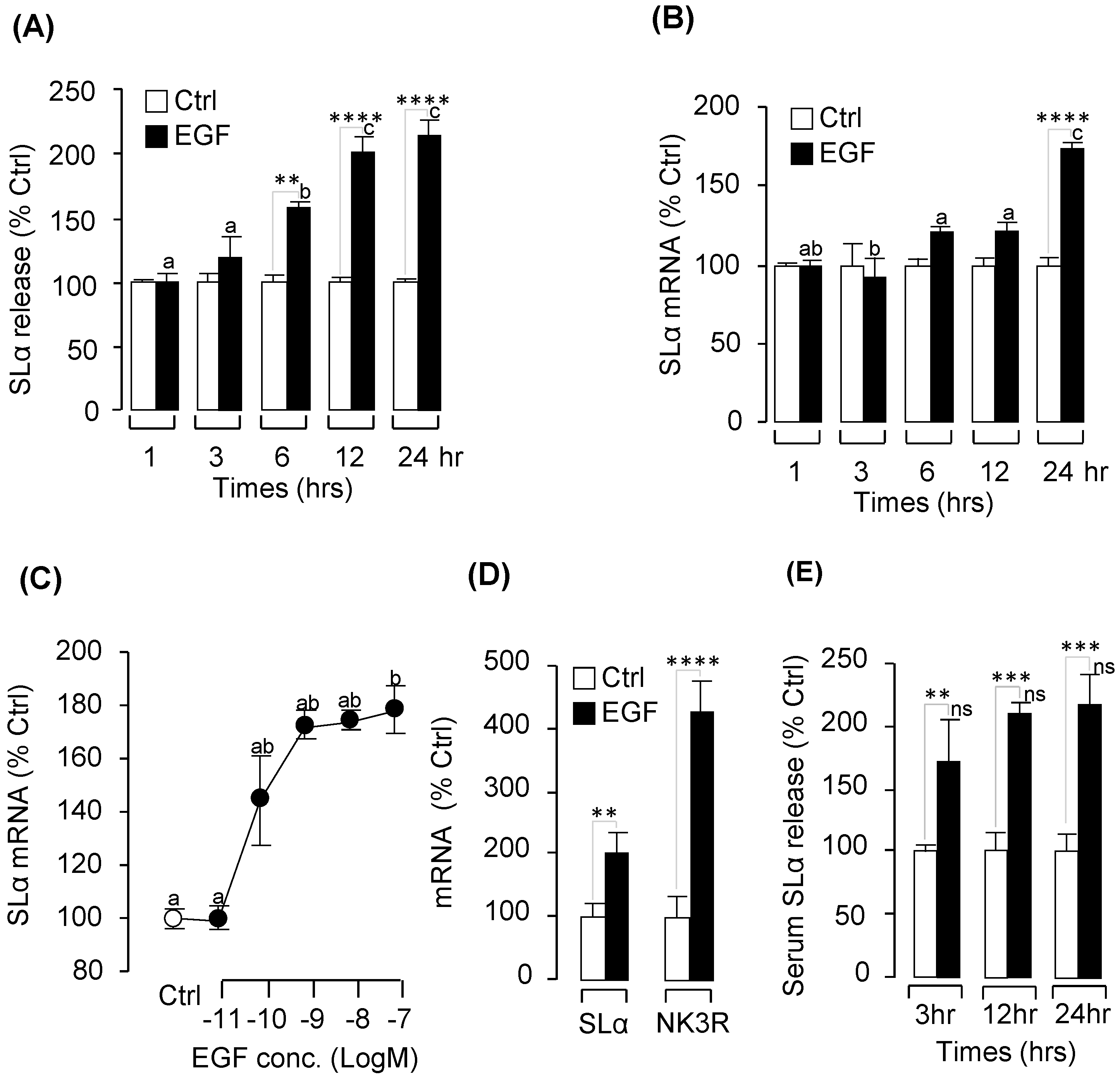
2.2. Pituitary Hormone Regulation by EGF In Vitro and In Vivo
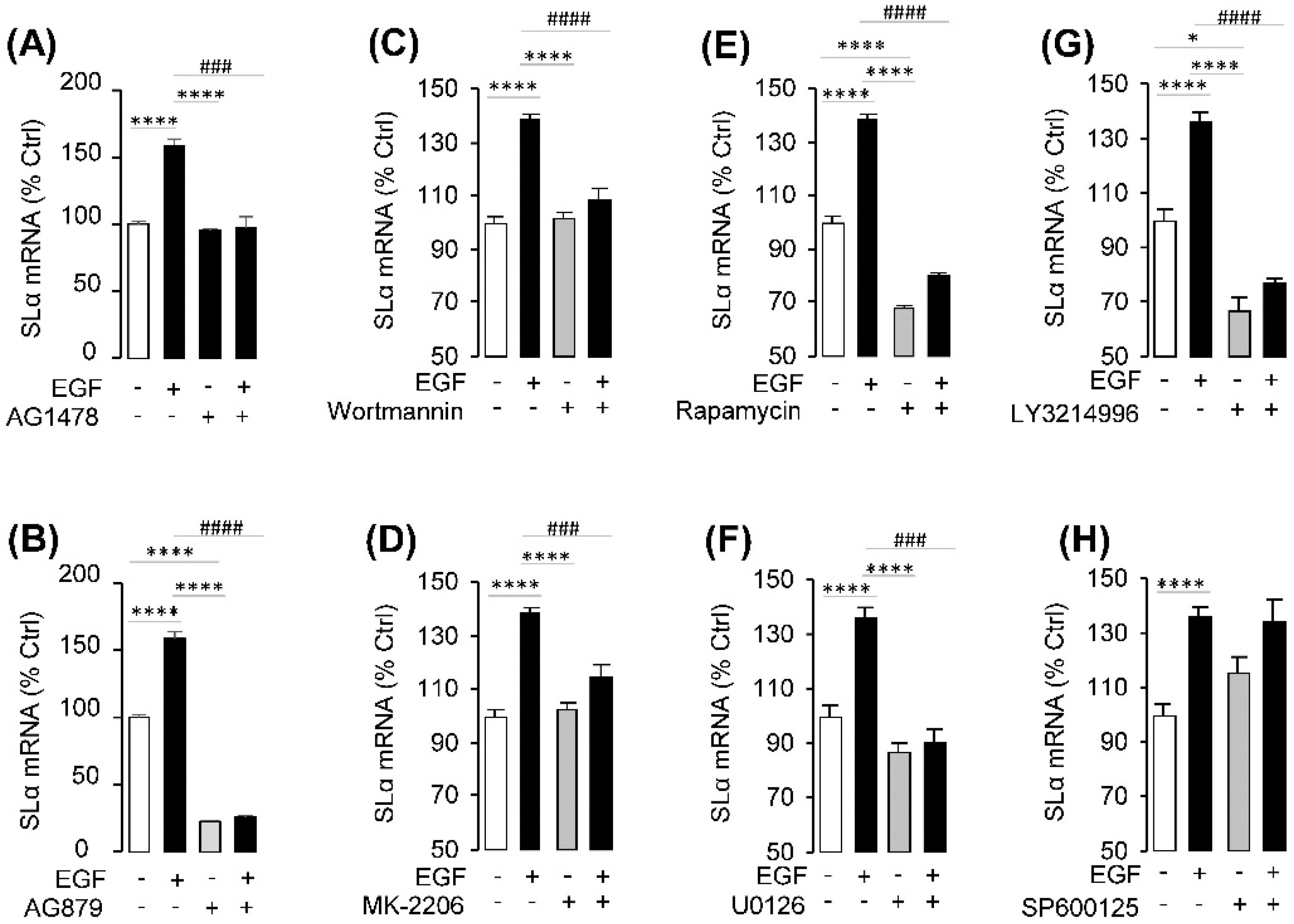
2.3. Receptor Specificity and Signal Pathway for SLα Regulation by EGF
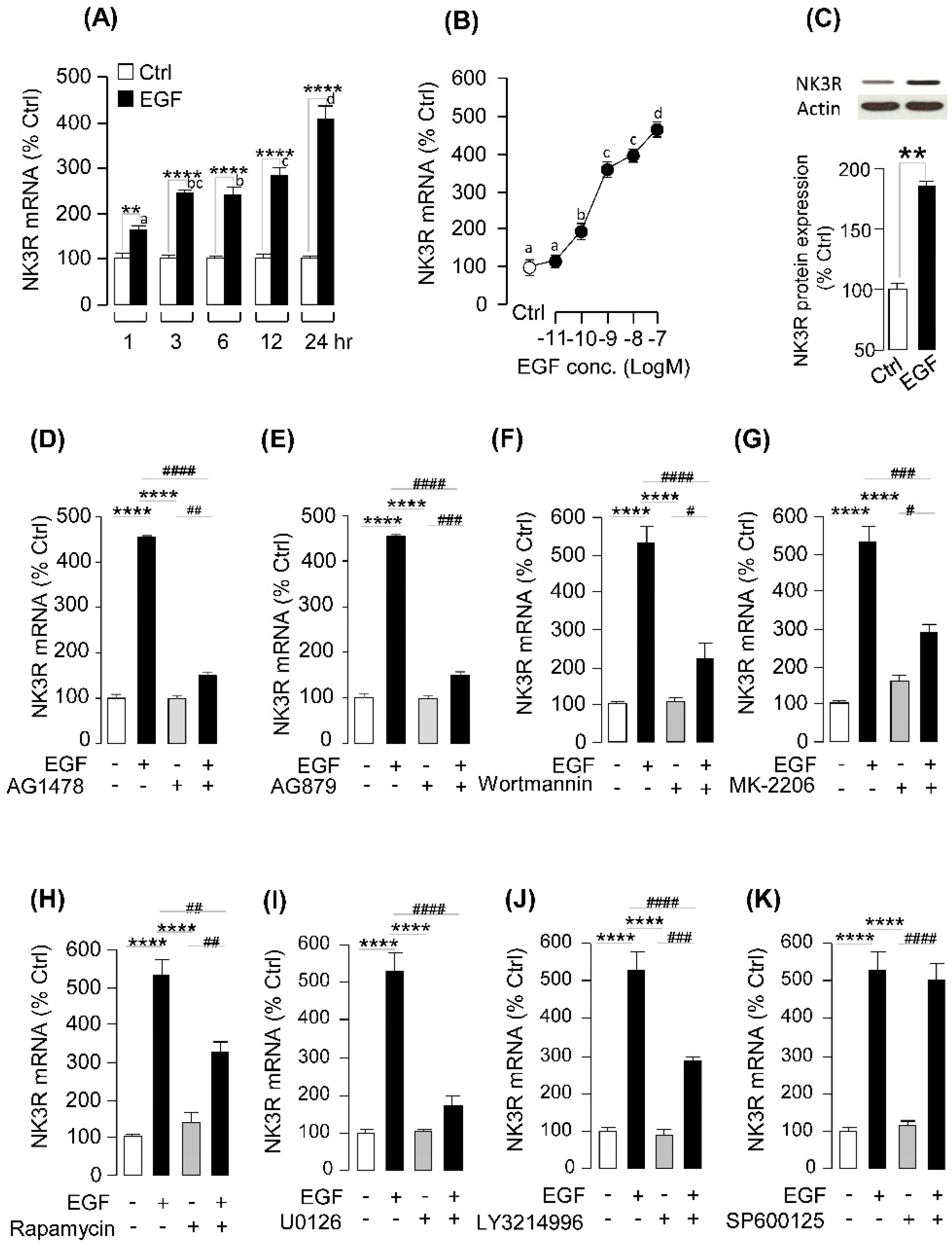
2.4. EGF-Induced NK3R Expression in Grass Carp Pituitary Cells
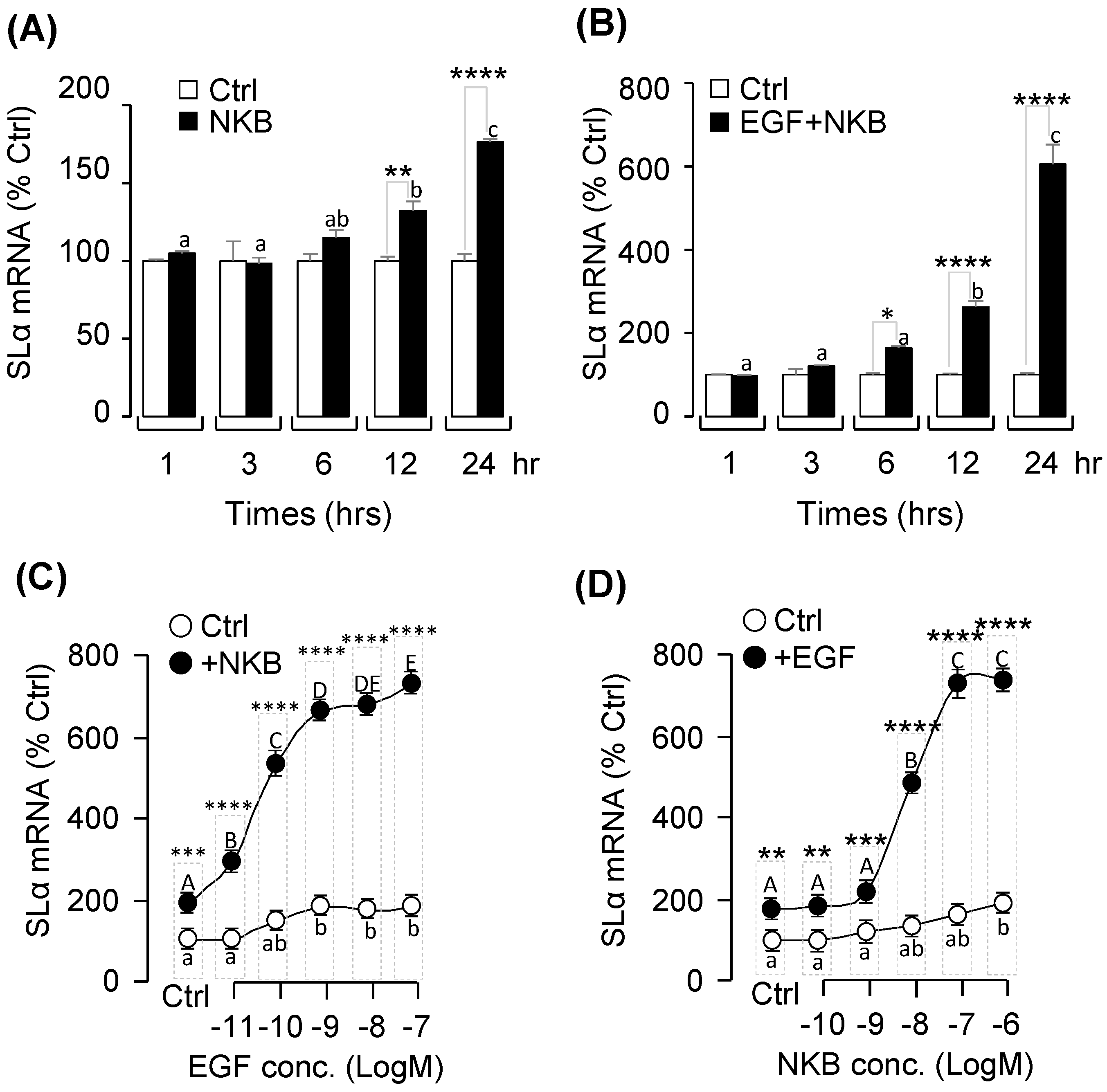
2.5. Synergistic Effects of EGF and NKB on SLα mRNA Expression
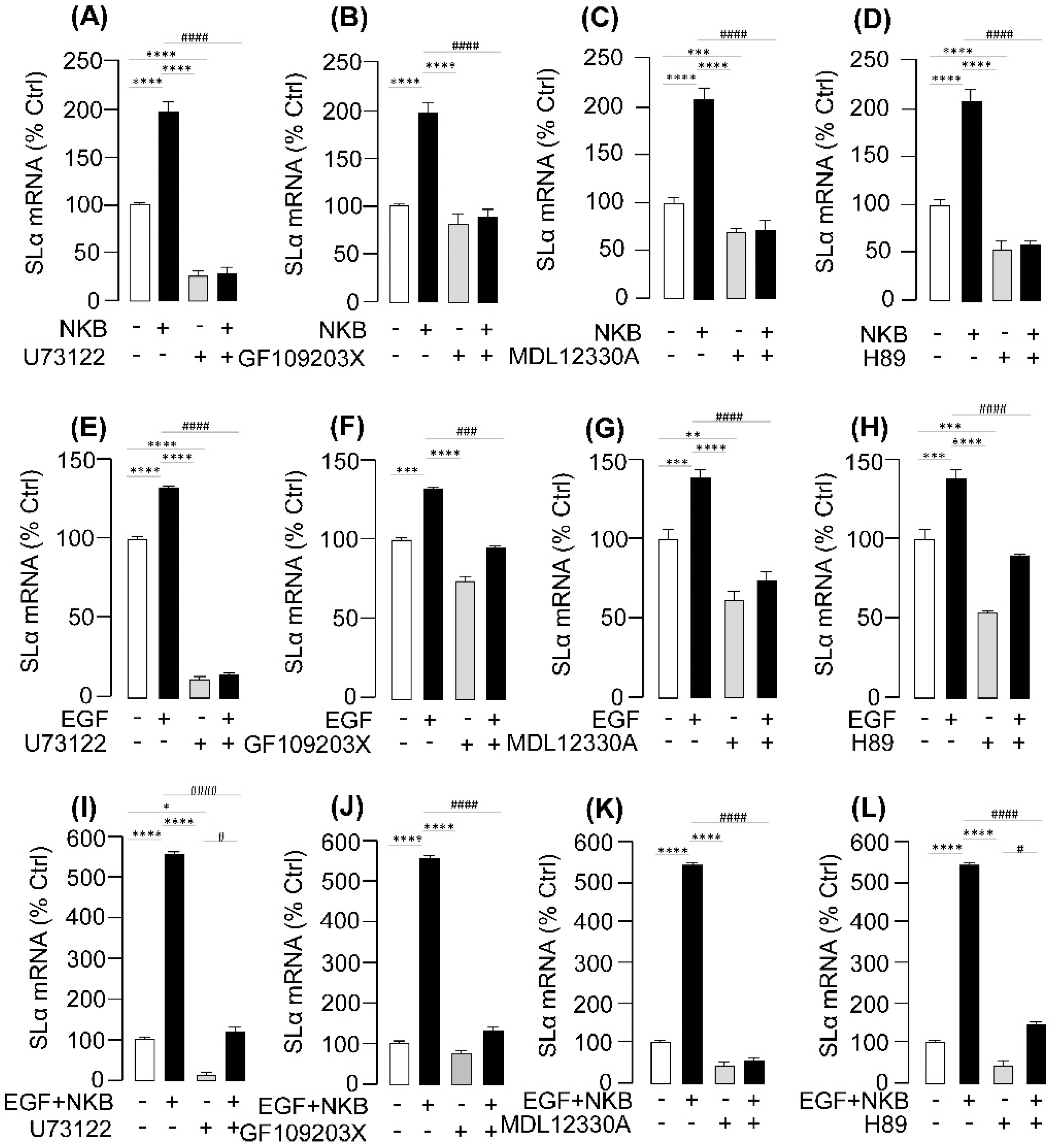
2.6. Signal Transduction for the Synergistic Effects of EGF and NK3R Activation
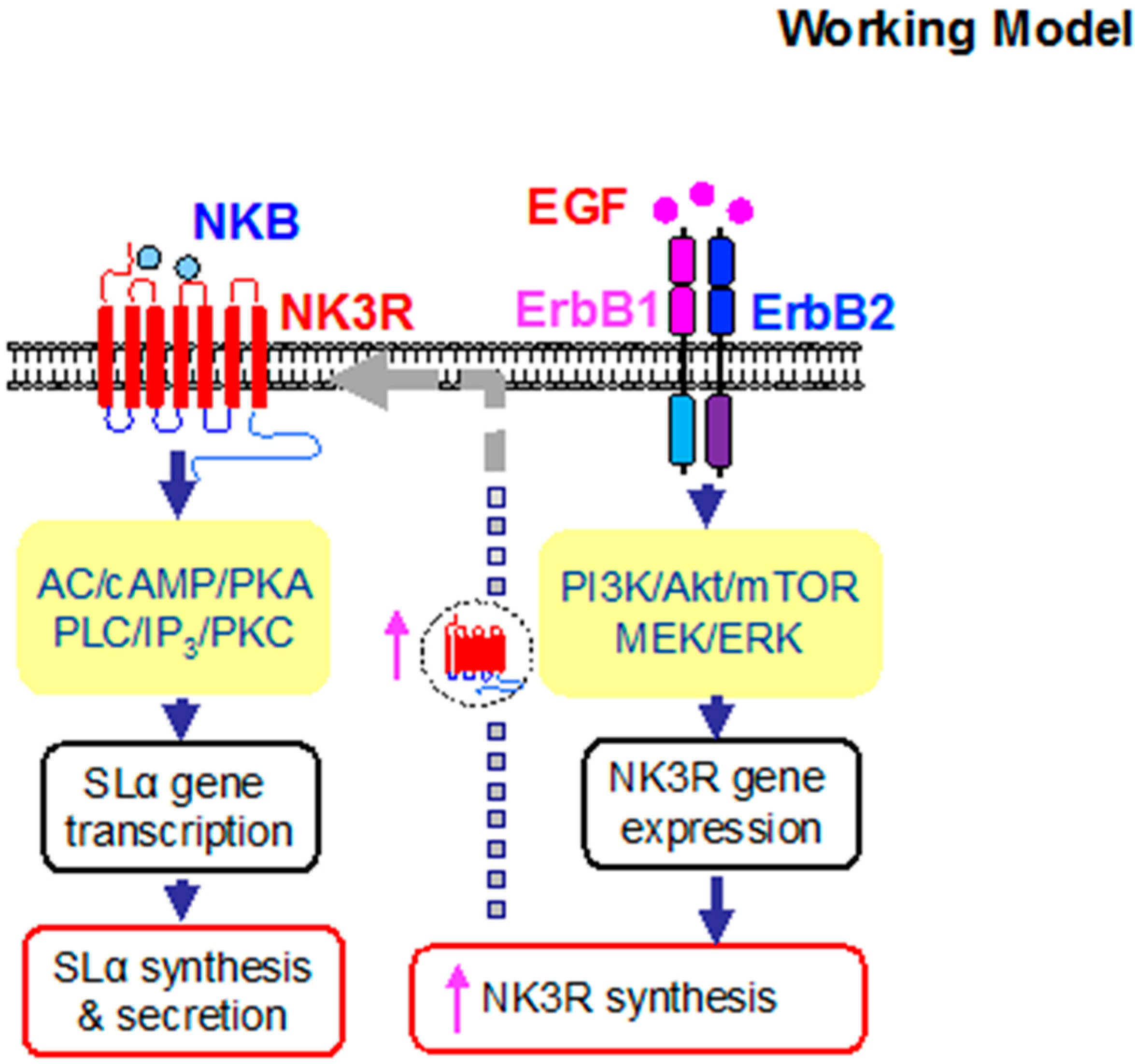
3. Discussion
4. Materials and Methods
4.1. Animals and Reagents
4.2. Molecular Cloning, Tissue Expression and Structural Analysis of carp EGF and EGFR
4.3. Measurement of Pituitary Hormone mRNA Expression
4.4. Measurement of SLα Secretion by Fluorescence-Based ELISA
4.5. In Vivo EGF Treatments and Sampling Procedure
4.6. Western Blot for Signaling Kinases
4.7. Data Transformation and Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AC | Adenylyl cyclase |
| Akt | protein kinase B |
| ERK | extracellular signal-regulated kinase |
| EGF | epidermal growth factor |
| NK3R | Type 3 NK receptor |
| NKB | neurokinin B |
| PI3K | phosphatidylinositol-3-kinase |
| PKC | Protein kinase C |
| PKA | Protein kinase A |
| PLC | phospholipase C |
| SLα | Somatolactin α |
References
- White, B.A.; Bancroft, F.C. Epidermal growth factor and thyrotropin-releasing hormone interact synergistically with calcium to regulate prolactin mRNA levels. J. Biol. Chem. 1983, 258, 4618–4622. [Google Scholar] [PubMed]
- Childs, G.V.; Rougeau, D.; Unabia, G. Corticotropin-releasing hormone and epidermal growth factor: Mitogens for anterior pituitary corticotropes. Endocrinology 1995, 136, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Chabot, J.G.; Walker, P.; Pelletier, G. Distribution of epidermal growth factor binding sites in the adult rat anterior pituitary gland. Peptides 1986, 7, 45–50. [Google Scholar] [CrossRef]
