MePHD1 as a PHD-Finger Protein Negatively Regulates ADP-Glucose Pyrophosphorylase Small Subunit1a Gene in Cassava
Abstract
1. Introduction
2. Results
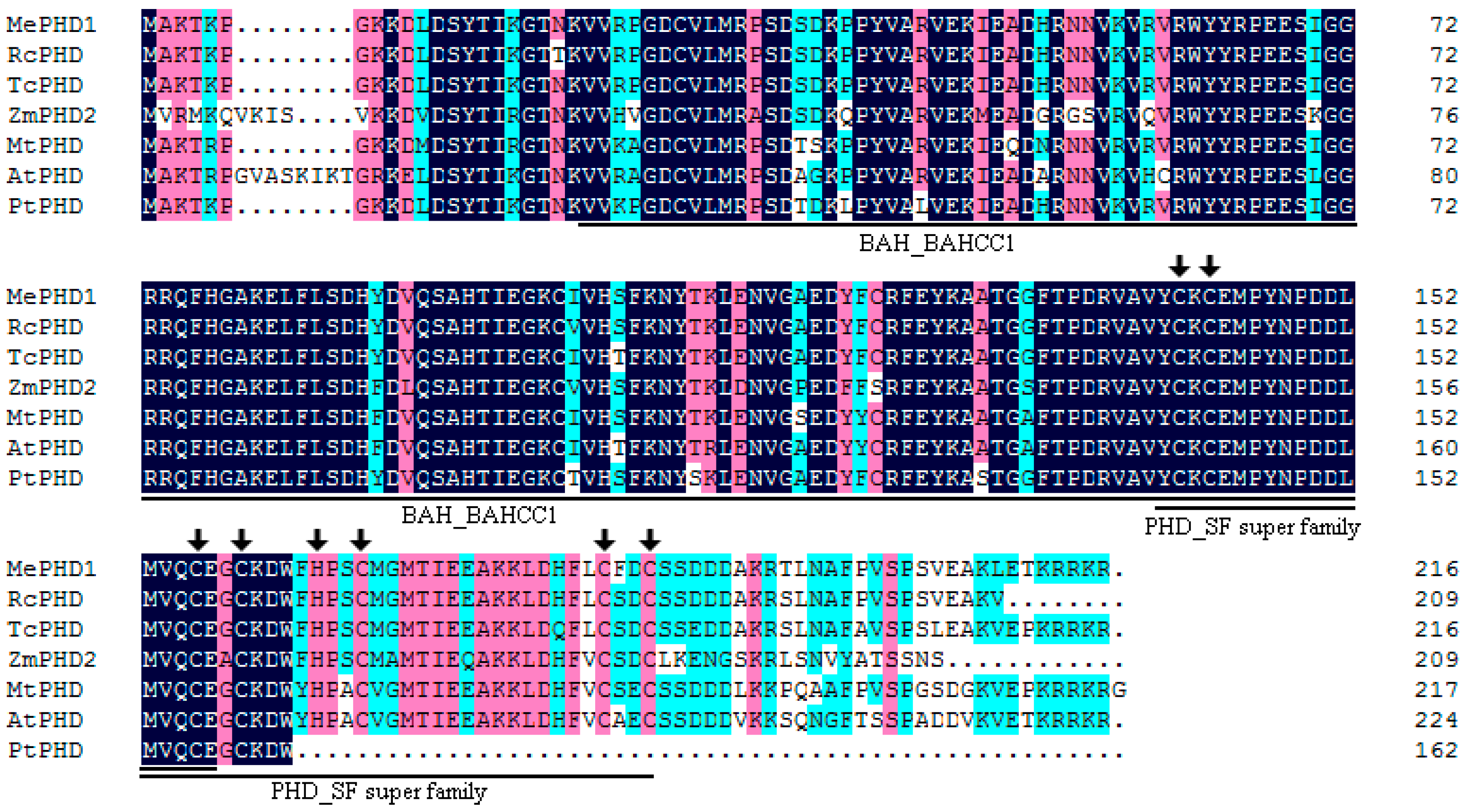
2.1. Isolation and Molecular Characterization of MePHD1
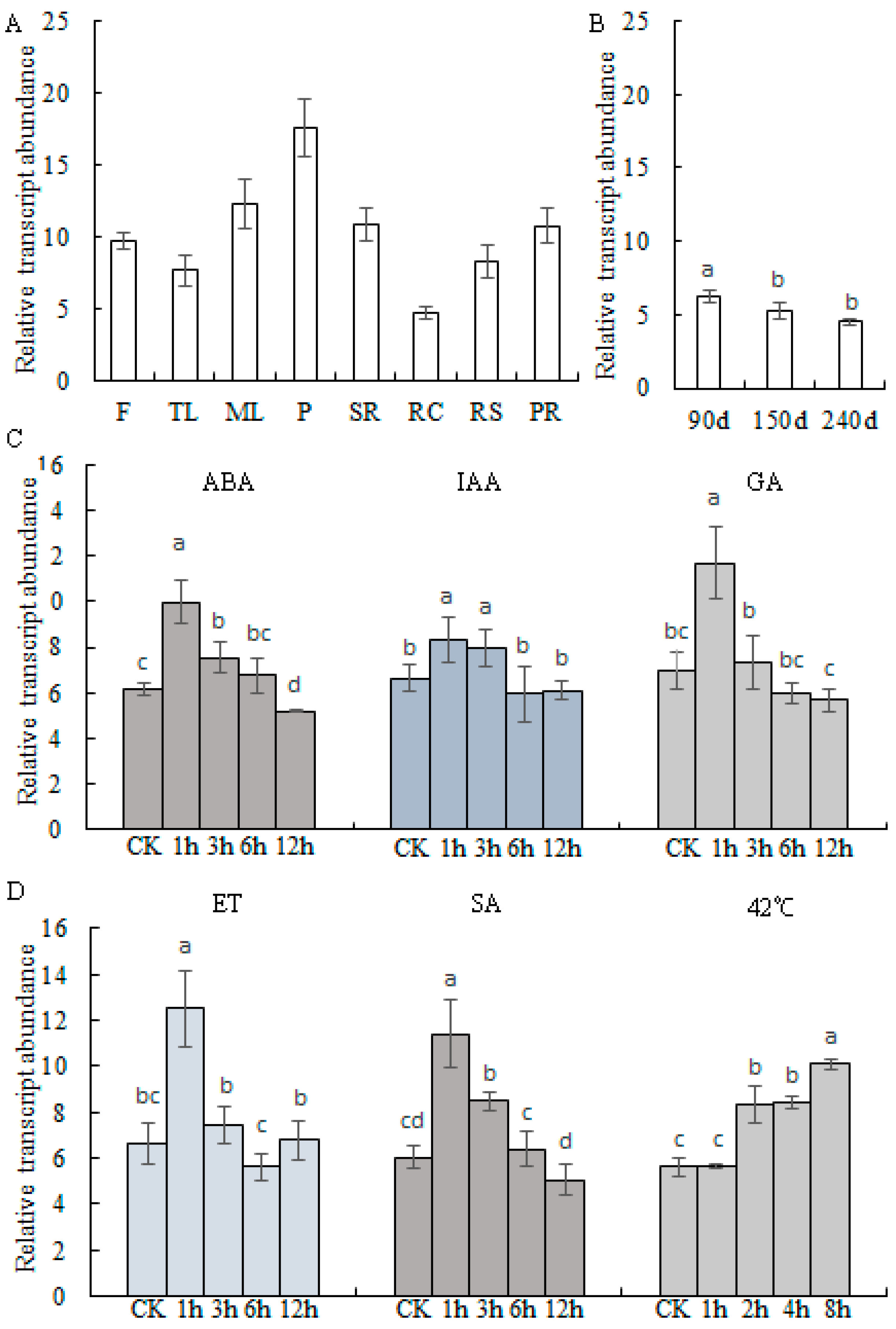
2.2. Transcript Profiles of MePHD1
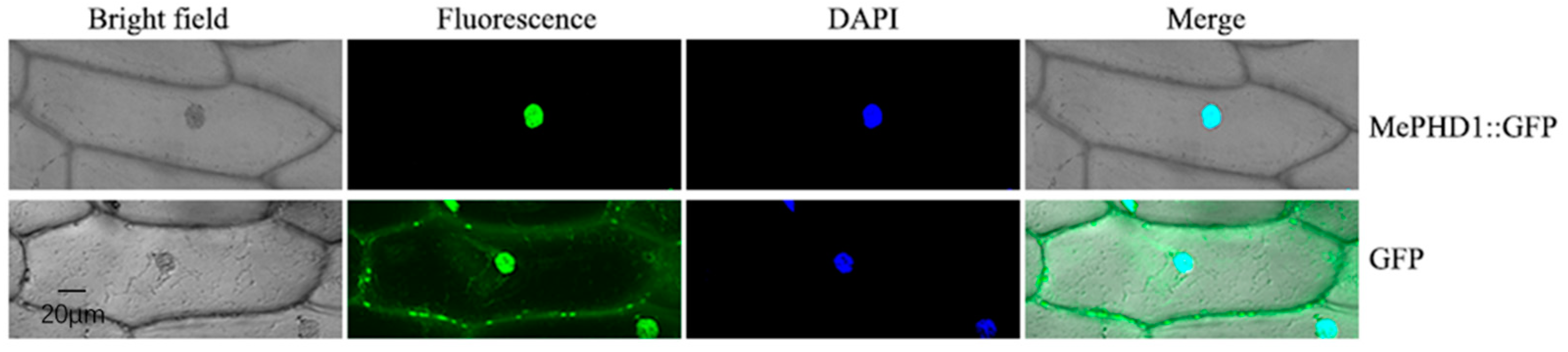
2.3. Subcellular Location of MePHD1 Protein
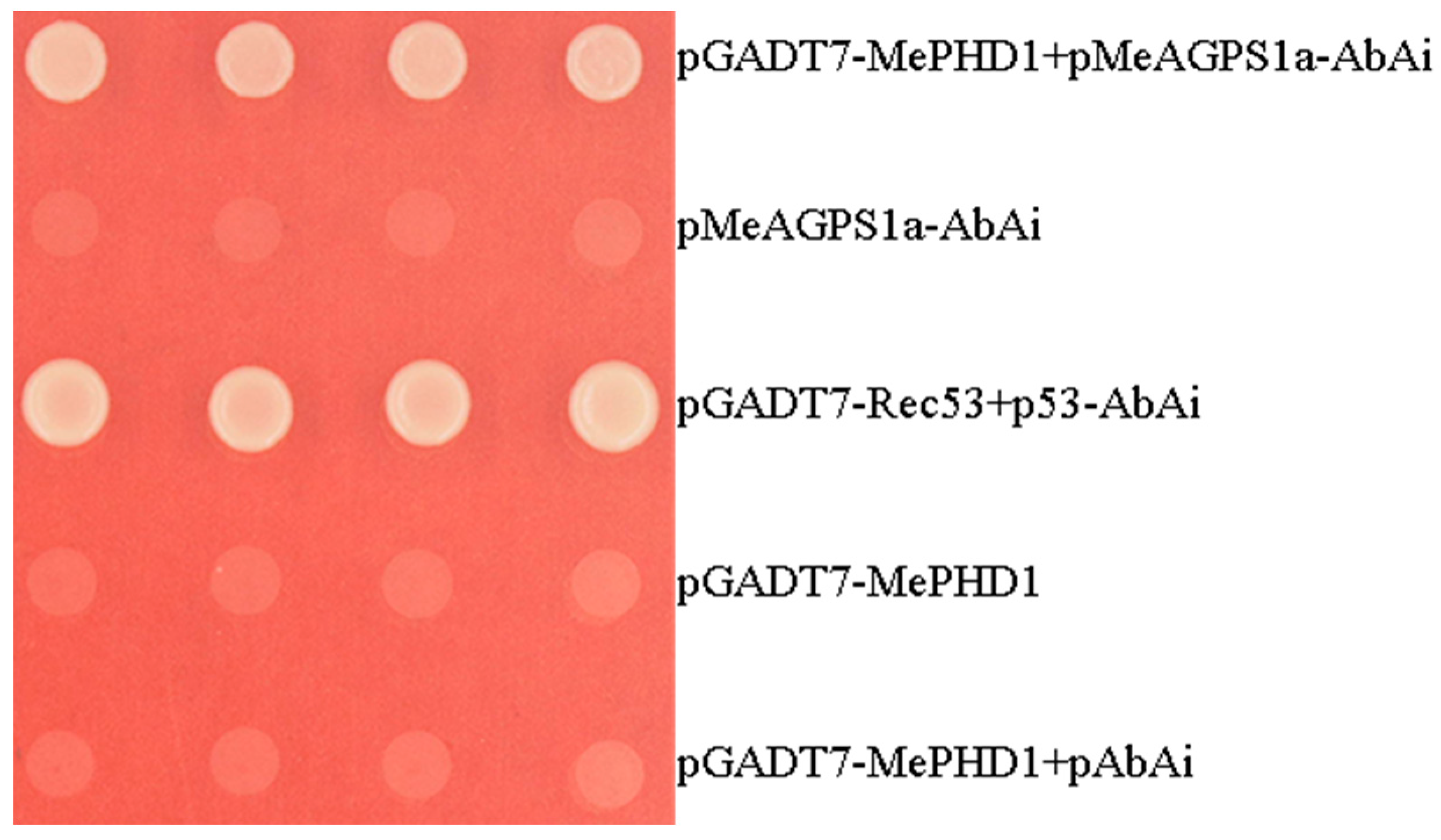
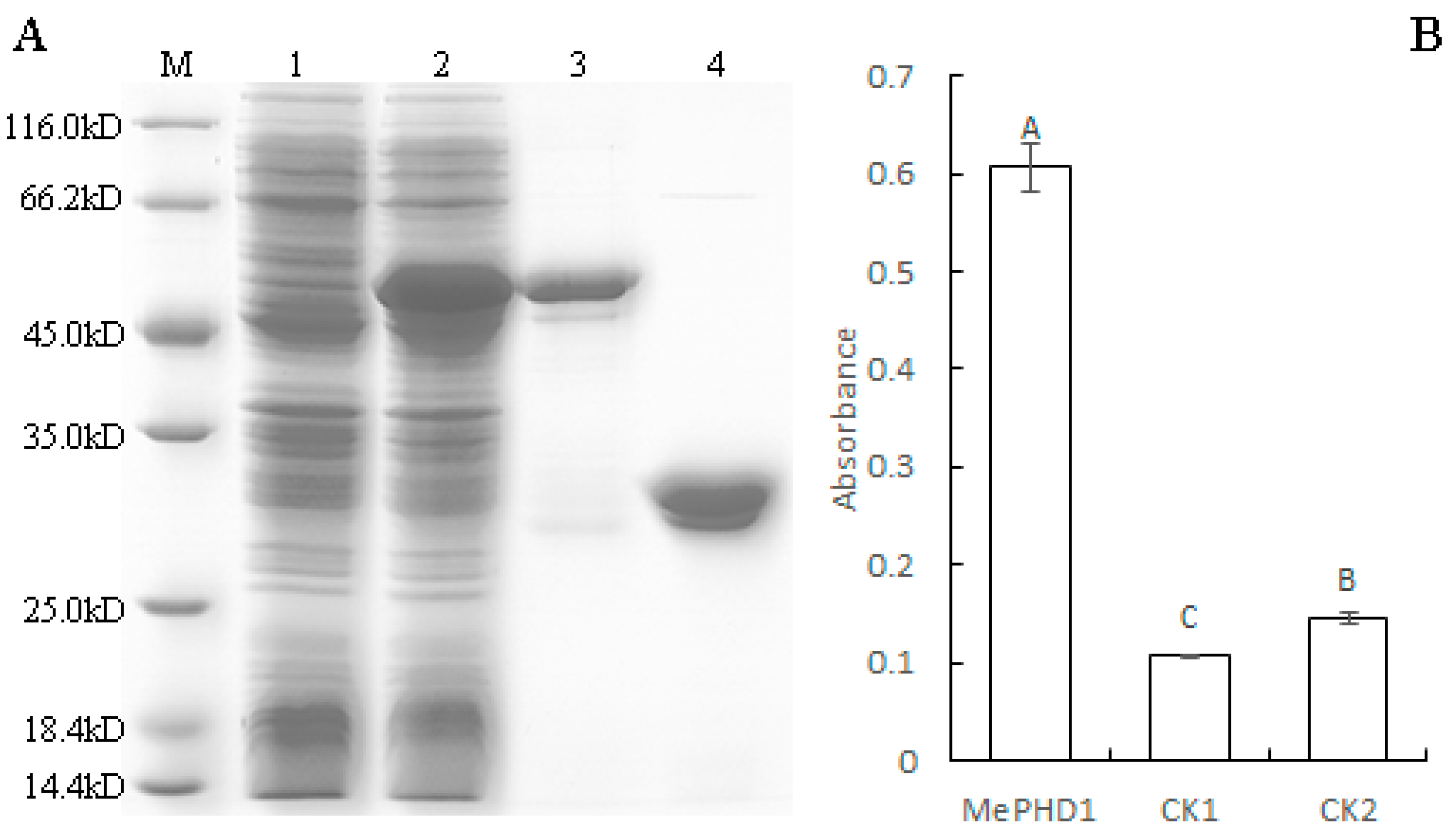
2.4. MePHD1 Binds to MeAGPS1a Promoter In Vitro
2.5. MePHD1 Binds to the −400 bp to −201 bp Region of MeAGPS1a Promoter
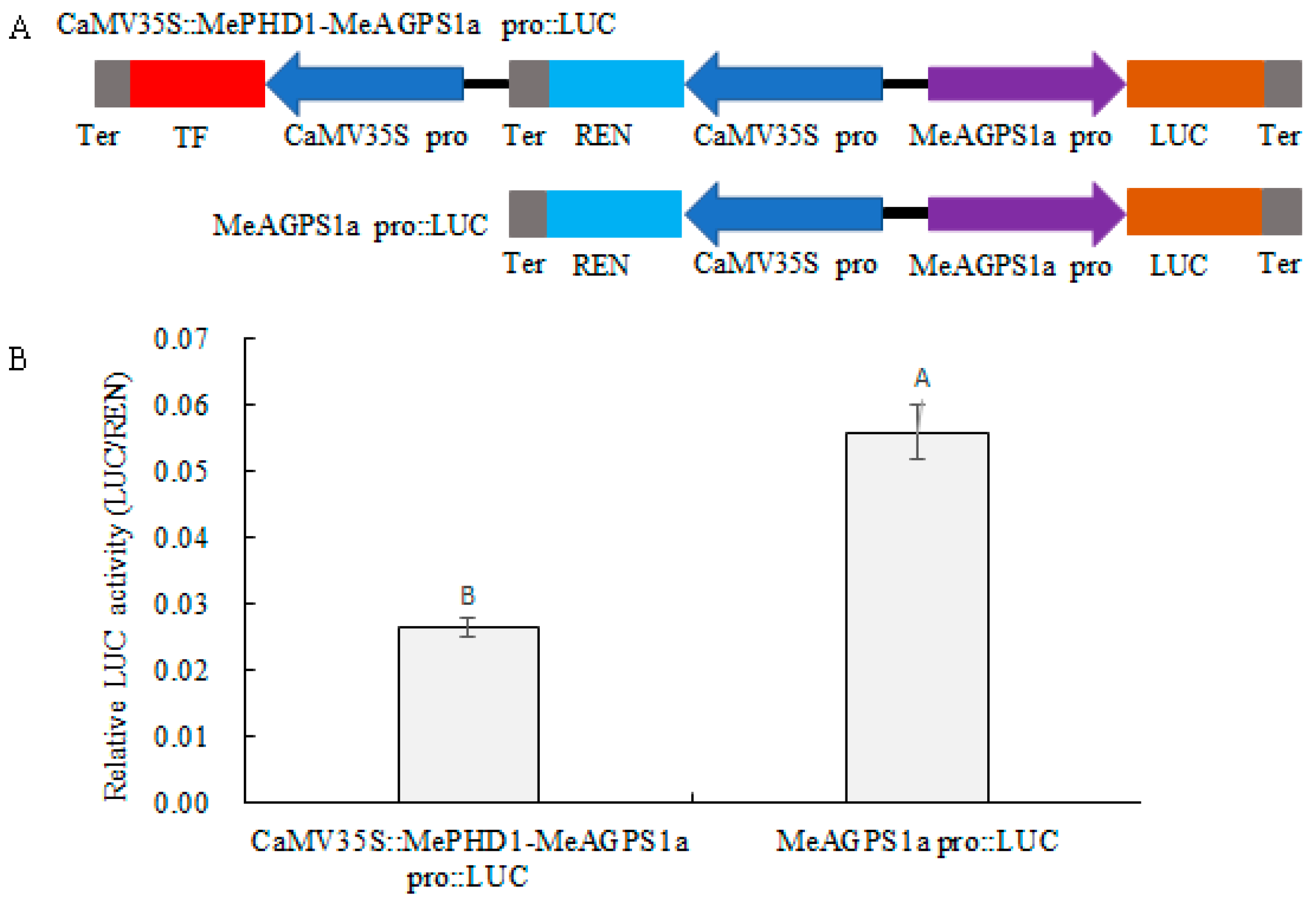
2.6. Repression of MeAGPS1a Promoter by MePHD1 Protein
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Yeast One-Hybrid
4.3. Chromosomal Location and Bioinformatic Analyses of the MePHD Gene Family
4.4. Quantitative Polymerase Chain Reaction (qPCR)
4.5. Nuclear Localization Analysis