- Birman, P.; Michard, M.; Li, J.Y.; Peillon, F.; Bression, D. Epidermal growth factor-binding sites, present in normal human and rat pituitaries, are absent in human pituitary adenomas. J. Clin. Endocrinol. Metab. 1987, 65, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Childs, G.V. Epidermal growth factor and transforming growth factor-α messenger ribonucleic acids and their receptors in the rat anterior pituitary: Localization and regulation. Endocrinology 1995, 136, 2284–2293. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Nagle, G.T.; Collins, T.J.; Childs, G.V. Differential regulation of epidermal growth factor and transforming growth factor-α messenger ribonucleic acid in the rat anterior pituitary and hypothalamus induced by stresses. Endocrinology 1995, 136, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Finley, E.L.; King, J.S.; Ramsdell, J.S. Human pituitary somatotropes express transforming growth factor-α and its receptor. J. Endocrinol. 1994, 141, 547–554. [Google Scholar] [CrossRef]
- Driman, D.K.; Kobrin, M.S.; Kudlow, J.E.; Asa, S.L. Transforming growth factor-α in normal and neoplastic human endocrine tissues. Hum. Pathol. 1992, 23, 1360–1365. [Google Scholar] [CrossRef]
- Kontogeorgos, G.; Stefaneanu, L.; Kovacs, K.; Cheng, Z. Localization of epidermal growth factor (EGF) and epidermal growth factor receptor (EGFr) in human pituitary adenomas and nontumorous pituitaries: An immunocytochemical study. Endocr. Pathol. 1996, 7, 63–70. [Google Scholar] [CrossRef]
- Cooper, O.; Vlotides, G.; Fukuoka, H.; Greene, M.I.; Melmed, S. Expression and function of ErbB receptors and ligands in the pituitary. Endocr. Relat. Cancer 2011, 18, R197–R211. [Google Scholar] [CrossRef]
- Mouihate, A.; Lestage, J. Epidermal growth factor: A potential paracrine and autocrine system within the pituitary. Neuroreport 1995, 6, 1401–1404. [Google Scholar] [CrossRef] [PubMed]
- Przylipiak, A.; Kiesel, L.; Rabe, T.; Helm, K.; Przylipiak, M.; Runnebaum, B. Epidermal growth factor stimulates luteinizing hormone and arachidonic acid release in rat pituitary cells. Mol. Cell. Endocrinol. 1988, 57, 157–162. [Google Scholar] [CrossRef]
- Mouihate, A.; Lestage, J. Estrogen increases the release of epidermal growth factor from individual pituitary cells in female rats. J. Endocrinol. 1995, 146, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Mouihate, A.; Verrier, D.; Lestage, J. EGF release by rat gonadotroph cells: Characteristics and effects of LHRH. Life Sci. 1996, 58, 107–114. [Google Scholar] [CrossRef]
- Mouihate, A.; Verrier, D.; Lestage, J. Identification of epidermal growth factor-secreting cells in the anterior pituitary of lactating female rats. J. Endocrinol. 1996, 148, 319–324. [Google Scholar] [CrossRef]
- Lewis, M.D.; Ham, J.; Rees, D.A.; Lewis, B.M.; Scanlon, M.F. Mitogen-activated protein kinase mediates epidermal growth factor-induced morphogenesis in pituitary GH3 cells. J. Neuroendocrinol. 2002, 14, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.W.; Ge, W. Differential regulation of gonadotropins (FSH and LH) and growth hormone (GH) by neuroendocrine, endocrine, and paracrine factors in the zebrafish-An in vitro approach. Gen. Comp. Endocrinol. 2009, 160, 183–193. [Google Scholar] [CrossRef]
- Ono, M.; Takayama, Y.; Rand-Weaver, M.; Sakata, S.; Yasunaga, T.; Noso, T.; Kawauchi, H. cDNA cloning of somatolactin, a pituitary protein related to growth hormone and prolactin. Proc. Natl. Acad. Sci. USA 1990, 87, 4330–4334. [Google Scholar] [CrossRef]