4.6. MePHD1 Protein Expression and Purification
4.7. DNA-Protein-Interaction Enzyme-Linked Immunosorbent Assay (DPI-ELISA)
4.8. Dual-Luciferase (Dual-LUC) Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-Glucose Pyrophosphorylase: A Regulatory Enzyme for Plant Starch Synthesis. Photosynth. Res. 2004, 79, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Chen, X.; Liu, C.; Meng, Y.; Xia, Z.; Zeng, C.; Lu, C.; Wang, W. MeSAUR1, Encoded by a Small Auxin-Up RNA Gene, Acts as a Transcription Regulator to Positively Regulate ADP-Glucose Pyrophosphorylase Small Subunit1a Gene in Cassava. Front. Plant. Sci. 2017, 8, 1315. [Google Scholar] [CrossRef] [PubMed]
- Bienz, M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006, 31, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Kaadige, M.R.; Ayer, D.E. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J. Biol. Chem. 2006, 281, 28831–28836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, Z.N.; Zhang, S. Genome-wide Analysis of PHD-finger Protein Family in Arabidopsis thaliana. Acta Botanica. Yunnanica. 2009, 31, 227–238. [Google Scholar] [CrossRef]
- Wu, X.J.; Li, M.Y.; Que, F.; Wang, F.; Xu, Z.S.; Xiong, A.S. Genome-wide analysis of PHD family transcription factors in carrot (Daucus carota L.) reveals evolution and response to abiotic stress. Acta Physiol. Plant. 2016, 38, 67. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Wang, Y.; Zhao, Y.; Jiang, H.; Cheng, B. Systematic Analysis of the Maize PHD-Finger Gene Family Reveals a Subfamily Involved in Abiotic Stress Response. Int. J. Mol. Sci. 2015, 16, 23517–23544. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wu, M.; Dong, Q.; Jiang, H.; Cai, R.; Xiang, Y. Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa. Gene 2016, 575, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Huang, J.; Hao, Y.J.; Zou, H.F.; Wang, H.W.; Zhao, J.Y.; Liu, X.Y.; Zhang, W.K.; Ma, B.; Zhang, J.S. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE 2009, 4, e7209. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Pi, E.X.; Tsai, S.N.; Lam, H.M.; Sun, S.M.; Kwan, Y.W.; Ngai, S.M. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant. Biol. 2011, 11, 178. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Makaroff, C.A.; Ma, H. The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant. Cell. 2003, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Fernandez Gomez, J.; Wilson, Z.A. A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant. Biotechnol. J. 2014, 12, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; Zhu, T.; et al. Construction of a multi-control sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhang, Y.Q.; Tao, J.J.; Chen, H.W.; Li, Q.T.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; Chen, S.Y. The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J. 2015, 81, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Mena, C.; Pineiro, M.; Franco-Zorrilla, J.M.; Salinas, J.; Coupland, G.; Martinez-Zapater, J.M. Early bolting in short days: An Arabidopsis mutation that causes early flowering and partially suppresses the floral phenotype of leafy. Plant. Cell. 2001, 13, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Pineiro, M.; Gomez-Mena, C.; Schaffer, R.; Martinez-Zapater, J.M.; Coupland, G. EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant. Cell. 2003, 15, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Narro-Diego, L.; Lopez-Gonzalez, L.; Jarillo, J.A.; Pineiro, M. The PHD-containing protein EARLY BOLTING IN SHORT DAYS regulates seed dormancy in Arabidopsis. Plant. Cell. Environ. 2017, 40, 2393–2405. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.H.; Kirchler, T.; Hummel, S.; Chaban, C.; Wanke, D. DPI-ELISA: A fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 2010, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Wei, L.R.; Guo, D.; Wang, Y.; Zhu, J.H.; Chen, X.T.; Peng, S.Q. HbMADS4, a MADS-box Transcription Factor from Hevea brasiliensis, Negatively Regulates HbSRPP. Front. Plant. Sci. 2016, 7, 1709. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Hu, W.; Xia, Z.; Zhou, X.; Wang, W. Validation of Reference Genes for Relative Quantitative Gene Expression Studies in Cassava (Manihot esculenta Crantz) by Using Quantitative Real-Time PCR. Front. Plant. Sci. 2016, 7, 680. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, P.; Chen, X.; Liu, C.; Xia, Z.; Song, Y.; Zeng, C.; Li, Y.; Wang, W. MePHD1 as a PHD-Finger Protein Negatively Regulates ADP-Glucose Pyrophosphorylase Small Subunit1a Gene in Cassava. Int. J. Mol. Sci. 2018, 19, 2831. https://doi.org/10.3390/ijms19092831
Ma P, Chen X, Liu C, Xia Z, Song Y, Zeng C, Li Y, Wang W. MePHD1 as a PHD-Finger Protein Negatively Regulates ADP-Glucose Pyrophosphorylase Small Subunit1a Gene in Cassava. International Journal of Molecular Sciences. 2018; 19(9):2831. https://doi.org/10.3390/ijms19092831
Chicago/Turabian StyleMa, Ping’an, Xin Chen, Chen Liu, Zhiqiang Xia, Yu Song, Changying Zeng, Youzhi Li, and Wenquan Wang. 2018. "MePHD1 as a PHD-Finger Protein Negatively Regulates ADP-Glucose Pyrophosphorylase Small Subunit1a Gene in Cassava" International Journal of Molecular Sciences 19, no. 9: 2831. https://doi.org/10.3390/ijms19092831
APA StyleMa, P., Chen, X., Liu, C., Xia, Z., Song, Y., Zeng, C., Li, Y., & Wang, W. (2018). MePHD1 as a PHD-Finger Protein Negatively Regulates ADP-Glucose Pyrophosphorylase Small Subunit1a Gene in Cassava. International Journal of Molecular Sciences, 19(9), 2831. https://doi.org/10.3390/ijms19092831