- Zhu, Y.; Stiller, J.W.; Shaner, M.P.; Baldini, A.; Scemama, J.L.; Capehart, A.A. Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J. Endocrinol. 2004, 182, 509–518. [Google Scholar] [CrossRef]
- Benedet, S.; Bjornsson, B.T.; Taranger, G.L.; Andersson, E. Cloning of somatolactin alpha, beta forms and the somatolactin receptor in Atlantic salmon: Seasonal expression profile in pituitary and ovary of maturing female broodstock. Reprod. Biol. Endocrinol. 2008, 6, 42. [Google Scholar] [CrossRef]
- Jiang, Q.; He, M.L.; Wang, X.Y.; Wong, A.O.L. Grass carp somatolactin: II. Pharmacological study on postreceptor signaling mechanisms for PACAP-induced somatolactin α and β gene expression. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E477–E490. [Google Scholar] [CrossRef] [PubMed]
- Khalil, N.A.; Hashem, A.M.; Ibrahim, A.A.; Mousa, M.A. Effect of stress during handling, seawater acclimation, confinement, and induced spawning on plasma ion levels and somatolactin-expressing cells in mature female Liza ramada. J. Exp. Zool. A 2012, 317, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Sasano, Y.; Yoshimura, A.; Fukamachi, S. Reassessment of the function of somatolactin alpha in lipid metabolism using medaka mutant and transgenic strains. BMC Genet. 2012, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Sudo, R.; Suetake, H.; Suzuki, Y.; Aoyama, J.; Tsukamoto, K. Profiles of mRNA expression for prolactin, growth hormone, and somatolactin in Japanese eels, Anguilla japonica: The effect of salinity, silvering and seasonal change. Comp. Biochem. Physiol. A 2013, 164, 10–16. [Google Scholar] [CrossRef]
- Hu, G.F.; He, M.L.; Ko, W.K.; Lin, C.Y.; Wong, A.O.L. Novel pituitary actions of TAC3 gene products in fish model: -Receptor specificity and signal transduction for prolactin and somatolactin alpha regulation by neurokinin B (NKB) and NKB-related peptide in carp pituitary cells. Endocrinology 2014, 155, 3582–3596. [Google Scholar] [CrossRef] [PubMed]
- Guran, T.; Tolhurst, G.; Bereket, A.; Rocha, N.; Porter, K.; Turan, S.; Gribble, F.M.; Kotan, L.D.; Akcay, T.; Atay, Z.; et al. Hypogonadotropic hypogonadism due to a novel missense mutation in the first extracellular loop of the neurokinin B receptor. J. Clin. Endocrinol. Metab. 2009, 94, 3633–3639. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu, A.K.; Reimann, F.; Guclu, M.; Yalin, A.S.; Kotan, L.D.; Porter, K.M.; Serin, A.; Mungan, N.O.; Cook, J.R.; Ozbek, M.N.; et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet. 2009, 41, 354–358. [Google Scholar] [CrossRef]
- Young, J.; Bouligand, J.; Francou, B.; Raffin-Sanson, M.L.; Gaillez, S.; Jeanpierre, M.; Grynberg, M.; Kamenicky, P.; Chanson, P.; Brailly-Tabard, S.; et al. TAC3 and TACR3 defects cause hypothalamic congenital hypogonadotropic hypogonadism in humans. J. Clin. Endocrinol. Metab. 2010, 95, 2287–2295. [Google Scholar] [CrossRef]
- Biran, J.; Palevitch, O.; Ben-Dor, S.; Levavi-Sivan, B. Neurokinin Bs and neurokinin B receptors in zebrafish-potential role in controlling fish reproduction. Proc. Natl. Acad. Sci. USA 2012, 109, 10269–10274. [Google Scholar] [CrossRef]
- Zhou, W.; Li, S.; Liu, Y.; Qi, X.; Chen, H.; Cheng, C.H.; Liu, X.; Zhang, Y.; Lin, H. The evolution of tachykinin/tachykinin receptor (TAC/TACR) in vertebrates and molecular identification of the TAC3/TACR3 system in zebrafish (Danio rerio). Mol. Cell. Endocrinol. 2012, 361, 202–212. [Google Scholar] [CrossRef]
- Qi, X.; Salem, M.; Zhou, W.; Sato-Shimizu, M.; Ye, G.; Smitz, J.; Peng, C. Neurokinin B exerts direct effects on the ovary to stimulate estradiol production. Endocrinology 2016, 157, 3355–3365. [Google Scholar] [CrossRef] [PubMed]
- Liebmann, C. EGF receptor activation by GPCRs: An universal pathway reveals different versions. Mol. Cell. Endocrinol. 2011, 331, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Ge, W. Cloning of epidermal growth factor (EGF) and EGF receptor from the zebrafish ovary: Evidence for EGF as a potential paracrine factor from the oocyte to regulated activin/follistatin system in the follicle cells. Biol. Reprod. 2004, 71, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, G.H.; Potter, E.; Nicolaisen, A.K.; Evans, R.M.; Rosenfeld, M.G. Epidermal growth factor rapidly stimulates prolactin gene transcription. Nature 1982, 300, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jonathan Chen, S.L.; Dunckely, J.A.; LaPensee, C.; Kansra, S. Estrogen receptor-α mediates the epithermal growth factor-stimulated prolactin expression and release in lactotrophs. Endocrinology 2009, 150, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Mitsuhashi, T.; Kubota, K.; Kuzuya, N.; Uchimura, H. Epidermal growth factor stimulates growth hormone secretion from superfused rat adenohypophyseal fragments. Endocrinology 1984, 115, 556–558. [Google Scholar] [CrossRef]
- Kawauchi, H.; Sower, S.A.; Moriyama, S. The neuroendocrine regulation of prolactin and somatolactin secretion in fish. Fish Physiol. 2009, 28, 197–234. [Google Scholar]
- Kumagai, T.; Davis, J.G.; Horie, T.; O’Rourke, D.M.; Greene, M.I. The role of distinct p185neu extracellular subdomains for dimerization with the epidermal growth factor (EGF) receptor and EGF-mediated signaling. Proc. Natl. Acad. Sci. USA 2001, 98, 5526–5531. [Google Scholar] [CrossRef]
- Kumagai, T.; Katsumata, M.; Hasegawa, A.; Furuuchi, K.; Funakoshi, T.; Kawase, I.; Greene, M.I. Role of extracellular subdomains of p185c-neu and the epidermal growth factor receptor in ligand independent association and transactivation. Proc. Natl. Acad. Sci. USA 2003, 100, 9220–9225. [Google Scholar] [CrossRef]
- Qian, X.; LeVea, C.M.; Freeman, J.K.; Dougall, W.C.; Greene, M.I. Heterodimerization of epidermal growth factor receptor and wild-type or kinase-deficient Neu: A mechanism of interreceptor kinase activation and transphosphorylation. Proc. Natl. Acad. Sci. USA 1994, 91, 1500–1504. [Google Scholar] [CrossRef]
- Riese, D.J.; Stern, D.F. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays 1998, 20, 41–48. [Google Scholar] [CrossRef]
- Beerli, R.R.; Graus-Porta, D.; Woods-Cook, K.; Chen, X.; Yarden, Y.; Hynes, N.E. New differentiation factor activation of ErbB-3 and ErbB-4 is cell specific and displays a differential requirement for ErbB-2. Mol. Cell. Biol. 1995, 15, 6496–6505. [Google Scholar] [CrossRef] [PubMed]
- Graus-Porta, D.; Beerli, R.R.; Hynes, N.E. Single-chain antibody-mediated intracellular retention of ErbB-2 impairs Neu differentiation factor and epidermal growth factor signaling. Mol. Cell. Biol. 1995, 15, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Karunagaran, D.; Tzahar, E.; Beerli, R.R.; Chen, X.; Graus-Porta, D.; Ratzkin, B.J.; Seger, R.; Hynes, N.E.; Yarden, Y. ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: Implications for breast cancer. EMBO J. 1996, 15, 254–264. [Google Scholar] [CrossRef]
- Wada, T.; Qian, X.L.; Greene, M.I. Intermolecular association of the p185neu protein and EGF receptor modulates EGF receptor function. Cell 1990, 61, 1339–1347. [Google Scholar] [CrossRef]
- Sliwkowski, M.X.; Schaefer, G.; Akita, R.W.; Lofgren, J.A.; Fitzpatrick, V.D.; Nuijens, A.; Fendly, B.M.; Cerione, R.A.; Vandlen, R.L.; Carraway, K.L., III. Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J. Biol. Chem. 1994, 269, 14661–14665. [Google Scholar]
- Satake, H.; Aoyama, M.; Sekiguchi, T.; Kawada, T. Insight into molecular and functional diversity of tachykinins and their receptors. Protein Pept. Lett. 2013, 20, 615–627. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Wong, A.O.; Ng, S.; Lee, E.K.; Leung, R.C.; Ho, W.K. Somatostatin inhibits (D-Arg6, Pro9-NEt) salmon gonadotropin-releasing hormone- and dopamine D1-stimulated growth hormone release from perifused pituitary cells of Chinese grass carp, Ctenopharyngodon idellus. Gen. Comp. Endocrinol. 1998, 110, 29–45. [Google Scholar] [CrossRef]
- Qin, X.F.; Xiao, Y.Q.; Ye, C.; Jia, J.Y.; Liu, X.J.; Liang, H.W.; Zou, G.W.; Hu, G.F. Pituitary action of E2 in prepubertal grass carp: Receptor specificity and signal transduction for luteinizing hormone and follicle-stimulating hormone regulation. Front. Endocrinol. 2018, 9, 308. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Ye, C.; Zhou, X.; Jia, J.; Xu, S.; Hu, Q.; Hu, G. NK3R Mediates the EGF-Induced SLα Secretion and mRNA Expression in Grass Carp Pituitary. Int. J. Mol. Sci. 2019, 20, 91. https://doi.org/10.3390/ijms20010091
Qin X, Ye C, Zhou X, Jia J, Xu S, Hu Q, Hu G. NK3R Mediates the EGF-Induced SLα Secretion and mRNA Expression in Grass Carp Pituitary. International Journal of Molecular Sciences. 2019; 20(1):91. https://doi.org/10.3390/ijms20010091
Chicago/Turabian StyleQin, Xiangfeng, Cheng Ye, Xiaoyun Zhou, Jingyi Jia, Shaohua Xu, Qiongyao Hu, and Guangfu Hu. 2019. "NK3R Mediates the EGF-Induced SLα Secretion and mRNA Expression in Grass Carp Pituitary" International Journal of Molecular Sciences 20, no. 1: 91. https://doi.org/10.3390/ijms20010091
APA StyleQin, X., Ye, C., Zhou, X., Jia, J., Xu, S., Hu, Q., & Hu, G. (2019). NK3R Mediates the EGF-Induced SLα Secretion and mRNA Expression in Grass Carp Pituitary. International Journal of Molecular Sciences, 20(1), 91. https://doi.org/10.3390/ijms20010091




